Abstract
Directed mutation is a proposed process that allows mutations to occur at higher frequencies when they are beneficial. Until now, the existence of such a process has been controversial. Here we describe a novel mechanism of directed mutation mediated by the transposon, IS5 in Escherichia coli. crp deletion mutants mutate specifically to glycerol utilization (Glp+) at rates that are enhanced by glycerol or the loss of the glycerol repressor (GlpR), depressed by glucose or glpR overexpression, and RecA-independent. Of the four tandem GlpR-binding sites (O1–O4) upstream of the glpFK operon, O4 specifically controls glpFK expression while O1 primarily controls mutation rate in a process mediated by IS5 hopping to a specific site on the E. coli chromosome upstream of the glpFK promoter. IS5 insertion into other gene activation sites is unaffected by the presence of glycerol or the loss of GlpR. The results establish an example of transposon-mediated directed mutation, identify the protein responsible, and define the mechanism involved.
Keywords: Directed mutation, insertion sequence, transposition regulation, transposon, transposable genetic element
Introduction
65 years ago, Luria and Delbruck reported that bacterial mutations from virus sensitivity to virus resistance arose randomly (Luria & Delbrück, 1943). They generalized their results, concluding that genetic mutations occur in the absence of selection, rather than arising in response to selection. It has thus become a basic principle of genetics that the likelihood of a particular mutation occurs independently of its phenotypic consequences (Rosenberg, 2001). The concept of directed mutation, defined as genetic change that is specifically induced by the stress conditions that the mutation relieves (Cairns et al., 1988), challenges this principle (Foster, 1999, Rosenberg, 2001, Wright, 2004). The topic of directed mutation is controversial, and its existence, as defined above, has been altogether questioned (Foster, 1997, Hendrickson et al., 2002, Roth et al., 2006).
The most studied potential example of directed mutation is the E. coli lac frameshift reversion system, in which cells carrying a lac +1 frameshift allele on an F’ episome mutate to Lac+ at a rate that is elevated by lactose during starvation (Cairns & Foster, 1991). A subsequent genome-wide analysis showed that such Lac+ point mutations (i.e., −1 deletions) are not ‘directed’ since higher frequencies of mutation also occur simultaneously in other chromosomal genes unrelated to lactose metabolism, and these unrelated mutations are usually not detrimental (Torkelson et al., 1997). A similar observation was examined in another study showing that mutations in a tet gene occurred in the same F’ episome concurrently with the Lac+ mutation (Foster, 1997). Two mechanisms were proposed to explain the increased rates of reversion to Lac+, one involving double-strand break repair mediated by error-prone DNA polymerase IV (Harris et al., 1994), the other involving amplification of the lac frameshift allele, thereby creating more targets for the occurrence of Lac+ point mutations (Andersson et al., 1998, Hendrickson et al., 2002). The first model was confirmed in a follow-up study (Ponder et al., 2005). The processes of both lac allele amplification and Lac+ point mutations were induced by lactose starvation and regulated by the stationary phase regulator, RpoS (Lombardo et al., 2004). These two processes have been shown to be two independent outcomes of genetic change during starvation, and both of them are adaptive processes (Hastings et al., 2004, Slack et al., 2006). This second model would be compatible with current neo-Darwinist dogma [constant genetic change followed by selection).
Amino acid biosynthetic auxotrophs of E. coli can be mutated to grow in the absence of the respective amino acids (Cashel et al., 1996). One such auxotroph, which contains a C-to-T point mutation (leading to a S286L mutation of the protein) at nucleotide 857 in the leuB gene, can be reverted to Leu+ when the Leu− cells are incubated under leucine-limited conditions (Wright & Minnick, 1997). 36 out of 53 Leu+ revertants analyzed were found to harbor a single nucleotide substitution that resulted in alteration of the 286th residue of LeuB from leucine to serine, valine or methionine (Wright & Minnick, 1997). These Leu+ reversion mutations proved to occur specifically due to leucine starvation, and the base substitutions appeared to be specific within the E. coli genome (Wright et al., 1999). The increased rate of Leu+ mutation is believed to result from increased rates of transcription elicited by guanosine tetraphosphate (ppGpp) (Reimers et al., 2004, Wright et al., 1999), whose synthesis is catalyzed by the ppGpp synthase during leucine starvation (Cashel et al., 1996). This transcription-promoted Leu+ mutation could be a potential example of directed mutation since most (but not all) of the identified mutations are directed to the leuB gene whose product is needed for the Leu+ phenotype.
Part of the justifiable skepticism concerning directed mutation resulted from experiments that supposedly demonstrated this phenomenon, but were subsequently shown to be explainable by classical genetics (Roth & Andersson, 2004, Roth et al., 2006). Mutation rates vary with environmental conditions [e.g., growth state) and genetic background (e. g., mutator genes) (Denamur & Matic, 2006, Foster, 2005, Wright, 2004), but this does not render the mutation “directed”. To establish the principle of directed mutation, it is necessary to demonstrate the phenomenon, identify the proteins involved, and characterize the mechanism responsible.
One frequently encountered type of mutation results from the hopping of transposable elements, transposons, which can activate or inactivate critical genes or operons when inserted into appropriate chromosomal loci (Chandler & Mahillon, 2002, Galas, 1989, Mahillon & Chandler, 1998) Transposon-mediated mutations occurring under stress conditions (e.g. toxin presence or starvation) have been shown to be usually beneficial to host organisms. In the presence of nitroaromatic compounds, NfsA and NfsB encoded by the E. coli nfsAB operon convert nitroaromatic compounds to toxic nitro-anion free radicals via nitroreduction (McCalla, 1979). When E. coli cells are exposed to nitroaromatic compounds contained in agar plates, resistant colonies appeared during incubation and all mutants were mutated in nfsA, nfsB or both by insertion of IS1, IS5 or other insertion elements (Whiteway et al., 1998). Activation of the normally cryptic β-glucoside (bgl) catabolic operon (Reynolds et al., 1981, Schnetz & Rak, 1992) and the ade gene encoding an adenine deaminase (Petersen et al., 2002) in E. coli can be accomplished by insertion of either IS1 or IS5 near the promoter. IS5 has also been found to activate the fucose/propandiol (PPD) fucAO promoter (Chen et al., 1989) and the flagellar motility master switch flhDC promoter (Barker et al., 2004).
The E. coli glp regulon consists of five operons, two of which (glpFK and glpD) are required for aerobic growth on glycerol (Lin, 1976). The glpFK operon encodes the glycerol facilitator, GlpF, and glycerol kinase (converting glycerol to glycerol-3-phosphate), GlpK, while the glpD gene encodes the aerobic glycerol-3-P dehydrogenase (Lin, 1976, 1996). Both operons are subject to negative control by the DNA-binding glp regulon repressor, GlpR (Zeng et al., 1996), which also binds glycerol-3-phosphate, the inducer of the glp regulon. The glpFK operon is additionally subject to positive regulation by Crp complexed with cAMP although glpD is not (Weissenborn et al., 1992). The glpFK regulatory region contains four GlpR binding sites, O1–O4, and two Crp binding sites which overlap O2 and O3 (Figure 1A). The strong Crp dependency of glpFK transcription is reflected by the fact that crp mutant cells are unable to utilize glycerol.
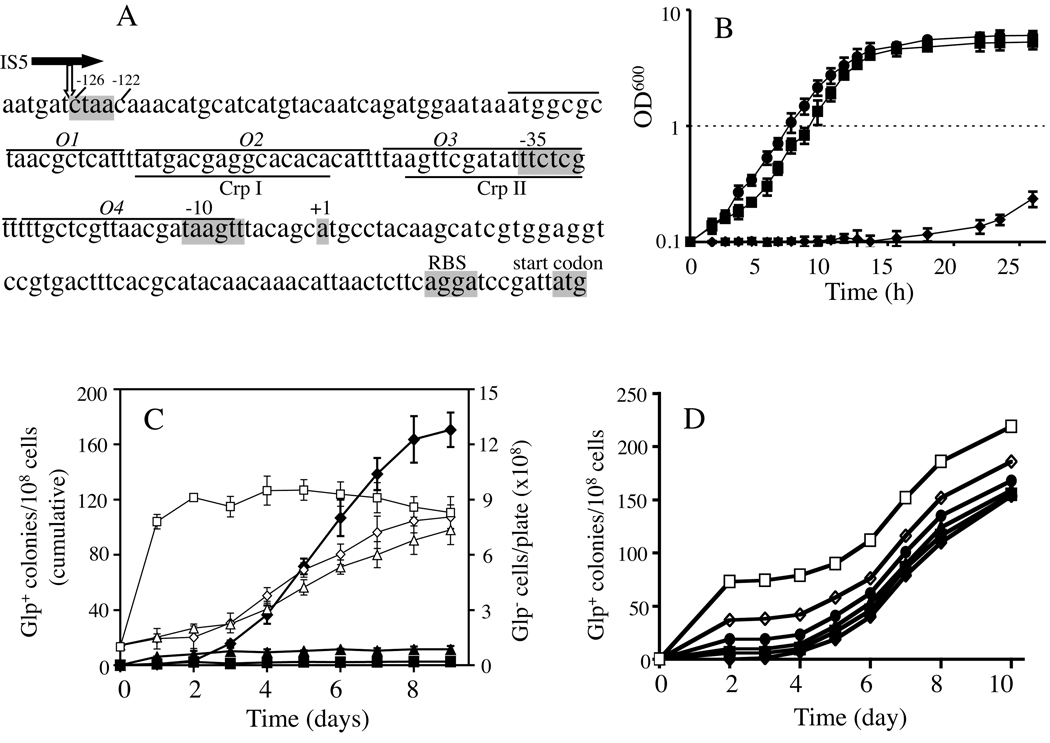
Figure 1. The appearance of Glp+ mutations in a crp genetic background.
(A) The glpFK promoter region. The transcriptional initiation site (+1), the −10 and −35 hexamers, the ribosome binding site (RBS) and the start codon for glpF translation are shaded. The GlpR binding sites (O1–O4; lines above the sequence) and Crp binding sites (CrpI and CrpII; lines under the sequence) are also shown. The location of the IS5 element upstream of the promoter in crp Glp+ cells is indicated by the vertical white arrow below the horizontal black arrow representing IS5. This arrow shows the orientation of IS5 with the 3’ end of its transposase gene linked to the downstream promoter. The 4-nucleotide IS5 target sequence (ctaa) at −126 to −122 is shaded. (B) Growth of E. coli wt (■), crp (◆), and crp Glp+ (●) cells in liquid glycerol M9 minimal medium. (C) Cumulative Glp+ mutations appearing per 108 cells (determined on the day measured) as a function of time in various media: solid M9 minimal media + 1% glycerol (♦), 0.01% glucose (■) or 1% sorbitol (σ). Total Glp− cell populations on these plates are denoted by the lines with ◊ (glycerol), □ (glucose) or △ (sorbitol). (D) The same minimal glycerol agar plates, where the crp mutant cells were plated together with various numbers of cells from a crp Glp+ strain: (□, 72; ◊, 38; ③, 19; σ, 10; ⑤, 5; and ♦, 0). The crp Glp+ cells were mixed with crp cells (108) and then applied onto M9 glycerol agar plates. Six independently isolated Glp+ mutant strains were tested and they behaved similarly. The results from one such Glp+ strain were shown in Figure 1D.
