Abstract
Hypoxanthine (H), the deamination product of adenine, has been implicated in the high frequency of A to G transitions observed in retroviral and other RNA genomes. Although H·C base pairs are thermodynamically more stable than other H·N pairs, polymerase selection may be determined in part by kinetic factors. Therefore, the hypoxanthine induced substitution pattern resulting from replication by viral polymerases may be more complex than that predicted from thermodynamics. We have examined the steady-state kinetics of formation of base pairs opposite template H in RNA by HIV-RT, and for the incorporation of dITP during first- and second-strand synthesis. Hypoxanthine in an RNA template enhances the k2app for pairing with standard dNTPs by factors of 10–1000 relative to adenine at the same sequence position. The order of base pairing preferences for H in RNA was observed to be H·C >> H·T > H·A > H·G. Steady-state kinetics of insertion for all possible mispairs formed with dITP were examined on RNA and DNA templates of identical sequence. Insertion of dITP opposite all bases occurs 2–20 times more frequently on RNA templates. This bias for higher insertion frequencies on RNA relative to DNA templates is also observed for formation of mispairs at template A. This kinetic advantage afforded by RNA templates for mismatches and pairing involving H suggests a higher induction of mutations at adenines during first-strand synthesis by HIV-RT.
INTRODUCTION
Deamination of bases in DNA and RNA is of great interest owing to the fact that conversion of a normal hydrogen bond donor (amine) to an acceptor (carbonyl) alters the base coding properties of nucleic acids. Thus, deamination events in DNA or RNA, if left unrepaired, could result in genetic alteration depending upon the identity of the nucleoside inserted opposite the deaminated bases by various polymerases.
In the special case of RNA genomes, the deamination of adenine to produce hypoxanthine (H) [the corresponding nucleo(t/s)ide of hypoxanthine is inosine] appears to be a common mechanism for hypermutagenesis. The evidence for H in RNA genomes derives principally from base substitutions observed in the corresponding cDNA libraries. The occurrence of A to G transitions is taken as evidence for deamination, owing to the well known thermodynamic preference for H·C base pairing (1). This transition in RNA cannot be readily explained by any other adenine modification. Although H·C pairs are known to be favored thermodynamically, H may pair with other bases, suggesting a wider variety of possible base substitutions upon replication of H-containing RNA. This base pairing degeneracy is best illustrated by the fact that H in tRNA wobble positions can pair with uracil, adenine or cytidine in mRNA. In light of this promiscuity, H has been proposed as an ‘inert’ base whose inclusion in hybridization probes could allow for the recognition of ambiguous nucleotides in complex genomic libraries (1,2).
In all, more than 15 published reports have used cDNA analysis as evidence for the presence of hypoxanthine in RNA (reviewed in 3). In the case of one retrovirus (avian leukosis virus), 30–50% of all adenines in a single clone were found to be modified, giving rise to A to G transitions (4). Similar results have been described for another avian retrovirus, Rous-associated virus 1 (5). RNA hypermutation ascribed to the presence of H in RNA has been described for the matrix mRNA of measles virus (6), the RNA genomes of respiratory syncytial virus (7), parainfluenza virus 3 (8), vesicular stomatitis virus (9) and polyomavirus (10). Examination of base substitutions in HIV env and TAR sequences using the database and alignment programs provided by the Los Alamos HIV Sequence Database (see Materials and Methods) similarly reveals a preponderance of adenine base substitutions. Among all possible base substitutions which could occur in these sequences, A to G transitions are overrepresented to the order of ∼20–30%.
Hypoxanthine in RNA thus appears to be widely distributed, and there are several contributing pathways which can account for its occurrence. The discovery of a ubiquitous enzyme with deaminating activity specifically for adenine in dsRNA, the double-stranded RNA adenosine deaminase (dsRAD; 3) has been shown to be important in both the extensive deaminations leading to hypermutagenesis as well as site-specific H modification. Significantly, the TAR sequence of HIV has been shown to be a substrate for the dsRAD protein (11). Deaminated adenines may also occur in RNA as a result of ITP incorporation during RNA synthesis. The concentration of ITP in human blood cells has been found to be ≥50 µM (12). This may be compared to average CTP levels, which are typically ∼100–200 µM. Spontaneous hydrolysis of adenine in RNA would also contribute to the presence of H. The rate for spontaneous hydrolytic purine deamination is ∼10-fold slower than that for deamination of pyrimidines (13–15), yet this pathway could contribute significantly since RNA is not repaired.
