Abstract
A ubiquitin ligase (E3) functions at the crossroad between ubiquitin activation and the attachment of ubiquitin to protein substrates. During this process, the E3 interacts with both a substrate and a ubiquitin-conjugating enzyme (E2). Although a major goal when investigating an E3 is to identify its substrates, recent evidence indicates that the E2 dictates the type of ubiquitin modification that will occur on the substrate. There are ~ 30 E2s identified in the human genome, many of which remain to be characterized. We found that the RING E3 BRCA1/BARD1 can interact with 10 different E2s. The ability of BRCA1 to interact with multiple E2s is likely to be a common feature among other RING and U-box E3s. We and others have also found that certain E2s show a preference for attaching either the first ubiquitin to a substrate lysine or ubiquitin to itself (chain building), suggesting that E2s may play a role in dictating product formation. Therefore, when investigating the functions of an E3 it is advisable to identify all E2s that interact with the E3 so that these can be used in E3-dependent substrate-ubiquitination assays. We describe a method used to identify all the E2s that interact with BRCA1. Defining the set of E2s that interact with other RING and U-box E3s will open the door for predictive models and lead to a better understand of substrate ubiquitination.
Keywords: BRCA1, NMR, protein-protein interactions, RING domain, UbcH5, Ubc13, ubiquitin ligase, ubiquitination, ubiquitin-conjugating enzyme, yeast two-hybrid
Introduction
Identifying the complement of molecules that interact with a given protein is an important step toward understanding the functional responsibilities of that protein in the cell. In addition, there are many protein domains which, although conserved and identifiable, are poorly understood or have unknown functions. Probing the functional properties of a protein by investigating possible interaction partners of its domains may illuminate functional properties utilized in the cell. We applied this principle to the ubiquitination system by looking for all possible interactions between a specific ubiquitin ligase (E3) and the family of ubiquitin-conjugating enzymes (E2s). Here, we review our strategy, which can be used for other E3s as well as for other weakly interacting systems.
Ubiquitination is a diverse signaling mechanism that involves multiple protein–protein interactions. Transfer of ubiquitin to a target substrate requires sequential protein–protein interactions among ubiquitin-activation (E1), E2 and E3 enzymes (Fig. 1). Although many of the basic principles of ubiquitination are well understood, the full functional extent of proteins involved in the ubiquitination pathway remains to be discovered. For example, there are over 30 different E2s and many hundreds of E3s encoded by the human genome, raising the question of what E2/E3 pairs are functional. Also, ubiquitin can be conjugated to a substrate in multiple forms, including monoubiquitination and polyubiquitin chains of up to seven different linkage types, raising the question of how the exact nature of the product is specified. These two properties are at the heart of the remarkable diversity of the ubiquitination system [1,2].
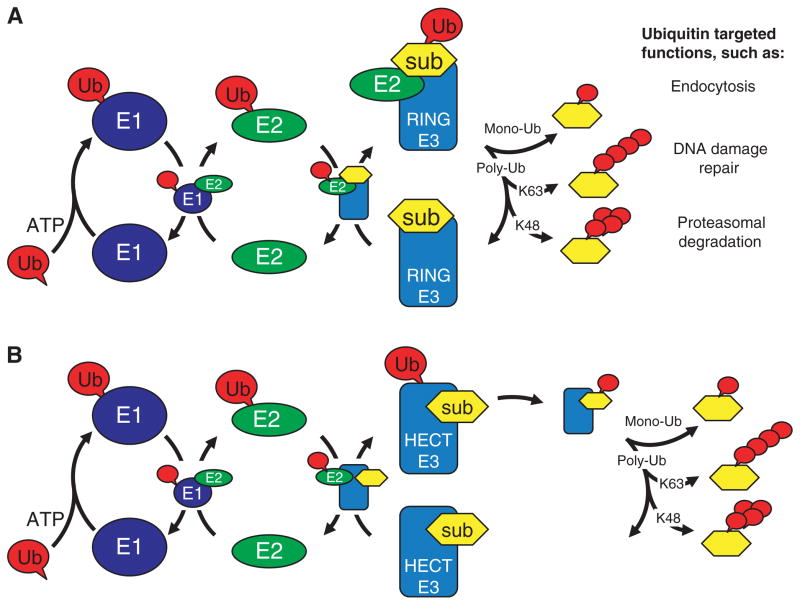
Fig. 1.
The ubiquitination pathway. Diagram illustrating protein–protein interactions that mediate the E3 dependent ubiquitination of substrates. (A) The pathway utilized by RING and U-box E3s and (B) the pathway utilized by HECT domain E3s. Proteins are noted as Ub (ubiquitin), E1 (ubiquitin-activating enzyme), E2 (ubiquitin-conjugating enzyme), E3 (ubiquitin ligase) and sub (substrate). The type of ubiquitin modification attached to the substrate dictates the function, activity, localization or even fate of the ubiquitinated protein.