We have shown that in the absence of Crp, the glpFK promoter can be activated by IS5 when this genetic element inserts upstream of the promoter (Zhang, submitted). High level expression of the resultant activated operon is nearly constitutive and relies on the DNA phase between the inserted IS5 and the promoter. A short region (IB) at the 3’ end of IS5, which contains an IHF binding site and A-tracts that generate a permanent bend (Muramatsu et al., 1988), is necessary and sufficient for such activation. Moreover, IS5 or IB, when inserted into appropriate sites, can activate other Crp-controlled promoters (Zhang, submitted).
Here, we demonstrate that this insertion event represents a genuine example of directed mutation. Mutation is mediated by IS5 insertion at a specific site upstream of the glpFK promoter. We describe the process and its mechanistic details. Mutation of a crp deletion mutant to Glp+ occurs with a ten fold higher frequency when glycerol is present or GlpR is lacking, but overexpression of glpR greatly depresses the mutation rate. Frequencies of IS5 insertion to other sites that activate dissimilar promoters were unaffected by glycerol or the loss of GlpR. We show that GlpR provides two distinct biological functions, one, recognized previously, to control gene expression by binding to the downstream operator, O4, and the other, described here, to control the IS5-dependent mutation rate by binding to the upstream operator, O1. This is the first example of transposon-mediated directed mutation where the molecular explanation, involving a DNA-binding protein, has been provided.
Results
Glp+ Mutations in A crp Genetic Background
E. coli cells that lack Crp can not utilize glycerol as the sole carbon and energy source since expression of an operon (glpFK) essential for glycerol uptake and metabolism is dependent on Crp. However, when crp cells were incubated on solid glycerol minimal medium, Glp+ colonies appeared after a prolonged incubation. Five crp Glp+ colonies (mutants) that were from different plates and arose on different days were purified and tested for growth on glycerol in defined liquid medium. These mutants all grew equally well. The growth curve for one such mutant is shown in Figure 1B. The lag phase for the crp Glp+ strain was shorter than, and its growth rate was at least equal to that of wild type (wt) E. coli. Using Biolog plates, we compared these two strains (parental crp Glp− and crp Glp+) for oxidation of other carbon, nitrogen, phosphorous and sulfur sources, but no obvious differences were observed. This fact suggests that the mutation that enables crp cells to utilize glycerol did not affect other phenotypes observed for parental crp cells.
The relative frequencies of Glp+ mutation were determined with 1% glycerol, 1% sorbitol or 0.01% glucose as the sole carbon source on minimal M9 agar plates (Figure 1C). On glycerol plates, colonies appeared first after 3 days although wt and crp Glp+ E. coli cells formed visible colonies in < 2 days. New colonies continued to appear at increasing frequencies thereafter. When the same crp cells were plated as before, but variable numbers of the cells from a crp Glp+ strain were included with the cells before plating, colonies appeared from the crp Glp+ cells within two days, and new Glp+ mutants arose at the same frequency as before (Figure 1D). We repeated this experiment using another five independent crp Glp+ mutant strains (isolated from different M9 + glycerol plates and arose on different days) by mixing their cells with crp cells on M9 + glycerol plates, and they all behaved similarly, i.e., colonies from added crp Glp+ cells appeared in two days. Thus, the Glp+ mutants arising from crp cells on these glycerol minimal medium plates were not present in the cell culture initially plated, and no growth inhibitor could account for the results. The ratio of Glp+ colonies (each colony represents an independent mutation that occurs after plating) to the total population increased with time on M9 glycerol plates (Figure 1C). On M9 agar plates containing sorbitol (a sugar that crp cells can not utilize appreciably), the frequency of appearance of Glp+ mutations was much lower than that on glycerol plates but substantially higher than that on glucose plates. On M9 glucose plates, the ratios of the Glp+ population to the total population did not increase with time (Figure 1C).
The total populations (i.e., the numbers of all cells on a plate with the numbers of Glp+ cells subtracted) on each of the three types of plates were determined (Figure 1C). The crp Glp− populations increased very slowly on the glycerol agar plates, from 108 cells per plate (day 0) to ~8×108 cells per plate during a 9-day incubation. Similar slow growth of crp Glp− cells (from 108 cells per plate at day 0 to ~7×108 cells per plate at day 9) was observed on the sorbitol agar plates. These slow growth rates may be due to (1) the presence of residual nutrients in M9 salts; and (2) the low-level leaky expression of the glpFK operon (for slow growth on the glycerol plates) and the srlAEBD operon (for slow growth on the sorbitol plates) in the absence of Crp. On the glucose agar plates, the total population increased to the maximal level at day 1 and maintained this level thorough out the experimental period.
IS5 Upstream of PglpFK in the crp Glp+ Mutants
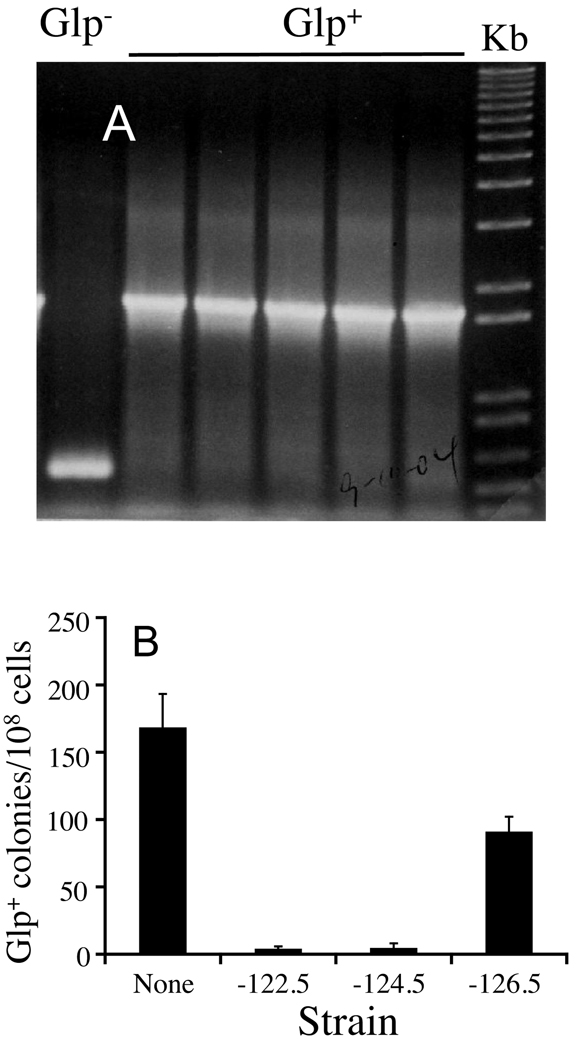
Independent crp Glp+ colonies that were isolated from different M9 + glycerol plates and arose on day 2 to day 9 after plating were purified, and the regulatory regions of the five glp operons were amplified by PCR and analyzed by gel electrophoresis and DNA sequencing. Four of these five operons were unchanged, but the glpFK regulatory region from 116 out of 116 independently isolated Glp+ colonies contained a ~1.1 kb insert. Shown in Figure 2A are the amplified products of the glpFK regulatory region from a small fraction of crp Glp+ colonies analyzed. Subsequent DNA sequencing analyses of all of these 116 mutants revealed that the insert is IS5, located upstream of the glpFK promoter at 126.5 nucleotides relative to the transcriptional start site and always orientated with the 3' end proximally upstream of the promoter (Figure 1A). The four base IS5 recognition sequence, CTAA, was repeated immediately adjacent to IS5 as expected (Mahillon & Chandler, 1998).
Figure 2. Gel electrophoresis of the PCR products of the glpFK regulatory regions in crp Glp− and crp Glp+ colonies (A) and effects of alteration of the IS5 insertion site (CTAA) on the appearance of Glp+ mutants (B).
In (A), the five crp Glp+ mutants shown were independently isolated. The glpFK regulatory regions were amplified using primers PglpFK-EcoR and PglpFK-Bam (Table S2). Size standards are shown on the far right. In (B) crp cells containing an 85-bp DNA fragment at −122.5, −124.5 and −126.5 relative to +1 of PglpFK, or lacking such an insert (none), as indicated on the X axis, were assayed for appearance of Glp+ colonies on M9 + glycerol agar plates. Numbers of colonies were scored after day 8.
To rule out the possibility that other mutations in addition to IS5 insertion had occurred, we transferred the Glp+ mutation from a crp Glp+ strain to the crp Glp− strain by P1 transduction. Colonies arose on glycerol minimal plates in 1.5 to 2 days (Note that spontaneous Glp+ mutants formed visible colonies only after 2.5 days). The colonies resulting from transduction were isolated, purified and examined for the presence of IS5 and for growth on glycerol. All 10 out of 10 Glp+ colonies (transductants) analyzed contained the IS5 element in the same position and orientation as in the donor crp Glp+ strains. They all grew on glycerol at the same rate as the donor crp Glp+ strain. We repeated the P1 transduction experiment using four other independently arising crp Glp+ strains as donors, and the same observations were made (i.e., Glp+ colonies from the transduction plates contained IS5 situated and oriented as for their derived crp Glp+ strains, and they grew on glycerol equally well). We sequenced the entire glpFK opeorn from three crp Glp+ strains and no other mutation was detected. These results show that the IS5 insert is both necessary and sufficient for the observed growth of crp cells on glycerol.
To determine if the Glp+ phenotype can result from insertion of other DNA elements upstream of PglpFK, we inserted a Kanamycin resistance (km) gene flanked by a FLP recognition target (FRT) (Datsenko & Wanner, 2000) at positions −126.5, −124.5 and −122.5 relative to +1 of PglpFK (see Figure 1A). After the km gene was deleted, an 85-bp FRT sequence remained at the same locations (i.e., immediately upstream, within or downstream of the CTAA insertion site, respectively). crp cells with the km gene or the 85-bp fragment at these three different locations could not grow on glycerol (data not shown). The Glp+ phenotype therefore resulted from the presence of IS5 and not a random DNA element. These results are consistent with our previous study showing that IS5 activation of PglpFK is not due to derepression (Zhang, submitted).