Purines in general are highly susceptible to deamination by nitric oxide (NO) and related oxidants (16–19). This reaction is similar to that caused by sodium nitrite (NaNO2), the classical agent for the induction of mutations due to purine deamination in a wide variety of organisms (20). NO is produced in large quantities by macrophages during HIV-1 infection (21). Thus, this pathway would be expected to contribute to deamination in the HIV genome. Perhaps significantly, deamination of purines by NO has been shown to be more extensive for RNA than DNA under similar conditions (18).
During first-strand DNA synthesis by HIV-1 reverse transcriptase (HIV-RT), the presence of unrepaired H in template RNA will give rise to base substitutions which will be propagated during subsequent DNA replication cycles. Thus, the fidelity of first-strand synthesis governs, to an appreciable extent, the creation of genetic diversity in retroviruses. What this base substitution pattern might look like and to what extent it would reflect base substitutions at adenines observed in clinical isolates has never been examined. Results presented here represent our efforts to model the formation of base pairs involving H using a kinetic, rather than a thermodynamic approach, in order to assess potential pairing interactions during the replication of HIV RNA which could contribute to retroviral mutagenesis.
Steady-state kinetic parameters were determined for the formation of base pairs opposite a uniquely placed H within an RNA template. We have also examined the incorporation of dITP using RNA and DNA templates in order to explore the mutation-inducing potential of H introduced from the nucleotide precursor pool during first- and second-strand DNA synthesis by HIV-RT. In contrast to dUTP, which is rapidly degraded in cells, dITP has been shown to persist in dNTP precursor pools (22) and to be efficiently incorporated during replication (23), suggesting a role in the generation of base substitutions.
MATERIALS AND METHODS
Deoxynucleotide triphosphates
All dNTPS except for deoxyinosine triphosphate were purchased from Pharmacia as 100 mM Ultrapure stocks. Deoxyinosine triphosphate was purchased from Sigma. [γ-33P]ATP was purchased from ICN. All dNTP stock solutions (2×) were made up in ddH2O.
RNA and DNA oligonucleotides
Modified RNA templates were synthesized by Dharmacon, Inc. (Boulder, CO) using 2′-O-bis(2-acetoxyethoxy)methyl (ACE) phosphoramidite technology (24). The template sequences and primers used for steady-state kinetic determinations are shown in Table 1. The template sequence corresponds to nucleotides 962–996 of the hypervariable region of the envelope gene (env) of the WMJ1 strain of HIV-1 (25). The DNA template was identical in sequence to the wild-type env gene, substituting thymidine for uracil. Primers were used for the single nucleotide ‘standing start’ polymerase assay and terminate immediately before C17, G19, A(H)20 or U(T)25. Oligonucleotides were purified by 20% polyacrylamide (1.5 mm) gel electrophoresis (PAGE). DNA(RNA) was visualized by UV shadowing, excised from the gel, and recovered using the crush and soak method. DNA primers were radiolabeled by incubating 100–200 pmol of primer with 10 µM ATP, 10 µCi [γ-33P]ATP and 10 U of T4 polynucleotide kinase (NEB) in buffer provided by the manufacturer. The labeled primers were purified by phenol-saturated chloroform extraction. Unincorporated nucleotides were removed by passage through G25 spin columns (Pharmacia) and recovered in 100–150 µl of ddH2O.
Table 1. DNA primers and RNA and DNA templates.
| Primers |
| 5′-CCTCTCGTAAAATATG-3′ |
| 5′-CCTCTCGTAAAATATGGC-3′ |
| 5′-CCTCTCGTAAAATATGGCC-3′ |
| 5′-CCTCTCGTAAAATATGGCCTCTTT-3′ |
| Templatesa |
| RNA 5 10 15 * ** * 30 35 |
| 3′-GGAGAGCAUUUUAUACCGGXGAAAUAAGAGGAAAU-5′ |
| DNA 5 10 15 * ** * 30 35 |
| 3′-GGAGAGCATTTTATACCGGAGAAATAAGAGGAAAT-5′ |
aThe templates are derived from a hypervariable region of the extra-cellular region of the envelope gene of the WMJ-1 strain of HIV-1. Template positions that accept incoming bases in primer extension assays are in bold and denoted by an asterisk. In the RNA template, the X denotes adenosine or hypoxanthine.