Although there are a growing number of examples of E3s that are active with more than one E2, the relationships have mostly been identified from activity assays that include only a limited number of E2s. Until our study involving the E3 ligase breast cancer susceptibility gene 1 (BRCA1), no exhaustive screen to identify all possible E2s for a given E3 had been performed [3]. Unexpectedly we found that 10 of the ~ 30 human E2s can bind to BRCA1 and that different E2s have different ubiquitin-transfer properties in conjunction with BRCA1. E2/E3 interactions pose several challenges that must be overcome in a successful screen. First, the complexes are of modest affinity and often do not survive conventional approaches such as pull-down assays or co-immunoprecipitation. Second, a significant number of E3s function as dimers (homo-and heterodimeric E3s are known) or as multicomponent protein complexes and are only functional within those contexts. In this minireview, we outline a strategy that addresses these challenges and we illustrate its use using the heterodimeric ubiquitin ligase encoded by BRCA1 and its partner, BRCA1-associated RING domain (BARD1).
There are two distinct classes of E3s that function in mechanistically different manners. HECT domain E3s contain a cysteine residue that, similar to the E1 and E2 enzymes, forms a thiolester intermediate with the C-terminus of activated ubiquitin. In this case, ubiquitin is transferred from an E2 to an E3 and finally to a lysine side chain of a substrate protein. RING and U-box E3s do not possess an active-site cysteine residue; these E3s act by bringing the E2–ubiquitin (Ub) complex and substrate in close proximity to mediate the transfer of ubiquitin from the E2 directly to the substrate. Identification of substrates of specific E3s is a major goal in the ubiquitination field, but this has proven to be a difficult task. We believe that one limitation in previous efforts to discover and/or confirm substrates using biochemical assays may be the absence of the correct E2.
The RING-domain E3s are by far the largest family of ubiquitin ligases, with > 600 RING-containing proteins encoded in the human genome [4]. RING-domain E3s come in four molecular architectures: single-chain, homodimeric, heterodimeric and multi-component. In all known cases, a RING domain interacts directly with an E2. However, in an increasing number of RING and U-box E3s, additional structural elements contribute significantly to the interaction. In some cases, elements proximal to the RING are responsible for either homo- or heterodimerization of the RING domains [5–8], which modulates their activity and substrate specificity, and for BRCA1/BARD1 only the dimeric species binds an E2 [3]. In gp78, a single-chain RING E3, a sequence outside the RING domain forms a second E2-binding site [9]. The E3 Rad18 also contains a second E2 binding region distal to its RING [10–12]. At present, it is not known how many or which E3s use and/or require additional elements for their E2 interactions, but given the number already known, it seems imprudent to assume that residues encompassed entirely within the zinc-chelating cysteine and histidine residues which define the RING domain, will be sufficient when investigating an unknown E3.
All E2s are recognizable by their conserved catalytic domains (referred to as Ubc) which contain the active cysteine residue, and 3D structures solved for dozens of E2 Ubcs reveal a conserved architecture. In addition, E2s are categorized into four classes, depending on whether they consist of only a Ubc (Class I), or have additional sequences N-terminal, C-terminal or both to the Ubc domain (Classes II, III and IV, respectively). Although several co-crystal structures and NMR mapping studies of E2/E3 complexes confirm that Ubcs interact directly with E3s, there are examples of non-Class I E2s that use elements outside the Ubc in their E3 interaction (UbcH10 [13] and UbcM2 [14]). Of the ~ 30 human E2s, 11 are Class I. Thus, although the functions of non-Ubc regions remain to be determined for most E2s, strategies that utilize only the Ubc may miss important interactions or features.
Below, we describe the method we devised to detect the human E2s that bind to BRCA1/BARD1. We took advantage of the 3D structure of the heterodimeric RING domain solved in 2001 [5]. Subsequent structures of other heterodimeric RINGs, homodimeric RINGs and U-box dimers are strikingly similar [1]. Therefore, approaches that are successful with BRCA1/BARD1 are likely to be generalizable to many other E3s. As illustrated in Fig. 2, the approach is comprised of: (a) a directed yeast two-hybrid assay to identify putative binding partners, (b) protein NMR to confirm direct binding and to map binding interfaces, (c) structure-directed sequence analyses to identify binding determinants, and (d) in vitro ubiquiti-nation assays to assess the activity of each E2/E3 pair.
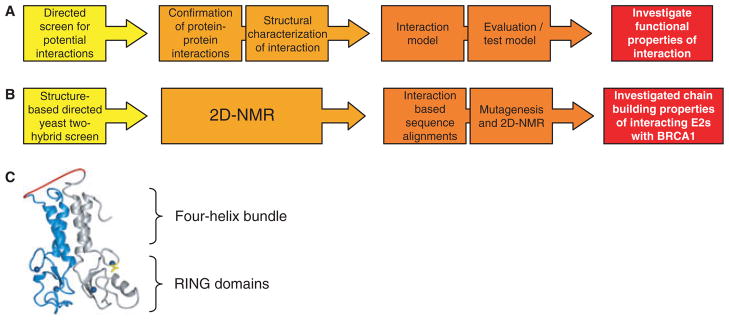
Fig. 2.