The three crp strains which carried the 85-bp fragment at −126.5, −124.5, and −122.5 were tested for the appearance of Glp+ mutations on M9 glycerol agar plates. Glp+ colonies from cells in which the IS5 CTAA insertion site was destroyed (−124.5) or where the insert was downstream of CTAA (−122.5) appeared with a frequency of <2% compared to that of the parental crp cells (Figure 2B). In the case where the CTAA (−126.5) was unchanged, but the 85-bp fragment was located immediately upstream of the site, the Glp+ mutation frequency decreased ~50% compared to the crp cells (Figure 2B). These results demonstrate that (i) the CTAA insertion site upstream of PglpFK is required for IS5 hopping to this region, (ii) an appropriate location of the CTAA element upstream of PglpFK is essential for glpFK operon activation, and (iii) the adjacent sequences upstream of the CTAA target site are important for maximization of the IS5 insertion rate. Sequencing analyses showed that the Glp+ mutants derived from the crp strain with the 85-bp insert at position −126.5 still harbored the IS5 element upstream of PglpFK, at the same location and orientation as in Glp+ mutants derived from the parental crp strain (see Figure 1A). However, the very low frequency of Glp+ mutations observed for the crp strains with inserts at positions −122.5 and −124.5 was due to IS2 insertion rather than IS5 insertion in the 85-bp FRT region. These results will be presented in more detail elsewhere.
Dependency of the Glp+ Mutation Rate on GlpR
Glycerol is phosphorylated by GlpK to glycerol-3-phosphate which binds to and releases GlpR from its operators (Lin, 1976, Weissenborn et al., 1992). When GlpR dissociates from its operators, a conformational change might be transmitted through the DNA, promoting insertion of IS5 at the CTAA site upstream of PglpFK. In other words, GlpR binding could have two functions: repression of gene expression and suppression of IS5 transposition to the upstream activating site.
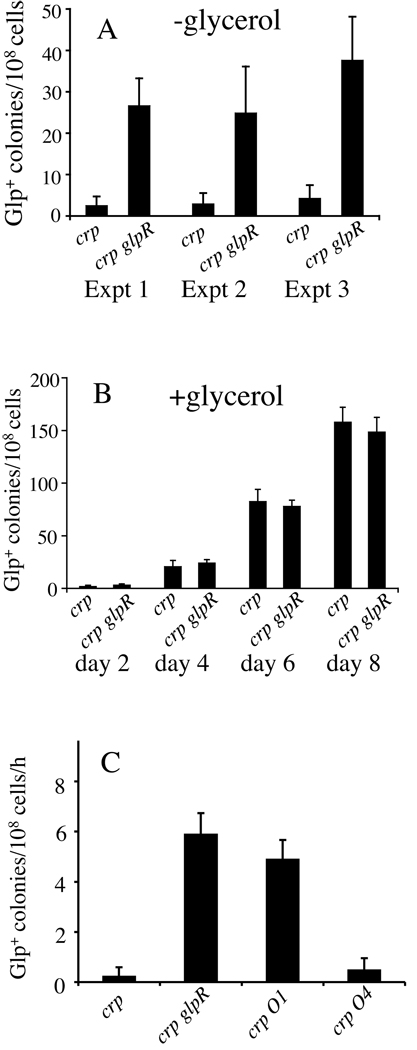
To test this possibility, the glpR gene was deleted, and the frequencies of appearance of Glp+ mutations in the crp glpR double mutant background were measured in the absence and presence of glycerol. When some cells (~50 cells per plate) of crp or crp glpR strains were plated onto LB agar plates without glycerol, individual colonies formed on the plates were examined for the total and Glp+ populations. As shown in Figure 3A, the ratios of Glp+ cells arising to total cells were 10 fold higher in the crp glpR double mutant colonies than in the crp mutant colonies. To determine if the loss of GlpR affected Glp+ mutations of crp cells in the presence of glycerol, we inoculated crp and crp glpR cells onto minimal M9 agar plates with glycerol as the sole carbon source rather than LB + glycerol agar plates since Glp+ cells grew faster than the parental Glp− cells in glycerol containing LB medium. On these minimal M9 glycerol plates, similar frequencies of appearance of Glp+ mutations were observed for both crp and crp glpR strains, indicating that GlpR did not inhibit Glp+ mutations of crp cells in the presence of glycerol (Figure 3B). Growth experiment showed that the crp glpR double mutant grew about 10% slower than the parental crp strain in LB medium (Figure S1A), indicating that the elevated frequency of Glp+ mutations by the loss of GlpR is not due to an increased growth rate of the crp glpR strain in this medium. Growth experiment also showed that both crp and crp glpR strains virtually could not grow in liquid glycerol minimal medium (not shown) since the Crp protein is essential for expression of glpFK. These experiments demonstrate that deletion of glpR is equivalent to inclusion of excess glycerol in the growth medium to enhance the appearance of Glp+ mutations. In a separate experiment, we compared the rates of mutation appearing per 108 cells per hour between crp and crp glpR cells in liquid LB media. As shown in Figure 3C, a much greater rate was observed for crp glpR cells than for crp cells.
Figure 3. Dependency of Glp+ mutation rate on GlpR.
(A) Effect of the loss of GlpR on the appearance of Glp+ mutants in the absence of glycerol. crp or crp glpR cells (<100 cells/plate) were applied to LB agar plates without glycerol. After two days of incubation at 30°C, ~50 colonies of each strain from different plates were used for determination of the total number of cells and the total number of Glp+ cells. Glp+ mutation frequency refers to the ratio of Glp+ cell number to total cell number. (B) Effect of the loss of GlpR on Glp+ mutation in crp cells in the presence of glycerol. Glp+ mutations were assayed on M9 + glycerol agar plates. (C) Effects of the losses of GlpR and its operators, O1 and O4, on mutation rates (mutations/108 cells/h) when grown in liquid LB medium at 30°C. Fresh overnight cultures were inoculated into 20 ml of fresh LB in 125 ml flasks (initial OD600 = 0.1). The flasks were shaken at 30°C. Samples were taken at one hour intervals for five hours for determination of total populations and Glp+ cell populations. The frequencies of Glp+ mutations relative to the total cell populations were plotted versus time, and the mutation rates were determined from the slopes of the curves.
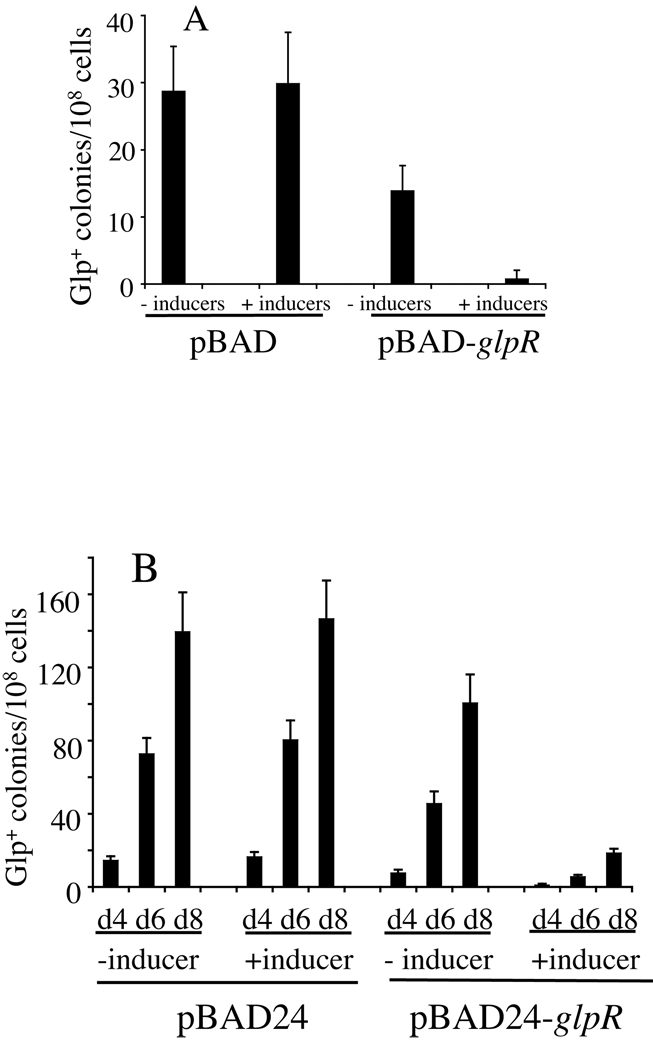
To further demonstrate that GlpR binding to the DNA inhibits the appearance of Glp+ mutations, the glpR gene was cloned into a pBAD vector (i.e., pBAD-glpR) and expressed under the control of the araBAD promoter. We first determined if expression of the glpR gene in pBAD inhibited growth of the crp glpR strain. As shown in Figure S1B, crp glpR/pBAD-glpR cells and Glp+ crp glpR/pBAD-glpR cells grew as well as the same cells carrying the empty pBAD plasmid in LB medium when glpR overexpression was induced by arabinose and indole acetic acid (IAA). The Glp+ crp glpR/pBAD-glpR cells also grew well on M9 + glycerol plates when glpR expression was induced as above. Thus, once Glp+ mutations in these crp glpR cells arise, visible colonies should appear on minimal M9 glycerol plates. Using crp glpR cells, we examined the effect of glpR overexpression on the appearance of Glp+ mutations on both LB agar plates (without glycerol) and minimal M9 agar plates (plus glycerol). When the cells carrying pBAD-glpR were grown on LB agar under inducing conditions (i.e., plus arabinose and IAA) and individual colonies were examined for Glp+ and total populations, the frequencies of Glp+ mutations (i.e., the ratios of Glp+ populations to total populations) decreased below the background rate observed for the crp strain (Figure 4A). Similar results were observed when experiments were conducted on minimal M9 agar plates containing glycerol (Figure 4B). In the absence of inducers, lower mutation frequencies were observed for cells with pBAD-glpR than cells with the control plasmid, presumably due to leaky expression of glpR in the high-copy number plasmid. However, in the presence of the inducers, further reduction of the mutation frequency was observed (Figure 4B). Control experiments showed that overexpression of glpR did not affect cell growth in M9 + glycerol medium (not shown).
Figure 4. Effects of glpR overexpression on the appearance of Glp+ mutants in a crp glpR genetic background when cells were grown on LB agar plates without glycerol (A) or on minimal M9 plates plus glycerol (B).
glpR expression in pBAD24 was induced by adding L-arabinose and IAA (inducers) to the media at 1 mM and 0.5 mM final concentrations, respectively. For the LB agar plate assay, ~100 cells were applied onto each plate. After incubation at 30°C for two days, individual colonies were examined for total and Glp+ cell populations. For the M9 + glycerol plate assay, Glp+ mutations and total populations were determined as described in Methods.