Purification of HIV-RT
Plasmid pHIV-RT(His)Prot, containing the genes for C-terminal hexahistidine tagged HIV-RT and HIV protease was the generous gift of Dr Paul Boyer (NCI). Co-expression of these genes in Escherichia coli results in cleavage of the HIV-RT C-terminus. The larger 51 kDa fragment resulting from this cleavage dimerizes with uncleaved HIV-RT to provide a p66/p51 heterodimer (26).
The plasmid was transformed into BL21(DE3)pLysS cells (Novagen, Madison, WI) by electroporation using a Bio-Rad Gene Pulser according to the manufacturer’s instructions, using chloramphenicol and ampicillin selection. The transformed cells were then grown up in 6 l broth (10 g bactotryptone, 5 g yeast extract, 10 g NaCl, pH 7.5, 25 mg/l chloramphenicol, 20 mg/l ampicillin) to an A600 of 0.6. This was followed by induction with isopropylthiogalactopyranoside (1.5 mM). Cell suspensions were incubated for 5 h at 37°C, harvested by centrifugation and stored at –80°C. The yield of cells was ∼5 g/l. Cells (∼10 g) were resuspended in 20 ml of 50 mM NaPO4, pH 8.0, 50 mM NaCl, 1.5 mM phenylmethylsulfonyl fluoride and 0.2 mg/ml lysozyme. The lysate was then centrifuged at 85 × 103 g for 1 h. The supernatant was diluted with an equal volume of 66 mM NaPO4, pH 6.8 and 300 mM NaCl, and loaded at a flow rate of 1 ml/min onto a 5 ml Ni-NTA superflow (Qiagen) column equilibrated with 100 ml of 50 mM NaPO4, pH 7.0, 300 mM NaCl and 10 mM imidazole. Elution was carried out according to the standard protocol, and the fractions were analyzed by SDS–PAGE. Fractions containing p66 and p51 were pooled, concentrated to 1 ml using Centricon®-30 concentrators (Amicon). Dithiothreitol (DTT) was added to a final concentration of 1 mM, and samples were loaded onto a 300 ml Sephacryl-300 (Pharmacia) gel filtration column equilibrated with 50 mM Tris, pH 8.0, 150 mM NaCl and 1 mM DTT. Fractions containing p66 and p51 were pooled, and the sample was dialyzed overnight against 50 mM Tris, pH 8.0, 2 mM EDTA and 50% glycerol. DTT was added to a concentration of 1 mM. The final enzyme concentration was determined by using an extinction value (ɛ280) of 520 mM–1cm–1 (27). The unit activity was determined by incorporation of [α-32P]TTP (0.5 mM, 0.4 Ci/mmol) into a poly(rA)·p(dT)12–18 template (0.4 mM) in a 50 mM Tris, pH 7.2, reaction buffer containing 12.5 mM MgCl2 and 40 mM KCl. Incorporation of radioactive nucleotides into acid insoluble product was determined by trichloroacetic acid precipitation.
Primer extension reactions
The primer extension protocol was adapted from the method of Boosalis et al. (28). Primer/template annealing was accomplished by combining 200 nM primer, 300 nM template in 20× buffer (1 M Tris, pH 7.2, 250 mM MgCl2, 400 mM KCl), 2 mM DTT and ∼0.5 µCi labeled primer in 75 µl. This mixture was heated to 90°C, allowed to cool slowly to ∼40°C and then placed on ice. Next, 25 µl of 1 µM HIV-RT was added, and 4 µl of enzyme/template/primer mixture was aliquoted into each of a set of 0.5 ml Eppendorf tubes. Individual reaction tubes were warmed to 37°C and 4 µl of prewarmed 2× dNTP solution was added to each tube. Reactions proceeded for 5 min and were quenched with 16 µl of 90% formamide, 20 mM EDTA and 0.05% each of xylene cyanol FF and bromphenol blue, before placing on ice. The reactions were analyzed using 20% PAGE (17 × 15 cm). Gels were dried and exposed to a phosphorimager screen. The images were visualized using a PhosphorImager (Molecular Dynamics).