Strategy to identify all E2s that interact with an E3. (A) Generalized flow chart for identification, confirmation, modeling and evaluation of interactions between members of two domain families. (B) Methods used to identify new E2 interactions with BRCA1/BARD1. (C) Model for the fused BRCA1/BARD1 bait molecule, with BRCA1 in gray, BARD1 in blue, five residue Gly2-Ser-Gly2 linker in red, zinc atoms in dark blue, and BRCA1 RING residue Ile26 in yellow.
A structure-based directed yeast two-hybrid assay
To identify all possible E2s that interact with BRCA1/BARD1 we chose a yeast two-hybrid assay because it is capable of giving a positive readout for short-lived interactions that are not amenable to other biochemical binding assays. The yeast two-hybrid system is based on splitting an essential transcription factor into two domains, usually a DNA-binding domain and an activation domain [15,16]. Two proteins can be tested for a possible interaction by fusing one to the DNA-binding domain and the other to the activation domain of the transcription factor yielding ‘bait’ and ‘prey’ molecules, respectively. Reconstitution of transcription factor activity results in yeast viability or growth on selective media and indicates a positive interaction between the bait and prey fusion proteins. This method has been widely utilized to identify thousands of important protein–protein interactions [17].
To design a bait molecule that recapitulates the functionally important features of our E3, we relied on the structure of the BRCA1/BARD1 RING domains [5] (Fig. 2C) which revealed that the complex is held together by α helices that flank each RING domain and form an antiparallel four-helix bundle. The helical bundle places the N- and C-termini of each polypeptide chain in proximity, so we made a single-chain bait molecule in which the first ~110 residues of BARD1 are fused to the first ~110 residues of BRCA1, joined by a short five-residue linker. Thus, the bait, referred to as ‘BDfBC’, contains ~50 residues of BRCA1 that are outside its RING plus the sequence of the essential partner of BRCA1, BARD1, known to be necessary for its E3 ligase activity [3]. In vitro ubiquitination assays using purified BDfBC confirmed that it retains E3 ligase activity and therefore BDfBC was fused to the Gal4–DBD to create the yeast two-hybrid bait. As a further demonstration of the necessity of using our fused RING–RING bait molecule, we found that only BDfBC supported a positive interaction with UbcH5c, whereas the BRCA1 RING alone did not in the yeast two-hybrid assay [3]. Therefore, in the context of BDfBC, BARD1 residues comprising its RING and flanking α helices position the BRCA1 RING in the structural conformation necessary to support interactions with E2s.
Through a detailed investigation of the interaction between UbcH5c and the RING of BRCA1 our laboratory has identified a key BRCA1 RING residue, Ile26 [18]. This residue is found within the first zinc-binding cysteine motif of the BRCA1 RING domain (Cys-Pro-Ile-Cys) and is at the interface of the interaction between BRCA1 and UbcH5c. Mutation of Ile26 to alanine (I26A) completely disrupts the interaction between BRCA1 and both UbcH5c and UbcH7 [18]. A hydrophobic residue at a position analogous to BRCA1 Ile26 is highly conserved among the family of RING domains. Mutation of this hydrophobic residue in other RING domains, such as POSH [19], MDM2 [7] and Bmi1/RING1b [6] also eliminates ubiquitin ligase activity. Therefore, we hypothesize that mutation of other RING domains in a manner analogous to BRCA1 I26A can be used as a control when searching for specific E2 interactions.
An E2 prey library was generated by fusing 30 full-length E2 or E2-like human sequences with the Gal4 AD sequence. The E2s chosen for our screen were based on Lorick et al. [20]. However, some E2s were specifically eliminated based on the following criteria: (a) the E2 domain was part of a much larger protein (BRUCE and Ube2o), (b) a highly similar E2 was used as a representative (Rad6a is 95% identical to Rad6b and therefore likely binds to the same E3s), and (c) E2s localize to a cellular compartment were BRCA1 is not found (Ube2j1 and Ube2j2 localize to the endoplasmic reticulum [21,22] and BRCA1 to the nucleus). Each E2–activation domain fusion was co-transformed with BDfBC–DNA binding domain into the yeast two-hybrid strain AH109 (Clontech, Mountain View, CA, USA). Colonies were selected for the presence of both bait and prey vectors and spotted on selective media (synthetic dropout media lacking histidine, leucine and tryptophan) with increasing stringency (increasing concentration of 3-amino-1,2,4-triazole). Positive interactions were rescreened against empty vector controls. By replica-plating colonies expressing specific bait and prey pairs along with empty vector controls on selective media with increasing stringency, auto-activation (false positives) and specific interactions can be easily distinguished. Essentially, any bait and prey pair that supports yeast growth at a stringency level above that where growth is detected with an empty vector control is scored as a positive interaction. In our screen, some bait–prey pairs only supported growth at the lowest level of stringency, whereas other interactions supported growth even at the highest level of stringency tested. Thus, performing empty vector controls on the same plates as true bait–prey screens under a gradient of stringency levels increases the confidence level and allows for the quick elimination of false positives. Unfortunately, false positives can still arise. For example, Ubc9 passed this initial empty-vector control test and was only later determined to be a false positive using a second method (2D NMR titration experiments) aimed at confirming each positive interacting partner. Hence, in our screens Ubc9 was a false positive. We note that Ubc9 is reported as a ‘positive’ in many two-hybrid screens and would caution care before concluding a real interaction with this E2. Yeast two-hybrid screens can also suffer from false negatives presumably because not all interactions can support the appropriate activation of the reporter cassette, impeding their detection in a yeast two-hybrid context. Thus, care should be taken before concluding anything about the negatives in a screen. To safeguard against these two experimental possibilities, all interactions detected should be confirmed using a second method that can detect direct yet weak protein–protein interactions.