Effects of the Presence of Glycerol or the Loss of GlpR on IS5 Hopping Into Other Known Recognition Sites
As introduced above, activation of flhDC, bglGFB and fucAO operons can result from IS5 insertion into the appropriate locations in their promoter regions. The conditions (i.e., the presence of glycerol or loss of GlpR) that induced Glp+ mutations were examined for their effects on IS5 insertions into these known insertion sites. Using crp and crp glpR cells, assays were performed for swarming mutation, Bgl+ mutation and PPD+ mutation on minimal M9 agar plates with 1% glycerol, 1% sorbitol, or 0.01% glucose as sole carbon source. For the swarming mutation assay, soft-agar (0.35%) plates were used, and the plates were examined for appearance of outgrowths (i.e., fast moving subpopulations, each of which is from a single mutated cell) from the streaked cells. For Bgl+ and PPD+ mutation assays, cells were washed off the plates and appropriate dilutions were applied to M9 + 1% arbutin agar plates and M9+ 1% propanediol agar plates for determination of Bgl+ and PPD+ cell populations, respectively. The total populations for both Bgl+ and PPD+ assays were determined by applying appropriate dilutions onto LB agar plates. For each of three mutation assays, no mutant arose from each strain (crp or crp glpR) and from each type of media (M9 ± glycerol, sorbitol or glucose) after 5 days (for swarming assays) or 15 days of incubation (for Bgl+ and PPD+ assays). This is not surprise since expression of flhDC (Soutourina et al., 1999), bglGFB (Gulati & Mahadevan, 2000) and fucAO (Zhu & Lin, 1988) is heavily dependent on Crp although other regulators are required for their expression as well.
To determine if Crp is required for increased motility, cell growth on arbutin, or growth on propanediol caused by IS5 insertions upstream of the promoter regions of flhDC, bglGFB or fucAO, respectively, the crp gene was deleted from each type of these IS5 insertion mutants. The resultant double mutants were tested for swarming, growth on arbutin and growth on propanediol, respectively compared to the respective parental crp+ IS5 insertion mutants. The loss of Crp abolished increased motility of swarming mutants, abolished growth of Bgl+ cells on arbutin, and abolished growth of PPD+ cells on propanediol. From these results, we conclude that (1) both IS5 insertion and the Crp protein are required for respective phenotypes of each of these three types of mutants (i.e., Crp is essential for IS5 mediated activation of each of these operons); and (2) E. coli cells lacking Crp can not be used for assays of mutations in these operons. Thus, in the following experiments, crp+ cells were used.
We first tested the swarming mutation using crp+ cells on minimal M9 soft agar plates with glycerol or sorbitol as the sole carbon source. Glucose was not included in experiments using crp+ cells since glucose represses synthesis of cAMP. Similar frequencies (mutations per 9-cm cell streak) of appearance of swarming mutants were seen from glycerol and sorbitol plates after incubation for 13 to 35 h (Figure 5A). When experiments were conducted on nutrient broth (NB) soft agar plates ± glycerol or sorbitol, again no difference in mutation frequency was observed among these three types of plates (Figure 5A). Forty independently arising hypermotile mutants from minimal M9 ± glycerol or sorbitiol soft agar plates, and 40 mutants from NB ± glycerol or sorbitiol plates, were analyzed by PCR amplification of the flhDC control region and sequencing. They all harbored insertion sequences, ~50% of which were IS5 and ~50% of which were IS1 or IS3. All proved to be upstream of PflhDC. IS5 and IS1 were located and oriented as reported in (Barker et al., 2004). IS3 was found to be capable of activating of PflhDC as well. It was located at either −200.5 or −207.5 relative to +1 of PflhDC and was oriented with its transposase gene (encoded by orfAB) (Sekine et al., 1994) transcribed in an opposite direction as flhDC.
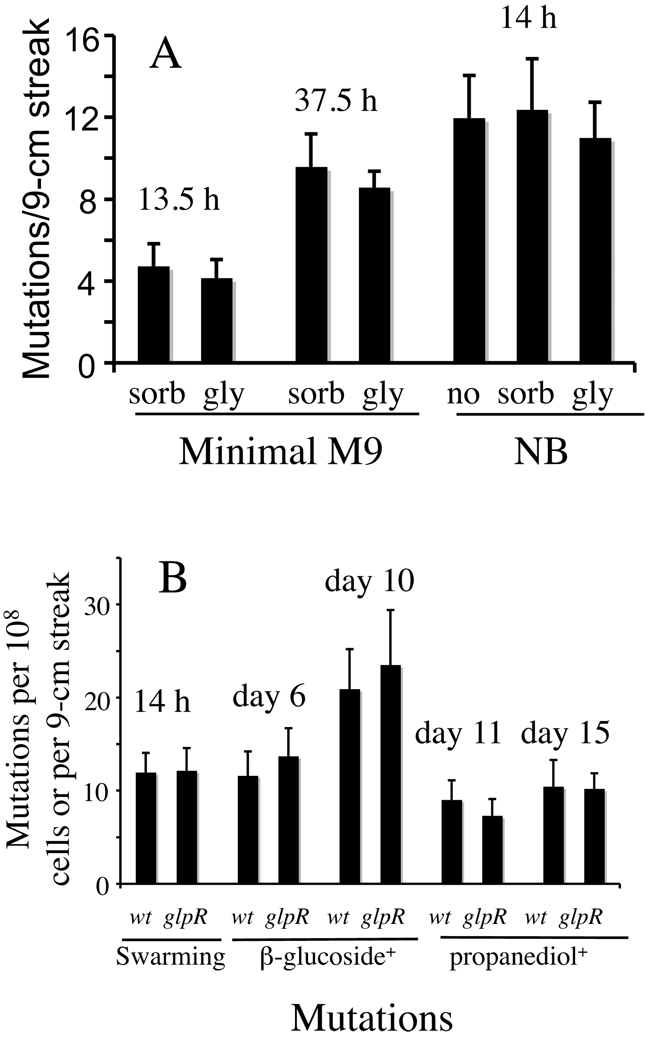
Figure 5. Effects of glycerol and GlpR on IS5 insertions at other IS5 insertional hot spots.
(A) Effect of glycerol and sorbitol on IS5 insertion upstream of the flhDC promoter. Swarming mutation assay was conducted on soft-agar plates of minimal M9 + sorbitol or glycerol and soft-agar plates of nutrient broth ± sorbitol or glycerol. The swarming mutations were normalized as outgrowing subpopulations (mutations) per 9-cm cell streak. ‘sorb’: sorbitol; and ‘gly’: glycerol. (B) Effects of the loss of GlpR on IS5 hopping into its insertion sites in the flhDC master switch operon controlling motility, the bglGFB β-glucoside utilization operon, and the fucAO fucose/propanediol operon. The swarming mutation assay was conducted on nutrient broth soft agar plates, and the mutations were normalized as outgrowing subpopulations (mutations) per streak. Bgl+ or propanediol+ mutation assays were conducted on minimal M9 arbutin plates or M9 minimal propanediol plates, and colonies (mutants) were counted daily and were normalized as mutations (colonies) per 108 cells.
To test if glycerol affects IS5 insertions into its recognition sites in the regulatory regions of the bglGFB and fucAO operons, crp+ cells were incubated on M9 minimal glycerol plates, and the cells were washed off daily and applied onto M9 + arbutin agar plates or M9 + propanediol plates for determination of Bgl+ or PPD+ cell populations, respectively. No colonies were found in ~2 days on these plates, indicating that Bgl+ or PPD+ mutations did not occur on the glycerol plates. These results indicate that the presence of glycerol did not affect insertion of IS5 upstream of the flhDC operon in E. coli cells no matter what medium was used.
The results described in the previous section show that glycerol elevation of Glp+ mutations in crp cells is due to relief of GlpR from the glpFK promoter region by glycerol-3-P synthesized from glycerol. Thus, the presence of glycerol and the loss of GlpR are equivalent in enhancing Glp+ mutations mediated by IS5 insertion upstream of PglpFK. We therefore determined the effect of the loss of GlpR on IS5 insertion into its recognition sites in the control regions of the flhDC, bglGFB and fucAO operons as described above. There were no obvious differences in the frequencies of each of these three types of mutations between glpR− cells and glpR+ cells (Figure 5B). For activation of the flhDC operon, all of the hypermotile mutants from both glpR+ and glpR− cells harbored insertion sequences, ~50% of which were IS5 and ~50% of which were IS1 or IS3 when 40 independent mutants were sequenced. They were situated and oriented the same as described above. In the case of the bglGFB operon, all Bgl+ mutants carried either IS5 (~60%) or IS1 (~40%) upstream of PbglGFB among 20 sequenced independently isolated mutants derived from each strain. In the case of the fucAO operon, 20 out of 20 propanediol+ mutants from either glpR+ or glpR− cells contained IS5 at the same location and in the same orientation upstream of PfucAO, as reported by (Chen et al., 1989). These results show that the presence of glycerol or GlpR binding to its operators in the glpFK promoter region does not affect IS5 insertion into target gene-activating sites other than the one upstream of PglpFK. These control experiments clearly indicate that GlpR specifically control IS5 insertion upstream of the glpFK promoter.
GlpR Operators Differentially Control glpFK Expression and Glp+ Mutation Rate
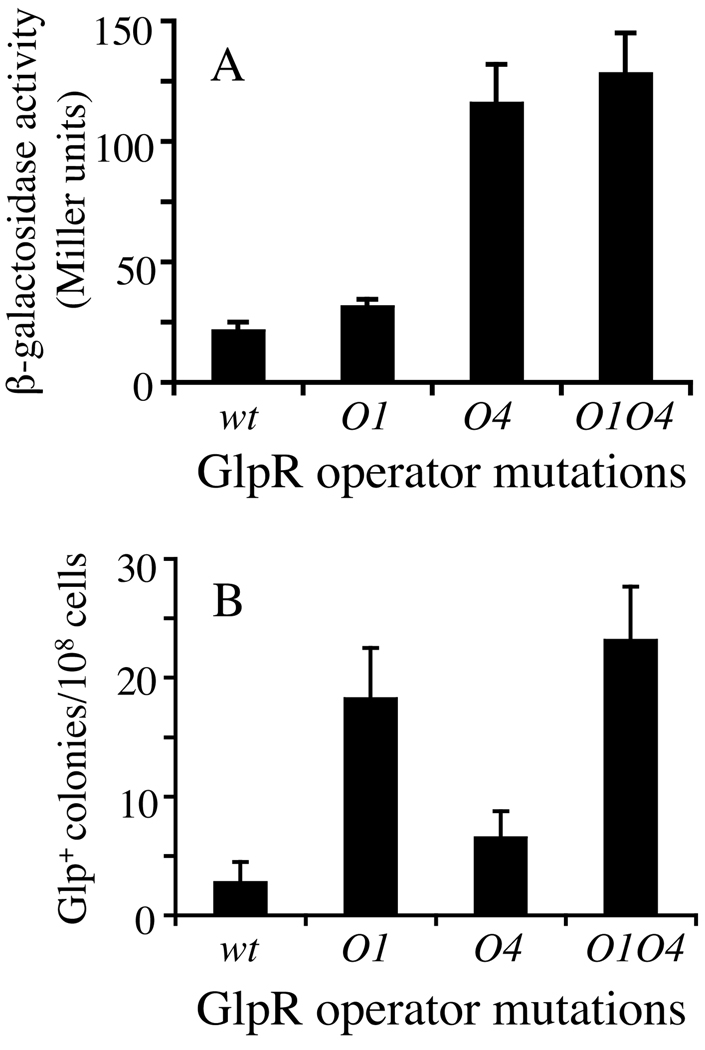
DNA footprinting identified four GlpR binding sites, O1–O4, in the upstream glpFK operon regulatory region (Weissenborn et al., 1992, see Figure 1A). We mutated the far upstream site (O1) and the far downstream site (O4) and compared the effects on glpFK expression using a lacZ reporter gene fusion construct with the frequency of mutation to Glp+ during growth in LB medium. As shown in Figure 6A, mutation of O4 increased glpFK operon expression about 5-fold although mutation of O1 was almost without effect. By contrast, loss of O1 yielded a 7-fold increase in mutation frequency although loss of O4 had only a 2-fold effect (Figure 6B). After sequencing the glpFK promoter region of 20 such Glp+ mutants from each strain, we confirmed that IS5 inserted in the same position and orientation as described above.