Data analysis
The amount of radioactivity in each band was determined using the ImageQuant software package (Molecular Dynamics). The velocity of each reaction was determined using the following equation:
v = I1[primer]/(I0 + 1/2I1)t
where v is velocity in pM/s, I0 is the intensity of the unextended primer band, I1 is the intensity of the extended primer band (or the sum of all such bands if there are more than one), and t is reaction time in seconds. The velocity data were plotted using direct linear plots (29) and KM and Vmax were determined for each base pairing reaction. The frequency of insertion for a mispair (fins) is defined at k2app(M)/k2app(WC); the ratio of apparent second order rate constants for a mispair and the corresponding Watson–Crick base pair.
Base substitution analyses of HIV sequences
Sequence analysis was conducted using the database tools available at the HIV Sequence Database site (http://hiv-web.lanl.gov). Consensus sequences were generated using 18 complete env subtype A and B sequences (∼4 kb). For construction of the TAR consensus sequence, 409 sequence entries were used. Alignments were achieved using SequencherTM 3.0.1 or Microsoft® Excel 98. The most common base at a given position was defined as the consensus sequence base. Sequences were compared to their respective consensus sequences using the Hypermut program. This returns a list of the differences between each sequence and the consensus. These were sorted by base changes and tabulated. Sequence differences resulting from insertions and deletions were omitted. Specific base substitutions expressed as percentages of all changes are provided in Table 4 for env subtypes A, B and TAR sequences.
Table 4. Base substitution frequencies for two sequences within the HIV genome.
| Base change |
env Subtype A
(%) |
env Subtype B
(%) |
TAR
(%) |
| A to C | 9.6 | 8.3 | 1.9 |
| A to G | 23.4 | 20.4 | 27.4 |
| A to T | 4.8 | 5.5 | 4.2 |
| C to A | 6.0 | 5.3 | 1.9 |
| C to G | 2.6 | 2.0 | 2.9 |
| C to T | 12.0 | 10.4 | 9.7 |
| G to A | 18.8 | 21.7 | 15.2 |
| G to C | 4.0 | 3.5 | 1.7 |
| G to T | 3.4 | 2.9 | 10.9 |
| T to A | 4.5 | 5.9 | 5.4 |
| T to C | 9.9 | 7.0 | 11.9 |
| T to G | 5.0 | 7.2 | 6.9 |
RESULTS
HIV-RT insertion of pyrimidine dNTPs opposite H or A in RNA
The sequences of primers and templates used in these studies are presented in Table 1. The 35mer RNA template, based upon a region of the HIV-1 hypervariable domain of the env glycoprotein, was uniquely substituted with hypoxanthine. The steady-state parameters that describe the insertion of all four unmodified nucleotides at this site were determined. The corresponding values obtained for RNA templates containing adenine at this same position were also measured. Thus, the mutation-inducing potential resulting from deamination of A to H in a particular sequence context could be evaluated. In order to compare the potential for mispairs at H relative to A, the frequency of insertion values (fins) were calculated and are presented graphically in Figure 1. These values are the apparent second-order rate constants (k2app = kcat/KM) for each possible base pair involving H20 and A20 in RNA divided by kcat/KM for the reaction A20 + dTTP. A complete set of kinetic data for all reactions involving RNA templates is compiled in Table 2.
Figure 1.
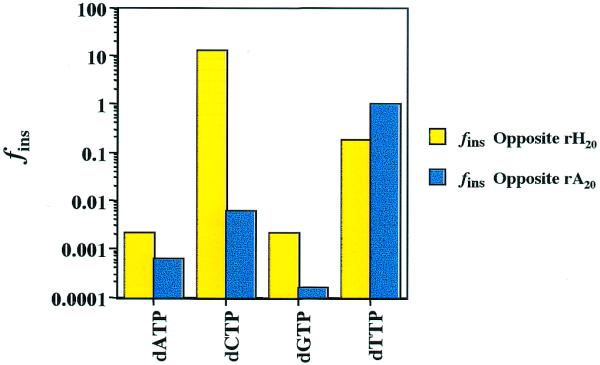
Fidelity (fins) values for insertion of dNTPs opposite H (yellow) and A (blue) in an RNA template catalyzed by HIV-RT. A(H) is located at position 20 in the oligonucleotide sequence shown in Table 1. The values of fins = [kcat/KM]A(H)·N/[kcat/KM]A·T, where N is any nucleotide. Complete steady state parameters describing the reactions on RNA templates are provided in Table 2.