2D NMR confirmation of protein interactions
Putative interactions detected in a yeast two-hybrid system should always be confirmed using another method that is capable of distinguishing direct protein–protein interactions. For the reasons discussed above, (1H, 15N)-2D NMR was our method of choice because it is sensitive to even weak protein interactions [23]. The approach requires purified recombinant proteins where one protein of the pair is labeled with 15N by growth and expression in media containing 15N-labeled ammonium chloride. In our investigation, the BRCA1/BARD1 E3 was 15N-labeled because our laboratory had previously assigned the BRCA1/BARD1 NMR spectrum. However, in the general case, it may be simpler to label the E2 components which are more generally amenable to NMR, because of their relatively small, monomeric structures. To detect a protein–protein interaction, an unlabeled component is added to the 15N-labeled component and spectra obtained in the presence and absence of the putative binding partner are compared. In all cases of E2/E3 interactions studied structurally to date, binding is not accompanied by a significant conformational change in either protein. Thus, the expected behavior in the NMR is for a limited number of protein NMR peaks to be perturbed if a direct binding event occurs. Another approach to confirm direct binding will be needed if at least one of the protein partners is not amendable to NMR analysis. Two other approaches that have successfully detected E2/E3 interactions are isothermal titration calorimetry [24] and surface plasmon resonance [25].
In addition to providing confirmation of a direct protein–protein interaction, the NMR binding assay can provide residue-specific information regarding the binding site. In cases where the NMR spectrum of the 15N-labeled protein component is assigned, affected peaks in the HSQC spectrum identify specific residues involved in binding and can be mapped onto the 3D structure, if available [26,27]. In the case of E2/E3 interactions, all RING and U-box domains that have been studied to date bind to the same structural elements within the Ubc of E2s, namely, α helix-1, and the loops L1 and L2 [8,28,29].
NMR binding experiments confirmed the interaction of six new E2 partners for BRCA1 and identified Ubc9 as a false positive in the yeast two-hybrid screen. The data revealed that each E2 interacts with essentially the same surface of the BRCA1 RING as previously mapped for the E2 UbcH5c. Notably, in the NMR spectrum the BRCA1 Ile26 peak is affected most strongly in each E2 titration. Furthermore, the I26A mutation abrogates or weakens each E2–BRCA1 interaction in the context of our fused bait yeast two-hybrid system. These results and our previous results with UbcH5c revealed that Ile26 of the BRCA1 RING plays a central role in each E2 interaction.
Identification of binding determinants
The overall objective described herein is to identify all interactions that occur between a specific bait protein domain and a conserved family of prey domains where at least one representative is already known to interact. The yeast two-hybrid and subsequent NMR binding assays can provide high confidence positive interactions and can identify false positives. However, screening methods may also suffer from false negatives, that is, real interactions that are not detected in the screen. Although use of the directed screen described here (as opposed to a large yeast two-hybrid screen involving a cDNA library as prey) will increase the chances of observing positive interactions that may be weak, it is impossible to be sure that the screen results are complete. When dealing with a family of related proteins or protein domains, it may be possible to identify important determinants of the interaction(s) from multiple sequence alignments and to use this information to identify other members of the family that share the determinants and may therefore be false negatives.
To focus specifically on shared features that relate directly to binding as opposed to those that are required to stabilize the domain structure or provide its catalytic activity, we generated alignments for the key structural elements in the E2 Ubcs known to be involved in E3 interactions. Our previous NMR studies identified six UbcH5c residues that are the most affected by BRCA1/BARD1 binding. The alignment revealed that three of those six are completely conserved among the 10 BRCA1-interacting E2s: two are in the N-terminal helix of the Ubc (Arg5 and Lys8) and the third is in a loop known as Loop L2 (Ala96). The N-terminal helix comprises part of both the E1 and E3 binding surfaces on E2s [30], so mutating residues in this structural feature may affect E2 activity in a manner that may not be related to E3 binding. A role for Ala96 was confirmed by mutagenesis of this residue in UbcH5c and the analogous residue in Ubc13 and Ube2k. In all cases, substitution of the Ala abrogated the interaction with BRCA1/BARD1 as evidenced by NMR and the associated ubiquitin transfer activity. Ala96 of UbcH5 is part of a Ser-Pro-Ala motif commonly observed in loop L2 of many E2s. We found that the Ala residue of this motif plays an important role in determining the E2s that interact with BRCA1. Interestingly, the Ser-Pro-Ala motif has also been found to be important for selection of E2s that interact with the U-box E3 CHIP [31].