Figure 6. Effects of mutations in GlpR operators on promoter activity and the appearance of Glp+ mutants.
The graphs show the effects of mutations in GlpR operators, O1 and O4 (see Figure 1A), on (A) PglpFK activity in crp cells grown in LB liquid medium, and (B) Glp+ mutation frequencies in crp cells grown on LB solid medium. The bars represent standard deviation values for three independently conducted experiments. wt: control with no mutation in either O1 or O4; O1: only operator 1 was mutated (see Methods); O4: only operator 4 was mutated; O1O4: both operators were mutated.
We also determined the effects of O1 and O4 mutations on Glp+ mutation rates (mutations per 108 cells per hour). Glp+ mutation rates were substantially elevated by loss of O1 but not by loss of O4 (Figure 3C). In both minimal and complex media, the O1 and O4 mutations exerted no measurable effect on growth rate (Figure S1A). These observations showed that while O1 primarily controls mutation rate, O4 primarily controls gene expression. Mutation rate is therefore not a function of glpFK expression level, and GlpR regulates expression and mutation rate independently.
IS5 Insertion Assay Using the Chloramphenicol (Cm) Resistance Gene (cat) As A Reporter
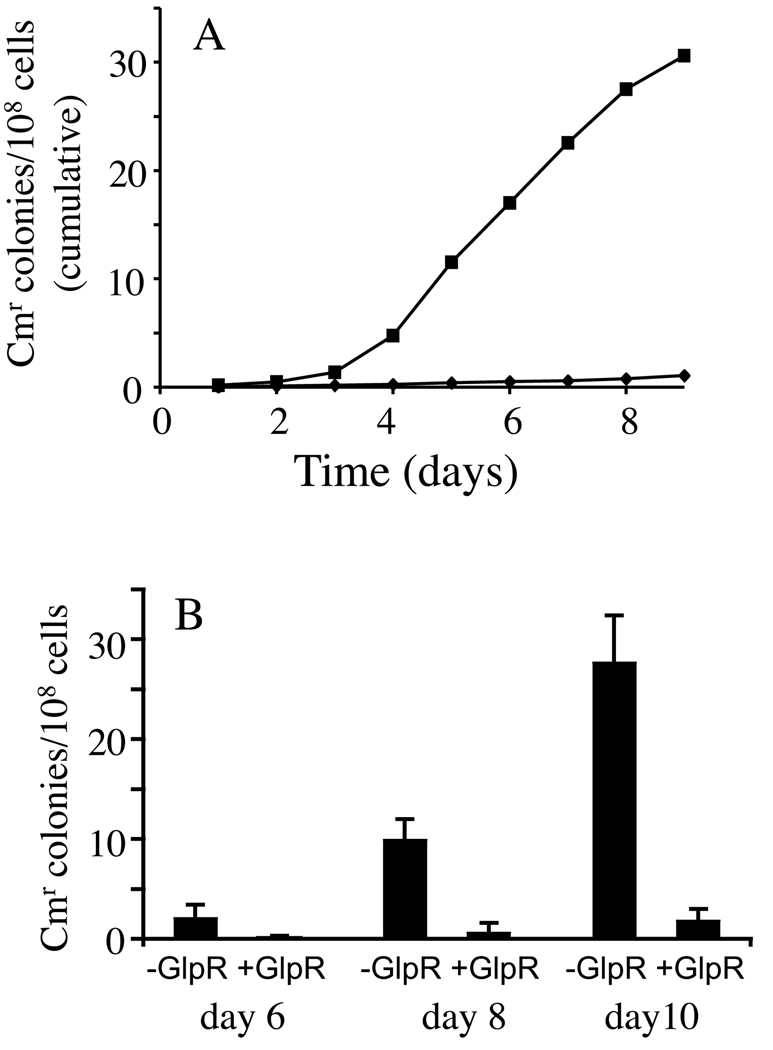
To confirm effects of glycerol and GlpR on IS5 insertion upstream of the glpFK promoter, we replaced the chromosomal glpFK operon with a cat structural gene so that cat is expressed solely from the glpFK promoter (i.e., PglpFK-cat at the glpFK locus). crp and crp glpR cells proved to be sensitive to Cm at < 25 µg/ml, but IS5 insertion as reported above rendered these cells resistant to Cm at >50 µg/ml. Using this chromosomal PglpFK-cat construct, IS5 insertion assays were performed by incubating cells on LB agar plates with Cm. Cm resistant (Cmr) colonies arose on both crp and crp glpR plates. However, the frequencies of appearance of Cmr colonies relative to the total populations were about 20 times higher for the crp glpR cells than for the crp cells (Figure 7A).
Figure 7. Effects of GlpR on the frequencies of IS5 insertion upstream of PglpFK when this promoter drives expression of a cat [chlorophenicol resistance (Cm)] gene.
The graph in A reveals the effect of the loss of GlpR on the frequency of IS5 insertion upstream of PglpFK in crp (◆) and crp glpR (■) genetic backgrounds. The graph in B shows the effect of GlpR overproduction on IS5 insertion using crp glpR cells that carry the pBAD24 vector alone (−GlpR) or pBAD24 expressing glpR (+GlpR). glpR expression is induced by inducers (1mM L-arabinose + 0.5 mM IAA). In both A and B, PglpFK is fused to a cat gene. Cells were incubated on LB + Cm (50 µg/ml) agar plates at 30°C, and Cm resistant colonies were counted after the time intervals indicated.
We confirmed that the total Cm sensitive populations on the LB + Cm plates increased slightly rather than declining during the experimental period (data not shown). This may be due to the low-level residual expression of cat driven by PglpFK, which allows low level metabolic activities in the cells. This residual cat expression may also contribute to synthesis of the transposase for IS5 transposition encoded by ins5A.
Using the same construct, we also determined the effect of glpR overexpression on IS5 insertion. When crp glpR cells carrying pBAD-glpR were incubated on LB + Cm agar plates, and glpR was overexpressed by addition of arabinose and IAA, the frequencies at which Cmr colonies arose decreased dramatically compared to the same cells carrying the empty plasmid (Figure 7B). Sequencing analysis showed that 20 out of 20 independently isolated Cmr mutants from either crp plates or crp glpR plates carried IS5 in the usual location and orientation. Using P1 transduction, we were able to transfer the Cm resistance from five Cmr mutants isolated from crp or crp glpR plates into the parental crp cells. The Cmr transductants harbored IS5 and cat at the same locations and orientations as in the donor strains. These results support our conclusions that (1) GlpR represses the appearance of Glp+ mutations in the absence of glycerol; and (2) IS5 insertion upstream of PglpFK is the sole cause of the Glp+ phenotype.
RecA Independence of Glp+ Mutation
The increased mutation rate in response to the loss of GlpR binding might have resulted from increased gene dosage accompanying partial chromosomal duplications (Petes & Hill, 1988, Roth & Andersson, 2004, Roth, 1996). Such duplications are RecA dependent (Roth, 1996). We therefore examined the dependency of Glp+ mutation on RecA. When cells were incubated on LB agar plates and individual colonies were examined for Glp+ and total populations, only a 15% apparent decrease in mutation frequency was observed in crp recA double mutant cells compared to crp cells (Figure S2A). Similarly, when cells were incubated on minimal glycerol agar plates, introduction of the recA mutation decreased the Glp+ mutation frequency by only about 15% (Figure S2B). We conclude that the presence of glycerol or the binding of GlpR to its operators in the glpFK promoter region does not appreciably influence a RecA-dependent process such as partial chromosomal duplication.
Discussion
Directed mutation has been defined as a genetic change that is specifically induced by the stress condition that the mutation relieves (see Introduction). Seldom have the mechanisms been determined (Massey & Buckling, 2002, Reimers et al., 2004, Rosenberg, 2001, Wright et al., 1999). In this paper, we demonstrate that mutations are directed to the glpFK control region, thereby allowing growth of E. coli crp mutants on glycerol. These mutations are specifically induced by the presence of glycerol. We identify the protein involved and elucidate the control mechanism. The glycerol regulon repressor, GlpR, which binds to its four operators (O1–O4) in front of the glpFK operon, is displaced from these sites when α-glycerol phosphate derived from glycerol by phosphorylation is bound (Weissenborn et al., 1992; Figure 1A). GlpR not only controls glpFK expression but also Glp+ mutation rate. Our results establish that O4, which overlaps the −10 region, primarily affects gene expression, O2 and O3, which overlap the two Crp binding sites and the −35 region, presumably antagonize activation by the cyclic AMP-Crp complex in wt cells, and O1 primarily affects IS5 hopping into the specific CTAA site, 126 base pairs upstream of the glpFK transcriptional start site. The mechanism may be indirect, involving changes in DNA conformation (e.g., supercoiling, secondary structure, etc). The results serve to dissociate the two functions of GlpR.
The adaptive Lac+ mutations that arise during lactose selection proved not to be directed to the gene in which mutation can relieve the stress (Foster, 1997, Torkelson et al., 1997). In the case of Glp+ mutations, we showed that (i) each of the 116 Glp+ mutants (isolated from different minimal M9 + glycerol agar plates and arose on different days) contain an IS5 copy, always present at the same location; (ii) only one glp operon (glpFK) is activated (see Results and Zhang, submitted); (iii) the presence of glycerol or the loss of GlpR did not affect IS5 hopping into three other gene activating sites in the genome; and (iv) the use of Biolog plates showed that there are no differences between Glp+ mutants and the parental crp cells in the utilization of other carbon, nitrogen, sulfur and phosphate sources. These results strongly suggest that mutation to Glp+ is directed to the glpFK operon while other IS5-mediated gene activating events do not occur at altered rates when glycerol is present or the glpR gene is deleted. Our results are therefore compatible to the definition of true directed mutation.