The multiple sequence alignment shown in Fig. 3 varies slightly from the one presented in our original publication [3]. In the intervening period, 3D structures have been deposited for the Ubc domains of almost every human E2 (http://www.rcsb.org). The alignment in Fig. 3 is structure based, rather than merely sequence based, and alignments of helix-1 have changed with this added information. The new alignment reveals three additional E2s, Ube2q1, Ube2q2 and Ube2z that appear to have the three conserved residues discussed above. Thus, these three E2s are good candidates for false negatives in the yeast two-hybrid screen and will be tested directly for binding to BRCA1/BARD1 in the future.
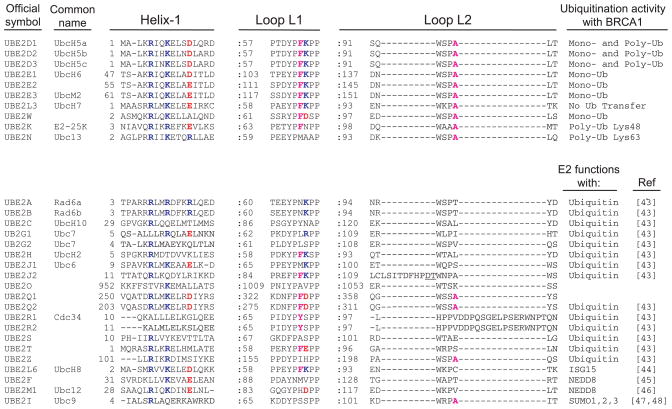
Fig. 3.
Structure-based multiple sequence alignment of human E2s. (Upper) Confirmed BRCA1-interacting E2s. (Lower) Non-BRCA1 interacting E2s. Sequences are shown only for E2 elements referred to as helix-1, loop L1 and loop L2, which are commonly found to interact with E3s. Ubiquitin transfer properties for E2s functioning in conjunction with BRCA1 are listed. The ubiquitin-like molecule used by each E2 is listed for non-BRCA1-interacting E2s.
In several other E2/E3 pairs that have been characterized structurally, the E2 loop L1 has been shown to be critical in E3 binding. In particular, the phenylalanine at position 62 (Fig. 3, UbcH5c numbering) and the lysine at position 63 have been implicated in binding to the RING E3s cCbl [29] and CNOT4 [28], respectively. Of particular note, substitution of Lys63 with a negatively charged residue abrogates interaction with CNOT4 [32]. As shown in Fig. 3, neither of these positions is conserved among the BRCA1-binding E2s and Ube2w has a negatively charged residue at the position analogous to Lys63. Thus, the limited data available concerning the binding determinants of E2s and RING domains indicate that although the same structural elements are used, different E3s use different positions within the structural elements to recognize and bind their cadre of E2s. Once complete sets of E2s have been identified for other RING E3s, patterns may emerge that can be used predictively.
Investigation of functional properties of newly identified interactions
Identification of all E2s that can interact with a given E3 ligase opens the door for an in-depth evaluation of the functional properties of each interacting pair. We evaluated the in vitro autoubiquitination activity of each E2 paired with BRCA1/BARD1 as a proxy for ubiquitin transfer activity. In contrast to the well-known polyubiquitination activity of BRCA1 with UbcH5c, four of the interacting E2s transfer a single ubiquitin to BRCA1. Two other interacting E2s, Ubc13 and Ube2k, do not transfer ubiquitin directly to BRCA1, but elongate polyubiquitin with specific chain linkages (Lys63 and Lys48, respectively) onto monoubiquitinated BRCA1. Thus, an assay designed to detect only (large) polyubiquitinated products in which a single E2 was present would have failed to identify activity for most of the E2/BRCA1 pairs, providing a cautionary note. Taken together, our results indicate a division of labor among the E2s that work with a given E3 ligase. The BRCA1-interacting E2s can be classified based on their activity: UbcH6, Ube2e2, UbcM2 and Ube2w transfer only a single ubiquitin to BRCA1 and are classified as substrate-specific (i.e. they transfer ubiquitin to a substrate lysine residue); Ubc13 and Ube2k can only extend a polyubiquitin chain and are classified as ubiquitin specific (i.e. they transfer ubiquitin to another ubiquitin lysine residue); UbcH5 transfers ubiquitin to either a substrate or another ubiquitin lysine and is therefore nonspecific, which may explain why this E2 is found to be active with most E3s; UbcH7 has no detectable ubiquitin transfer activity with BRCA1/BARD1, although it binds to the E3 ligase indistinguishably from UbcH5c. There is no reason to believe that BRCA1/BARD1 is unique in its activities and it is likely that other E3 ligases will be found to have different activity dependent upon the nature of the E2 or E2s present.
The ultimate goal to understand BRCA1 function and why mutations that eliminate ubiquitin ligase activity are associated with breast and ovarian cancer susceptibility requires the identification of substrates. Our results suggest that a strategy which includes all BRCA1-interacting E2s will be advantageous. The protocol in use in our laboratory is to first assay each substrate-specific E2 (and UbcH5) for its ability to transfer ubiquitin directly to a substrate. If monoubiquitination is observed, each ubiquitin-specific E2 is tested in combination with the active monoubiquitinating E2 for chain elongation. An alternate strategy in which a cocktail of E2s is assayed simultaneously can also be adopted. If a substrate is observed to become ubiquitinated, the identity of the E2 or E2s responsible can be quickly revealed.