Several mechanisms have been proposed to account for increased rates of mutation in E. coli cells under stress conditions (Galhardo et al., 2007). Two of these (point mutagenesis mechanism and adaptive gene amplification mechanism) have been shown to be responsible for lac frameshift reversion (Galhardo et al., 2007). Another is responsible for amino acid-starvation elicited adaptive mutation (Reimers et al., 2004, Wright et al., 1999). The point mutagenesis mechanism for lac frameshift reversion includes DNA double-strand breaks and their subsequent repair, executed by error-prone DNA polymerase IV. The normal high fidelity of DNA polymerase IV in growing cells is switched to an error-prone version during starvation (Harris et al., 1994, Ponder et al., 2005). The gene amplification mechanism involves increasing the copy number of the lac +1 allele to up to 50 tandem repeats per cell, thereby elevating the opportunity for mutation to occur in one of the copies (Hastings et al., 2000, Hendrickson et al., 2002, Slack et al., 2006). Another mechanism for increasing mutation rate involves the guanosine tetraphosphate (ppGpp)-mediated elevation of transcription of amino acid metabolic genes in response to nutritional stress, thereby enhancing the amount of local single-stranded DNA, which is more vulnerable to mutation (Reimers et al., 2004, Wright et al., 1999). The most studied example of transcription-elicited mutations is the Leu+ reversion of an E. coli leuB auxotroph during leucine starvation (Reimers et al., 2004, Wright et al., 1999, Wright & Minnick, 1997). A fraction (~25%) of the Leu+ mutants proved not to be true revertants, and they contained one or more mutations other than a nucleotide substitution in the leuB gene (Wright & Minnick, 1997).
These reported mechanisms appear not to apply to Glp+ mutation of crp cells during starvation on glycerol plates. In the case of Glp+ mutations, (i) no evidence suggests an involvement of DNA double-strand breaks or Pol IV-mediated error-prone repair; (ii) gene amplification proved not to be responsible for the elevated mutation rate induced by the presence of glycerol or the loss of GlpR (see Figure S2); (iii) the Glp+ mutation rate is not a function of glpFK expression level (see Figure 6); and (iv) a local protein (GlpR) that normally regulates expression of the glp regulon, depresses the Glp+ mutation rates in the absence of glycerol.
In several other cases, the insertion frequencies of transposons were reported to increase in the presence of specific carbon sources (Hall, 1998, Hall, 1999), but the mechanisms were not revealed. A series of host proteins have been identified to increase or decrease transposition of transposons in E. coli (Chaconas et al., 1996, Chandler & Mahillon, 2002, Galas, 1989, McCalla, 1979). Many of these proteins are either nucleoid-like structuring proteins such as H-NS, IHF and Fis or proteins related to DNA recombination and repairing such as RecG, Dcd, and DinD (Swingle et al., 2004, Twiss et al., 2005). Often, increased transposition occurs in response to nutritional stress (Twiss et al., 2005). The host DNA structuring proteins have been reported to be involved in assembly of the transposome and in target selection (Chalmers et al., 1998, Lavoie & Chaconas, 1993, Surette & Chaconas, 1989). The exact roles of these DNA recombination and repairing proteins in transposition have not been elucidated (Twiss et al., 2005). In some cases, the Dam DNA methylase also affects transposition, probably due to its effect on transposase transcription and activity (Roberts et al., 1985, Yin et al., 1988). Similar observations have been made in yeast, in which multiple host factors were found to be involved in transpositions of retrotransposons such as Ty1 and Ty3 (Aye et al., 2004, Scholes et al., 2001). It is not surprising that these host proteins affect the overall transpositions of transposons in the E. coli genome (Chandler & Mahillon, 2002, Galas, 1989, Mahillon & Chandler, 1998, Swingle et al., 2004, Twiss et al., 2005). It would be of interest to know if and how they host proteins influence GlpR-regulated, IS5-mediated Glp+ mutation rate in crp cells.
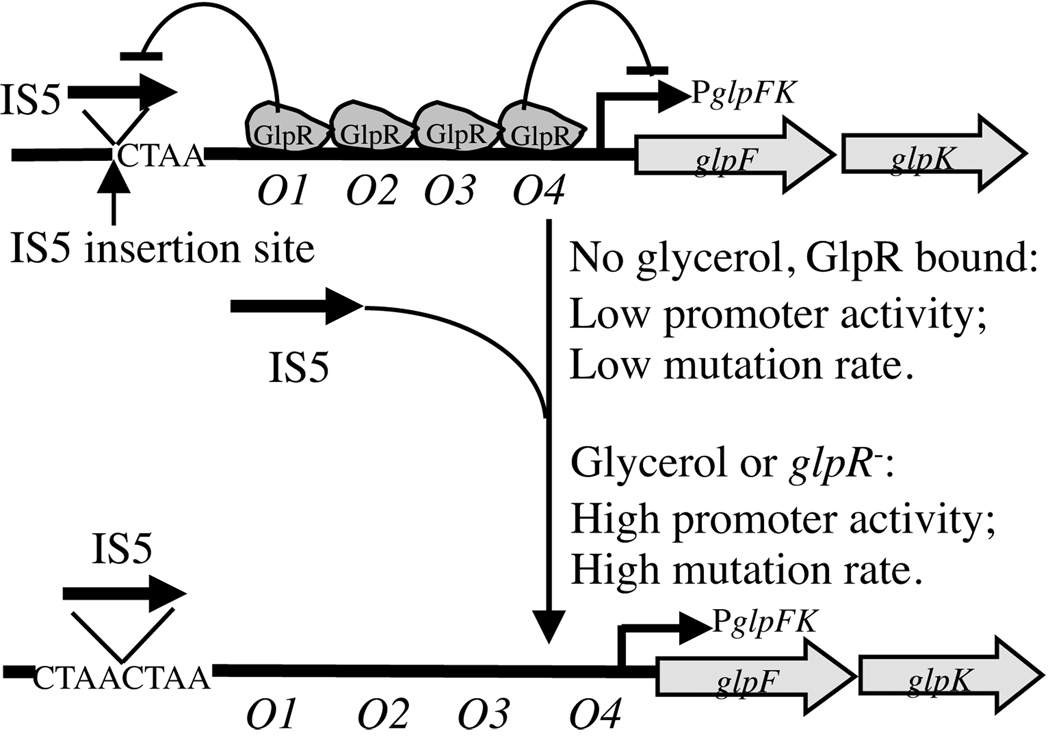
The mechanism identified here for PglpFK activation provides relief from starvation and therefore could have been selected for through evolution. It appears to be a genuine example of directed mutation, since mutation is “directed” to a specific operon and occurs at a greater rate under conditions that allow benefit to the organism. The fact that mutation rate is influenced by the presence of glycerol in a process mediated by the glycerol repressor provides a mechanistic explanation for IS5-mediated directed mutational control. It leads us to propose that GlpR has two functions, one in controlling glp regulon gene expression as recognized previously, and the other in controlling the conformational state of the upstream DNA so as to influence the rate of IS5 insertion. These mechanisms, illustrated in Figure 8, may provide a partially explanation for the presence of four GlpR binding sites in the control region of the glpFK operon.
Figure 8. Schematic diagram illustrating GlpR-mediated control of (right) glpFK transcription and (left) the rate of IS5 hopping (directed mutation) into the CTAA site upstream of the glpFK promoter.
With GlpR bound to its operators (O1–O4), transcription and IS5 hopping both occur at low rates. When GlpR is not bound to its operators, both transcriptional initiation and IS5 hopping increase about 10x. Binding of GlpR to operator O1 blocks IS5 insertion, while binding of GlpR to operator O4 blocks transcription as indicated.
Materials and Methods
Bacterial Strains and Growth Conditions
Strains and DNA oligonucleotides used in this study are described in Table S1 and Table S2, respectively. The crp deletion mutant was constructed as described in Gosset et al. (Gosset et al., 2004). glpR and recA single mutants were generated from the same parental strain (E. coli K-12 strain BW25113) using the method of Datsenko and Wanner [41]. For each mutant, a kanamycin resistance gene (km), flanked by the FLP recognition site (FRT) was amplified from the template plasmid pKD4 using a pair of mutation oligos (Table S2), each of which is composed of a ~20 bp region at the 3’ end that is complementary to the FRT-flanking km sequence, and a ~50 bp region at the 5’ end that is homologous to the gene to be mutated. The PCR products were gel purified using an EZgene™ purification kit (Biomiga), treated with DpnI, and then electroporated into BW25113 cells expressing the lamada-Red proteins encoded by plasmid pKD46. The pKD46 plasmid, which carries a temperature-sensitive origin of replication, was removed by growing the mutant cells overnight at 40 °C. The kmr mutants were verified for the replacement of the target gene by the FRT-flanking km gene by PCR. The km gene was subsequently eliminated (leaving an 85-bp FRT sequence) using plasmid pCP20 that bears the FLP recombinase. The crp glpR double mutant was made by transferring a km insertion mutation of the crp gene into the glpR deletion mutant background using P1 transduction. The crp recA double mutant was made by deleting the recA gene in the crp deletion background using the same approach as described above.
To mutate or relocate the CTAA IS5 insertion site located between −126 and −123 bps upstream of the transcriptional start site (Figure 1A), an FRT-flanking km gene was inserted at −122.5 (immediately downstream of CTAA), −124.5 (between CT and AA), and −126.5 (immediately upstream of CTAA) in the crp deletion mutant using the same method as presented above (Datsenko & Wanner, 2000). After the km gene was deleted, the 85-bp FRT sequence remained at the same locations (i. e. −122.5, −124.5, and −126.5 respectively), relative to the transcriptional start site. The resultant strains were named crp (−122.5), crp (−124.5), and crp (−126.5) (Table S1), respectively.
To replace the chromosomal glpFK operon with a cat gene encoding chloramphenicol (Cm) resistance, the cat gene with its own promoter was first cloned from pKD3 into the BamHI and SalI sites upstream of the FRT-flanking km gene in pKD13 (Datsenko & Wanner, 2000), yielding pKD13-cat. The cat structural gene [with no promoter and no ribosome binding site (RBS)], together with the FRT-flanking km gene, was amplified from pKD13-cat using primers glpFKI-P1 and glpFKI-P2 (Table S2). The PCR products were integrated into the crp chromosome to substitute for glpFK so that expression of cat is solely under the control PglpFK. The km gene was deleted, and the resultant strain was named crp (PglpFK-cat). Strain crp glpR (PglpFK-cat) was made by P1 transduction.
The strains were cultured in LB or minimal M9 media with various carbon sources at 37°C or 30°C. When appropriate, kanamycin (Km; 25 µg/ml), ampicillin (Ap; 100 µg/ml), or chloramphenicol (Cm; 25–50 µg/ml) was added to the media.
Mutations of Chromosomal GlpR Operators
To modify the chromosomal GlpR binding sites in PglpFK, the promoter was amplified from the wt genomic DNA and ligated into the SalI and BamHI sites of pKD13 so that PglpFK and km were oriented in opposite directions, yielding pKD13-PglpFK. Following the recommended protocol, the quick-change site-directed mutagenesis kit (Stratagene) was used for introducing mutations in PglpFK contained within pKD13-PglpFK. Operator O1 (see Figure 1A) was mutated by changing gcgcgat (−86 to −80 relative to +1 of PglpFK) to aggtacc using primers PglpFKO1-F and PglpFKO1-R. Operator O4 was mutated by changing gctcgtt (−24 to −18 relative to +1 of PglpFK) to agggccc using primers PglpFKO4-F and PglpFKO4-R (Table S2). The double O1O4 mutation was created by mutating O4 in the O1 mutated PglpFK. These yielded pKD13-PglpFKO1, pKD13-PglpFKO4, and pKD13-PglpFKO1O4, respectively. Using these plasmids as templates, the region containing the km gene and PglpFK, with or without operator mutations, was PCR amplified by the primers listed in Table S2. The PCR products were integrated into the crp mutant chromosome to replace the wt PglpFK. The km gene was removed, and the resultant strains were named crp(wt), crp(O1), crp(O4), and crp(O1O4) (Table S1), respectively.
Glp+ Mutation Assays Under Various Conditions
Using the crp deletion mutant, mutation to Glp+ was measured on minimal M9 agar plates with 1% glycerol, 1% sorbitol or 0.01% glucose as the sole carbon source. Cells of an overnight crp culture (from a single colony) in LB liquid medium were washed twice using carbon source-free M9 salts and inoculated onto plates (~108 cells/plate). The plates were then incubated in a 30°C incubator, inside of which moisture was provided by distilled water in a plastic pan. For glycerol plates, they were examined daily for the appearance of Glp+ colonies with each colony representing a Glp+ mutation. On these glycerol minimal agar plates, any colonies appearing by day 2 were considered to be from Glp+ cells initially presented and applied onto the plates. They were therefore subtracted from the subsequent measurements. The total numbers of cells were determined as described by Cairns and Foster (Foster, 1999). Briefly, agar plugs (diameter = 1 cm) were removed using a sterile borer from plates to glass tubes containing sterile M9 salts with no carbon source. The cells were washed off by vortexing. The washings were appropriately diluted and then applied onto LB plates and M9 + glycerol plates. The total Glp− cell population was determined by subtracting the populations obtained from M9 + glycerol plates from the populations obtained from LB plates. The frequencies of Glp+ mutations on glycerol M9 plates were determined by dividing total Glp− populations by the numbers of Glp+ colonies. For sorbitol or glucose plates, the cells were washed off daily, serially diluted and plated onto LB agar plates and M9 + glycerol plates for determination of total populations and Glp+ populations, respectively. The frequencies of Glp+ mutations on these plates were determined by dividing Glp+ populations by total populations.
To determine the effect of GlpR on Glp+ mutation frequency, the crp glpR double mutant was examined for Glp+ mutations on glycerol minimal agar plates as described above. Comparing crp and crp glpR cells, Glp+ mutation assays were carried out on LB agar plates. Overnight LB cultures were serially diluted with carbon source-free M9 salts. Appropriate dilutions were plated onto LB agar plates so that 10 to 100 colonies were formed per plate. After one and a half to two days of incubation at 30°C, ~50 colonies from five plates were individually transferred into sterile microcentrifugation tubes, each containing 200 µl of M9 salts. The total populations and the Glp+ cell populations were determined on LB agar plates and M9 + glycerol plates, respectively, as described above.
To determine the effect of glpR overexpression on Glp+ mutations, the glpR structural gene was amplified from the wt gnomic DNA using primers glpR-Nco and glpR-Hind (Table S2). The PCR products were digested with NcoI and HindIII, gel purified and then ligated to the same sites of pBAD24 [64], yielding pBAD24-glpR. crp glpR cells containing pBAD24 alone or pBAD24-glpR were tested for Glp+ mutations on both LB agar and M9 + glycerol agar plates as described above. As reported in Ebright and Beckwith (Ebright & Beckwith, 1985), indole-3-acetic acid (IAA), together with L-arabinose, is able to activate transcription of the araBAD operon, and such activation is not Crp dependent. The detailed mechanism for such activation is unknown. To induce expression of glpR in pBAD24-glpR, 1 mM L-arabinose and 0.5 mM IAA were added to the agar media. The pH values in the media were not changed.
To determine the effects of mutations in the GlpR operators in the PglpFK region on the appearance of Glp+ mutations, the crp cells with mutations in operator O1, O4 or both were examined for the appearance of Glp+ mutations on LB agar plates as described above.
Swarming Mutation Assay
The swarming mutation assay for the appearance of hyperswarming mutants (outgrowing subpopulations from the inoculated cells) was carried out using the method of Barker et al. (Barker et al., 2004). Briefly, overnight cell cultures in nutrient broth (NB) media were washed once with M9 salts, and diluted to an OD600 of 1.0 prior to use. 2 µl of the cell suspensions were streaked across the centers of semisolid (0.35% agar) plates (diameter = 9 cm) using a plastic transfer loop. NB or minimal M9 + 1% glycerol or sorbitol or 0.01% glucose were used. The plates were incubated at 30 °C. The swarming mutants, represented by outgrowths of motile subpopulations from the streaked cells, were counted. The mutation frequency was normalized as outgrowths (mutations) per 9-cm cell streak.
Bgl+ Mutation Assay
Overnight LB cultures were washed twice with carbon source-free M9 salts and resuspended in the same buffer. Cell suspensions (~108 cells/plate) were applied onto arbutin (0.5%) M9 minimal agar plates. The plates were incubated in a 30 °C incubator. Colonies (each representing a Bgl+ mutation) were counted daily, and total populations were determined as described under “Glp+ mutation assay”.
Propanediol (PPD) Growth Mutation Assay
Assay for PPD+ mutations was conducted by applying cell suspensions from fresh overnight cultures onto propanediol (1%) M9 minimal agar plates (~108 cells/plate). Growth positive mutations and total populations were determined as described above under “Glp+ mutation assay”.
Chromosomal lacZ Fusions and β-Galactosidase Assays
The glpFK promoter region (−204 to +66 relative to the transcriptional start site) was amplified from wt genomic DNA using primers PglpFK-EcoR and PglpFK-Bam (Table S2). Using the same primers, glpFK promoters with the O1, O4, or O1O4 mutations were amplified from pKD13-PglpFKO1, pKD13-PglpFKO4, pKD13-PglpFKO1O4 (Table S1), respectively. The PCR products were digested with EcoRI and BamHI, gel purified, and then ligated into the same sites of pRS551 (Simons et al., 1987). This yielded pRS551-PglpFK, pRS551-PglpFKO1, pRS551-PglpFKO4, and pRS551-PglpFKO1O4, in which the promoter is transcriptionally fused to a promoter-less lacZ gene. The promoter-lacZ fusions carried in these plasmids were subsequently integrated into the attB site of the crp chromosome using the method of Simons et al. (Simons et al., 1987). The resultant strains were named crp(PglpFKwt-lacZ), crp(PglpFKO1-lacZ), crp(PglpFKO4-lacZ), and crp(PglpFKO1O4-lacZ) (Table S1), respectively. For β-galactosidase assays, strains were cultured in liquid LB media at 30 °C. When cultures entered the exponential phase, samples were collected for measurement of β-galactosidase activities. β-Galactosidase assays were conducted as described by Miller (Miller, 1972).
Supplementary Material
(A) Growth in LB liquid medium. (B) Effects of glpR overexpression on growth of crp glpR and crp glpR Glp+ cells in LB with or without inducers (1 mM arabinose + 0.5 mM IAA). All growth experiments were conducted at 30°C. In (A), ◆ = crp, ν = crp glpR, σ = crp O1, △ = crp O4, and ● = crp Glp+. In (B), ◆ = crp glpR (pBAD), ◇ = crp glpR (pBAD-glpR), ν = crp glpR Glp+ (pBAD), □ = crp glpR Glp+ pBAD-glpR).
In (A), cells were incubated on LB agar plates, and the numbers of Glp+ mutants from 50 individual colonies were recorded after 36 hours. In (B), cells were incubated on M9 + glycerol plates, and colonies were counted after the time intervals indicated. Error bars represent standard deviations for experiments conducted in triplicate.
Acknowledgements
We thank Dr. Ming Ren Yen for assistance with some of the experiments described, Dr. Karen Schnetz for critically reviewing this manuscript, and Drs. Terry Hwa and David Meyer for helpful discussions. This work was supported by NIH grants GM 64368 and GM 077402.