The unexpected finding that BRCA1 and presumably other E3s can interact and function with numerous E2s begs the question, how does an E3 choose which E2 to use for ubiquitin transfer to specific substrates? It is unlikely that BRCA1 itself prefers a given E2 over other interacting E2s, because the binding affinities appear to be very similar. Furthermore, under assay conditions in which more than one E2 is present, products of both E2s are observed, indicating that one E2 does not out-compete the other. This is consistent with the modest E2/E3 affinities and fast off-rates reported [18,24,33,34], which allow for multiple E2s to access an E3’s RING to carry out their respective functions. It is likely that other factors present in a cell can affect or determine the relevant E2/E3 pairings. For example, BRCA1 is found in several multiprotein complexes in the cell [35–39]. Different E2s among the BRCA1-interacting set may be preferred depending on the BRCA1/BARD1-containing complex and substrate to be modified. Another factor may be the substrate itself, which must come in close proximity to the E2 at the moment of transfer. Post-translational modification of the E2 may also be a determining factor [40,41]. Other factors may include the subcellular localization or tissue distribution of the E2. To date, these features have not yet been rigorously determined for the E2s in any organism, but the information, when available, may well be informative.
Perspectives
With hundreds of ubiquitin ligases encoded in the human genome it is not surprising that many of these E3s have important biological functions. Loss of ubiquitin ligase activity can have devastating effects. For example, some of the most commonly observed cancer-associated missense mutations of BRCA1 are found in its RING domain and eliminate ubiquitin ligase activity [18,42]. This loss of activity suggests that loss of ubiquitin transfer to at least one critical substrate leads to breast and ovarian cancer susceptibility. Hence, the ultimate goal when investigating the important functions of an E3 has been to identify its substrates. Unfortunately, identifying substrates of specific E3s has had only limited success. We felt that a significant limitation may be lack of the proper E2. Therefore, we set out to identify all E2s that could function with a single E3. Unexpectedly, we found that BRCA1 could interact with multiple different E2s and displayed unique ubiquitin-transfer properties depending on the E2 used. It is unlikely that BRCA1 is unique in its ability to interact with so many different E2s. The screening of additional E3s for their cadre of interacting E2s will lead to further insight into the mechanisms of protein ubiquitination. Some foreseeable areas that will benefit include: identification of more E3s that function with the ubiquitin-like proteins, discovery of unique ubiquitin transfer properties for the remaining non-BRCA1 E2s, development of predictive models guiding E2 selection by an E3, and ideally, the ability to thoroughly investigate E3-dependent substrate ubiquitination.
Abbreviations
- BARD1
BRCA1-associated RING domain
- BRCA1
breast cancer susceptibility gene 1
- E1
ubiquitin-activating enzyme
- E2
ubiquitin-conjugating enzyme
- E3
ubiquitin ligase
- Ub
ubiquitin
References
- 1.Hibbert RG, Mattiroli F, Sixma TK. Structural aspects of multi-domain RING/Ubox E3 ligases in DNA repair. DNA Repair (Amst) 2009;8:525–535. doi: 10.1016/j.dnarep.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 2.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen DE, Brzovic PS, Klevit RE. E2–BRCA1 RING interactions dictate synthesis of mono- or specific polyubiquitin chain linkages. Nat Struct Mol Biol. 2007;14:941–948. doi: 10.1038/nsmb1295. [DOI] [PubMed] [Google Scholar]
- 4.Li W, Bengtson MH, Ulbrich A, Matsuda A, Reddy VA, Orth A, Chanda SK, Batalov S, Joazeiro CA. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PLoS ONE. 2008;3:e1487. doi: 10.1371/journal.pone.0001487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brzovic PS, Rajagopal P, Hoyt DW, King MC, Klevit RE. Structure of a BRCA1–BARD1 heterodimeric RING–RING complex. Nat Struct Biol. 2001;8:833–837. doi: 10.1038/nsb1001-833. [DOI] [PubMed] [Google Scholar]
- 6.Buchwald G, van der Stoop P, Weichenrieder O, Perrakis A, van Lohuizen M, Sixma TK. Structure and E3-ligase activity of the Ring–Ring complex of polycomb proteins Bmi1 and Ring1b. EMBO J. 2006;25:2465–2474. doi: 10.1038/sj.emboj.7601144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Linke K, Mace PD, Smith CA, Vaux DL, Silke J, Day CL. Structure of the MDM2/MDMX RING domain heterodimer reveals dimerization is required for their ubiquitylation in trans. Cell Death Differ. 2008;15:841–848. doi: 10.1038/sj.cdd.4402309. [DOI] [PubMed] [Google Scholar]
- 8.Zhang M, Windheim M, Roe SM, Peggie M, Cohen P, Prodromou C, Pearl LH. Chaperoned ubiquitylation – crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP–Ubc13–Uev1a complex. Mol Cell. 2005;20:525–538. doi: 10.1016/j.molcel.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Back JW, Notenboom V, de Koning LJ, Muijsers AO, Sixma TK, de Koster CG, de Jong L. Identification of cross-linked peptides for protein interaction studies using mass spectrometry and 18O labeling. Anal Chem. 2002;74:4417–4422. doi: 10.1021/ac0257492. [DOI] [PubMed] [Google Scholar]
- 11.Bailly V, Lauder S, Prakash S, Prakash L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J Biol Chem. 1997;272:23360–23365. doi: 10.1074/jbc.272.37.23360. [DOI] [PubMed] [Google Scholar]
- 12.Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Summers MK, Pan B, Mukhyala K, Jackson PK. The unique N terminus of the UbcH10 E2 enzyme controls the threshold for APC activation and enhances checkpoint regulation of the APC. Mol Cell. 2008;31:544–556. doi: 10.1016/j.molcel.2008.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plafker KS, Singer JD, Plafker SM. The ubiquitin conjugating enzyme, UbcM2, engages in novel interactions with components of cullin-3 based E3 ligases. Biochemistry. 2009;48:3527–3537. doi: 10.1021/bi801971m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chien CT, Bartel PL, Sternglanz R, Fields S. The two-hybrid system: a method to identify and clone genes for proteins that interact with a protein of interest. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fields S, Sternglanz R. The two-hybrid system: an assay for protein–protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 17.Parrish JR, Gulyas KD, Finley RL., Jr Yeast two-hybrid contributions to interactome mapping. Curr Opin Biotechnol. 2006;17:387–393. doi: 10.1016/j.copbio.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 18.Brzovic PS, Keeffe JR, Nishikawa H, Miyamoto K, Fox D, III, Fukuda M, Ohta T, Klevit RE. Binding and recognition in the assembly of an active BRCA1/BARD1 ubiquitin-ligase complex. Proc Natl Acad Sci USA. 2003;100:5646–5651. doi: 10.1073/pnas.0836054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alroy I, Tuvia S, Greener T, Gordon D, Barr HM, Taglicht D, Mandil-Levin R, Ben-Avraham D, Konforty D, Nir A, et al. The trans-Golgi network-associated human ubiquitin-protein ligase POSH is essential for HIV type 1 production. Proc Natl Acad Sci USA. 2005;102:1478–1483. doi: 10.1073/pnas.0408717102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorick KL, Jensen JP, Weissman AM. Expression, purification, and properties of the Ubc4/5 family of E2 enzymes. Methods Enzymol. 2005;398:54–68. doi: 10.1016/S0076-6879(05)98006-3. [DOI] [PubMed] [Google Scholar]
- 21.Oh RS, Bai X, Rommens JM. Human homologs of Ubc6p ubiquitin-conjugating enzyme and phosphorylation of HsUbc6e in response to endoplasmic reticulum stress. J Biol Chem. 2006;281:21480–21490. doi: 10.1074/jbc.M601843200. [DOI] [PubMed] [Google Scholar]
- 22.Tiwari S, Weissman AM. Endoplasmic reticulum (ER)-associated degradation of T cell receptor subunits. Involvement of ER-associated ubiquitin-conjugating enzymes (E2s) J Biol Chem. 2001;276:16193–16200. doi: 10.1074/jbc.M007640200. [DOI] [PubMed] [Google Scholar]
- 23.Vaynberg J, Qin J. Weak protein–protein interactions as probed by NMR spectroscopy. Trends Biotechnol. 2006;24:22–27. doi: 10.1016/j.tibtech.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 24.Eletr ZM, Kuhlman B. Sequence determinants of E2–E6AP binding affinity and specificity. J Mol Biol. 2007;369:419–428. doi: 10.1016/j.jmb.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marteijn JA, van der Meer LT, Smit JJ, Noordermeer SM, Wissink W, Jansen P, Swarts HG, Hibbert RG, de Witte T, Sixma TK, et al. The ubiquitin ligase Triad1 inhibits myelopoiesis through UbcH7 and Ubc13 interacting domains. Leukemia. 2009;23:1480–1489. doi: 10.1038/leu.2009.57. [DOI] [PubMed] [Google Scholar]
- 26.Gao G, Williams JG, Campbell SL. Protein–protein interaction analysis by nuclear magnetic resonance spectroscopy. Methods Mol Biol. 2004;261:79–92. doi: 10.1385/1-59259-762-9:079. [DOI] [PubMed] [Google Scholar]
- 27.Zuiderweg ER. Mapping protein–protein interactions in solution by NMR spectroscopy. Biochemistry. 2002;41:1–7. doi: 10.1021/bi011870b. [DOI] [PubMed] [Google Scholar]