References
- Andersson DI, Slechta ES, Roth JR. Evidence that gene amplification underlies adaptive mutability of the bacterial lac operon. Science. 1998;282:1133–1135. doi: 10.1126/science.282.5391.1133. [DOI] [PubMed] [Google Scholar]
- Aye M, Irwin B, Beliakova-Bethell N, Chen E, Garrus J, Sandmeyer S. Host factors that affect Ty3 retrotransposition in Saccharomyces cerevisiae. Genetics. 2004;168:1159–1176. doi: 10.1534/genetics.104.028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker CS, Pruss BM, Matsumura P. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol. 2004;186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Foster PL. Adaptive reversion of a frameshift mutation in Escherichia coli. Genetics. 1991;128:695–701. doi: 10.1093/genetics/128.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cairns J, Overbaugh J, Miller S. The origin of mutants. Nature. 1988;335:142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Cashel M, Gentry DR, Hernandez VD, Vinella D. The stringent response. In: Neidhardt FCea., editor. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1458–1496. [Google Scholar]
- Chaconas G, Lavoie BD, Watson MA. DNA transposition: jumping gene machine, some assembly required. Curr Biol. 1996;6:817–820. doi: 10.1016/s0960-9822(02)00603-6. [DOI] [PubMed] [Google Scholar]
- Chalmers R, Guhathakurta A, Benjamin H, Kleckner N. IHF modulation of Tn10 transposition: sensory transduction of supercoiling status via a proposed protein/DNA molecular spring. Cell. 1998;93:897–908. doi: 10.1016/s0092-8674(00)81449-x. [DOI] [PubMed] [Google Scholar]
- Chandler M, Mahillon J. Insertion sequences revised. In: Craig NL, Craigie RG, Gellert M, Lambowitz AM, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 305–366. [Google Scholar]
- Chen YM, Lu Z, Lin EC. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on L-1,2-propanediol. J Bacteriol. 1989;171:6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol. 2006;60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- Ebright RH, Beckwith J. The catabolite gene activator protein (CAP) is not required for indole-3-acetic acid to activate transcription of the araBAD operon of Escherichia coli K-12. Mol Gen Genet. 1985;201:51–55. doi: 10.1007/BF00397986. [DOI] [PubMed] [Google Scholar]
- Foster PL. Nonadaptive mutations occur on the F' episome during adaptive mutation conditions in Escherichia coli. J Bacteriol. 1997;179:1550–1554. doi: 10.1128/jb.179.5.1550-1554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Mechanisms of stationary phase mutation: a decade of adaptive mutation. Annu Rev Genet. 1999;33:57–88. doi: 10.1146/annurev.genet.33.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster PL. Stress responses and genetic variation in bacteria. Mutat Res. 2005;569:3–11. doi: 10.1016/j.mrfmmm.2004.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galas DJ. Bacterial insertion sequences. In: Berg D, Howe M, editors. Mobile DNA. Washington, DC: American Society for Microbiology; 1989. pp. 109–162. [Google Scholar]
- Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit Rev Biochem Mol Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosset G, Zhang Z, Nayyar S, Cuevas WA, Saier MH., Jr Transcriptome analysis of Crp-dependent catabolite control of gene expression in Escherichia coli. J Bacteriol. 2004;186:3516–3524. doi: 10.1128/JB.186.11.3516-3524.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulati A, Mahadevan S. Mechanism of catabolite repression in the bgl operon of Escherichia coli: involvement of the anti-terminator BglG, CRP-cAMP and EIIAGlc in mediating glucose effect downstream of transcription initiation. Genes Cells. 2000;5:239–250. doi: 10.1046/j.1365-2443.2000.00322.x. [DOI] [PubMed] [Google Scholar]
- Hall BG. Activation of the bgl operon by adaptive mutation. Mol Biol Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- Hall BG. Spectra of spontaneous growth-dependent and adaptive mutations at ebgR. J Bacteriol. 1999;181:1149–1155. doi: 10.1128/jb.181.4.1149-1155.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris RS, Longerich S, Rosenberg SM. Recombination in adaptive mutation. Science. 1994;264:258–260. doi: 10.1126/science.8146657. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Bull HJ, Klump JR, Rosenberg SM. Adaptive amplification: an inducible chromosomal instability mechanism. Cell. 2000;103:723–731. doi: 10.1016/s0092-8674(00)00176-8. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Slack A, Petrosino JF, Rosenberg SM. Adaptive amplification and point mutation are independent mechanisms: evidence for various stress-inducible mutation mechanisms. PLoS Biol. 2004;2:e399. doi: 10.1371/journal.pbio.0020399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson H, Slechta ES, Bergthorsson U, Andersson DI, Roth JR. Amplification-mutagenesis: evidence that "directed" adaptive mutation and general hypermutability result from growth with a selected gene amplification. Proc Natl Acad Sci U S A. 2002;99:2164–2169. doi: 10.1073/pnas.032680899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie BD, Chaconas G. Site-specific HU binding in the Mu transpososome: conversion of a sequence-independent DNA-binding protein into a chemical nuclease. Genes Dev. 1993;7:2510–2519. doi: 10.1101/gad.7.12b.2510. [DOI] [PubMed] [Google Scholar]
- Lin EC. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- Lin ECC. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt DC, editor. Escherichia coli and Salmonella Cellular and Molecular Biology. Washington, DC: ASM Press; 1996. pp. 307–342. [Google Scholar]
- Lombardo MJ, Aponyi I, Rosenberg SM. General stress response regulator RpoS in adaptive mutation and amplification in Escherichia coli. Genetics. 2004;166:669–680. doi: 10.1093/genetics/166.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria SE, Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey RC, Buckling A. Environmental regulation of mutation rates at specific sites. Trends Microbiol. 2002;10:580–584. doi: 10.1016/s0966-842x(02)02475-7. [DOI] [PubMed] [Google Scholar]
- McCalla DR. Mechanism of action of antibacterial agents. Berlin, Germany: Springer-Verlag; 1979. [Google Scholar]
- Miller JH. Experiments in molecular genetics. New York: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Muramatsu S, Kato M, Kohara Y, Mizuno T. Insertion sequence IS5 contains a sharply curved DNA structure at its terminus. Mol Gen Genet. 1988;214:433–438. doi: 10.1007/BF00330477. [DOI] [PubMed] [Google Scholar]
- Petersen C, Moller LB, Valentin-Hansen P. The cryptic adenine deaminase gene of Escherichia coli. Silencing by the nucleoid-associated DNA-binding protein, H-NS, and activation by insertion elements. J Biol Chem. 2002;277:31373–31380. doi: 10.1074/jbc.M204268200. [DOI] [PubMed] [Google Scholar]
- Petes TD, Hill CW. Recombination between repeated genes in microorganisms. Annu Rev Genet. 1988;22:147–168. doi: 10.1146/annurev.ge.22.120188.001051. [DOI] [PubMed] [Google Scholar]
- Ponder RG, Fonville NC, Rosenberg SM. A switch from high-fidelity to error-prone DNA double-strand break repair underlies stress-induced mutation. Mol Cell. 2005;19:791–804. doi: 10.1016/j.molcel.2005.07.025. [DOI] [PubMed] [Google Scholar]
- Reimers JM, Schmidt KH, Longacre A, Reschke DK, Wright BE. Increased transcription rates correlate with increased reversion rates in leuB and argH Escherichia coli auxotrophs. Microbiology. 2004;150:1457–1466. doi: 10.1099/mic.0.26954-0. [DOI] [PubMed] [Google Scholar]
- Reynolds AE, Felton J, Wright A. Insertion of DNA activates the cryptic bgl operon in E. coli K12. Nature. 1981;293:625–629. doi: 10.1038/293625a0. [DOI] [PubMed] [Google Scholar]
- Roberts D, Hoopes BC, McClure WR, Kleckner N. IS10 transposition is regulated by DNA adenine methylation. Cell. 1985;43:117–130. doi: 10.1016/0092-8674(85)90017-0. [DOI] [PubMed] [Google Scholar]
- Rosenberg SM. Evolving responsively: adaptive mutation. Nat Rev Genet. 2001;2:504–515. doi: 10.1038/35080556. [DOI] [PubMed] [Google Scholar]
- Roth JR, Andersson DI. Amplification-mutagenesis--how growth under selection contributes to the origin of genetic diversity and explains the phenomenon of adaptive mutation. Res Microbiol. 2004;155:342–351. doi: 10.1016/j.resmic.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu Rev Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- Roth JRea. Rearrangements of the bacterial chromosome: formation and application. In: Niedhardt FC, editor. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Washington, D.C: ASM Press; 1996. pp. 147–168. [Google Scholar]
- Schnetz K, Rak B. IS5: a mobile enhancer of transcription in Escherichia coli. Proc Natl Acad Sci U S A. 1992;89:1244–1248. doi: 10.1073/pnas.89.4.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholes DT, Banerjee M, Bowen B, Curcio MJ. Multiple regulators of Ty1 transposition in Saccharomyces cerevisiae have conserved roles in genome maintenance. Genetics. 2001;159:1449–1465. doi: 10.1093/genetics/159.4.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekine Y, Eisaki N, Ohtsubo E. Translational control in production of transposase and in transposition of insertion sequence IS3. J Mol Biol. 1994;235:1406–1420. doi: 10.1006/jmbi.1994.1097. [DOI] [PubMed] [Google Scholar]
- Simons RW, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Slack A, Thornton PC, Magner DB, Rosenberg SM, Hastings PJ. On the mechanism of gene amplification induced under stress in Escherichia coli. PLoS Genet. 2006;2:e48. doi: 10.1371/journal.pgen.0020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, Danchin A, Bertin P. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette MG, Chaconas G. A protein factor which reduces the negative supercoiling requirement in the Mu DNA strand transfer reaction is Escherichia coli integration host factor. J Biol Chem. 1989;264:3028–3034. [PubMed] [Google Scholar]
- Swingle B, O'Carroll M, Haniford D, Derbyshire KM. The effect of host-encoded nucleoid proteins on transposition: H-NS influences targeting of both IS903 and Tn10. Mol Microbiol. 2004;52:1055–1067. doi: 10.1111/j.1365-2958.2004.04051.x. [DOI] [PubMed] [Google Scholar]
- Torkelson J, Harris RS, Lombardo MJ, Nagendran J, Thulin C, Rosenberg SM. Genome-wide hypermutation in a subpopulation of stationary-phase cells underlies recombination-dependent adaptive mutation. EMBO J. 1997;16:3303–3311. doi: 10.1093/emboj/16.11.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twiss E, Coros AM, Tavakoli NP, Derbyshire KM. Transposition is modulated by a diverse set of host factors in Escherichia coli and is stimulated by nutritional stress. Mol Microbiol. 2005;57:1593–1607. doi: 10.1111/j.1365-2958.2005.04794.x. [DOI] [PubMed] [Google Scholar]
- Weissenborn DL, Wittekindt N, Larson TJ. Structure and regulation of the glpFK operon encoding glycerol diffusion facilitator and glycerol kinase of Escherichia coli K-12. J Biol Chem. 1992;267:6122–6131. [PubMed] [Google Scholar]
- Whiteway J, Koziarz P, Veall J, Sandhu N, Kumar P, Hoecher B, Lambert IB. Oxygen-insensitive nitroreductases: analysis of the roles of nfsA and nfsB in development of resistance to 5-nitrofuran derivatives in Escherichia coli. J Bacteriol. 1998;180:5529–5539. doi: 10.1128/jb.180.21.5529-5539.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE. Stress-directed adaptive mutations and evolution. Mol Microbiol. 2004;52:643–650. doi: 10.1111/j.1365-2958.2004.04012.x. [DOI] [PubMed] [Google Scholar]
- Wright BE, Longacre A, Reimers JM. Hypermutation in derepressed operons of Escherichia coli K12. Proc Natl Acad Sci U S A. 1999;96:5089–5094. doi: 10.1073/pnas.96.9.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright BE, Minnick MF. Reversion rates in a leuB auxotroph of Escherichia coli K-12 correlate with ppGpp levels during exponential growth. Microbiology. 1997;143(Pt 3):847–854. doi: 10.1099/00221287-143-3-847. [DOI] [PubMed] [Google Scholar]
- Yin JC, Krebs MP, Reznikoff WS. Effect of dam methylation on Tn5 transposition. J Mol Biol. 1988;199:35–45. doi: 10.1016/0022-2836(88)90377-4. [DOI] [PubMed] [Google Scholar]
- Zeng G, Ye S, Larson TJ. Repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K-12: primary structure and identification of the DNA-binding domain. J Bacteriol. 1996;178:7080–7089. doi: 10.1128/jb.178.24.7080-7089.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. A novel mechanism of transposon-mediated gene activation. PLoS Genet. 2009 doi: 10.1371/journal.pgen.1000689. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Lin EC. A mutant crp allele that differentially activates the operons of the fuc regulon in Escherichia coli. J Bacteriol. 1988;170:2352–2358. doi: 10.1128/jb.170.5.2352-2358.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Growth in LB liquid medium. (B) Effects of glpR overexpression on growth of crp glpR and crp glpR Glp+ cells in LB with or without inducers (1 mM arabinose + 0.5 mM IAA). All growth experiments were conducted at 30°C. In (A), ◆ = crp, ν = crp glpR, σ = crp O1, △ = crp O4, and ● = crp Glp+. In (B), ◆ = crp glpR (pBAD), ◇ = crp glpR (pBAD-glpR), ν = crp glpR Glp+ (pBAD), □ = crp glpR Glp+ pBAD-glpR).
In (A), cells were incubated on LB agar plates, and the numbers of Glp+ mutants from 50 individual colonies were recorded after 36 hours. In (B), cells were incubated on M9 + glycerol plates, and colonies were counted after the time intervals indicated. Error bars represent standard deviations for experiments conducted in triplicate.