- 28.Dominguez C, Bonvin AM, Winkler GS, van Schaik FM, Timmers HT, Boelens R. Structural model of the UbcH5B/CNOT4 complex revealed by combining NMR, mutagenesis, and docking approaches. Structure. 2004;12:633–644. doi: 10.1016/j.str.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 29.Zheng N, Wang P, Jeffrey PD, Pavletich NP. Structure of a c-Cbl–UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell. 2000;102:533–539. doi: 10.1016/s0092-8674(00)00057-x. [DOI] [PubMed] [Google Scholar]
- 30.Huang DT, Paydar A, Zhuang M, Waddell MB, Holton JM, Schulman BA. Structural basis for recruitment of Ubc12 by an E2 binding domain in NEDD8’s E1. Mol Cell. 2005;17:341–350. doi: 10.1016/j.molcel.2004.12.020. [DOI] [PubMed] [Google Scholar]
- 31.Xu Z, Kohli E, Devlin KI, Bold M, Nix JC, Misra S. Interactions between the quality control ubiquitin ligase CHIP and ubiquitin conjugating enzymes. BMC Struct Biol. 2008;8:26. doi: 10.1186/1472-6807-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winkler GS, Albert TK, Dominguez C, Legtenberg YI, Boelens R, Timmers HT. An altered-specificity ubiquitin-conjugating enzyme/ubiquitin-protein ligase pair. J Mol Biol. 2004;337:157–165. doi: 10.1016/j.jmb.2004.01.031. [DOI] [PubMed] [Google Scholar]
- 33.Eletr ZM, Huang DT, Duda DM, Schulman BA, Kuhlman B. E2 conjugating enzymes must disengage from their E1 enzymes before E3-dependent ubiquitin and ubiquitin-like transfer. Nat Struct Mol Biol. 2005;12:933–934. doi: 10.1038/nsmb984. [DOI] [PubMed] [Google Scholar]
- 34.Ulrich HD. Protein–protein interactions within an E2–RING finger complex. Implications for ubiquitin-dependent DNA damage repair. J Biol Chem. 2003;278:7051–7058. doi: 10.1074/jbc.M212195200. [DOI] [PubMed] [Google Scholar]
- 35.Chiba N, Parvin JD. Redistribution of BRCA1 among four different protein complexes following replication blockage. J Biol Chem. 2001;276:38549–38554. doi: 10.1074/jbc.M105227200. [DOI] [PubMed] [Google Scholar]
- 36.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 37.Sobhian B, Shao G, Lilli DR, Culhane AC, Moreau LA, Xia B, Livingston DM, Greenberg RA. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B, Matsuoka S, Ballif BA, Zhang D, Smogorzewska A, Gygi SP, Elledge SJ. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan J, Kim YS, Yang XP, Li LP, Liao G, Xia F, Jetten AM. The ubiquitin-interacting motif containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 2007;67:6647–6656. doi: 10.1158/0008-5472.CAN-07-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pichler A, Knipscheer P, Oberhofer E, van Dijk WJ, Körner R, Olsen JV, Jentsch S, Melchior F, Sixma TK. SUMO modification of the ubiquitin-conjugating enzyme E2-25K. Nat Struct Mol Biol. 2005;12:264–269. doi: 10.1038/nsmb903. [DOI] [PubMed] [Google Scholar]
- 41.Sadowski M, Mawson A, Baker R, Sarcevic B. Cdc34 C-terminal tail phosphorylation regulates Skp1/cullin/F-box (SCF)-mediated ubiquitination and cell cycle progression. Biochem J. 2007;405:569–581. doi: 10.1042/BJ20061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hashizume R, Fukuda M, Maeda I, Nishikawa H, Oyake D, Yabuki Y, Ogata H, Ohta T. The RING heterodimer BRCA1–BARD1 is a ubiquitin ligase inactivated by a breast cancer-derived mutation. J Biol Chem. 2001;276:14537–14540. doi: 10.1074/jbc.C000881200. [DOI] [PubMed] [Google Scholar]
- 43.Jin J, Li X, Gygi SP, Harper JW. Dual E1 activation systems for ubiquitin differentially regulate E2 enzyme charging. Nature. 2007;447:1135–1138. doi: 10.1038/nature05902. [DOI] [PubMed] [Google Scholar]
- 44.Zhao C, Beaudenon SL, Kelley ML, Waddell MB, Yuan W, Schulman BA, Huibregtse JM, Krug RM. The UbcH8 ubiquitin E2 enzyme is also the E2 enzyme for ISG15, an IFN-α/β-induced ubiquitin-like protein. Proc Natl Acad Sci USA. 2004;101:7578–7582. doi: 10.1073/pnas.0402528101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang DT, Ayrault O, Hunt HW, Taherbhoy AM, Duda DM, Scott DC, Borg LA, Neale G, Murray PJ, Roussel MF, et al. E2–RING expansion of the NEDD8 cascade confers specificity to cullin modification. Mol Cell. 2009;33:483–495. doi: 10.1016/j.molcel.2009.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gong L, Yeh ET. Identification of the activating and conjugating enzymes of the NEDD8 conjugation pathway. J Biol Chem. 1999;274:12036–12042. doi: 10.1074/jbc.274.17.12036. [DOI] [PubMed] [Google Scholar]
- 47.Desterro JM, Thomson J, Hay RT. Ubch9 conjugates SUMO but not ubiquitin. FEBS Lett. 1997;417:297–300. doi: 10.1016/s0014-5793(97)01305-7. [DOI] [PubMed] [Google Scholar]
- 48.Tatham MH, Jaffray E, Vaughan OA, Desterro JM, Botting CH, Naismith JH, Hay RT. Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem. 2001;276:35368–35374. doi: 10.1074/jbc.M104214200. [DOI] [PubMed] [Google Scholar]