Table 2. HIV-RT steady-state kinetic data for RNA templates.
| Template basea |
dNTP |
KM (µM) |
Vmax (pM/s) |
kcat (s–1) |
Second order rate
(s–1M–1) |
finsb |
| H20 | A | 14 | 60 | 4.8 × 10–4 | 3.4 × 101 | 5.0 × 10–3 |
| H20 | C | 0.024 | 206 | 2.1 × 10–3 | 8.7 × 104 | 1.3 × 101 |
| H20 | G | 83.000 | 150 | 1.2 × 10–3 | 1.4 × 101 | 2.1 × 10–3 |
| H20 | T | 0.590 | 88 | 7.0 × 10–4 | 1.2 × 103 | 1.8 × 10–1 |
| A20 | H | 530.000 | 110 | 8.8 × 10–4 | 1.7 × 100 | 2.4 × 10–4 |
| C17 | H | 0.043 | 104 | 8.3 × 10–4 | 1.9 × 104 | 8.0 × 100 |
| G19 | H | 92.000 | 76 | 6.1 × 10–4 | 6.6 × 100 | 8.3 × 10–4 |
| U25 | H | 125.000 | 44 | 3.5 × 10–4 | 2.8 × 100 | 5.9 × 10–4 |
| A20 | A | 97.000 | 51 | 4.1 x 10–4 | 4.2 x 10–6 | 6.2 × 10–4 |
| A20 | C | 6.200 | 32 | 2.6 × 10–4 | 4.1 × 101 | 6.1 × 10–3 |
| A20 | G | 120.000 | 16 | 1.3 × 10–4 | 1.1 × 100 | 1.6 × 10–4 |
| C17 | A | 41.000 | 60 | 4.8 × 10–4 | 1.2 × 101 | 4.9 × 10–3 |
| G19 | A | 78.000 | 18 | 1.4 × 10–4 | 1.8 × 100 | 2.3 × 10–4 |
| C17 | G | 0.099 | 30 | 2.4 × 10–4 | 2.4 × 103 | |
| G19 | C | 0.039 | 39 | 3.1 × 10–4 | 8.0 × 103 | |
| A20 | T | 0.052 | 44 | 3.5 × 10–4 | 6.8 × 103 | |
| U25 | A | 0.140 | 84 | 6.7 × 10–4 | 4.8 × 103 |
aNumbers correspond to template base positions from Table 1.
bFrequency of insertion (fins) is defined in the legend to Figure 1.
Inspection of Figure 1 reveals a dramatic preference for H·C base pair formation by HIV-RT. The insertion of dCTP opposite H20 was found to occur 2100 times more frequently than insertion of dCTP opposite A20. Relative to formation of the normal Watson–Crick base pair at this position (A20 + dTTP), the H20 + dCTP pairing occurs 13 times more frequently. In contrast, the incorporation of dCTP opposite A20 occurs 164-fold less frequently than the Watson–Crick pairing. Comparison of k2app values reveals that this reaction is favored even over the formation of G·C pairs by a factor of ∼10. The data in Table 2 demonstrate that this is due to both lower KM and increased Vmax values. In contrast to the situation observed with dCTP, the kinetic discrimination for insertion of dTTP at H20 versus A20 is not very large. Examination of the apparent second order rate constants for the reactions r(A/H)20 + dTTP reveal that the H·T pairing is only ∼5-fold less efficient than formation of the Watson–Crick base pair.
HIV-RT insertion of purine dNTPs opposite H or A in RNA
Comparison of fins values for the reactions at r(A/H)20 with dGTP reveals that H·G pairs are preferred, by a factor of 13. This preference is influenced primarily by a substantially larger Vmax (∼10-fold) rather than a significant lowering of KM (∼1.4-fold). Insertion of dATP opposite rH20 by HIV-RT is favored over the rA20 + dATP reaction by a factor of three, due solely to differences in KM. Thus, the data in Figure 1 and Table 2 reveal that substitution of H for A in template RNA significantly increases the probability for formation of base pairs leading to transversions.
H·N base pair formation efficiency: participation as template base or dITP
For any given H·N base pair, the efficiency of formation may vary depending upon whether H participates as an RNA template base or as dITP from the nucleoside triphosphate precursor pool. These situations are not kinetically equivalent. Steady-state kinetic differences for individual base pair combinations involving hypoxanthine were examined within this context (rH + dNTP versus rN + dITP). The apparent second order rate constants for H·N combinations are grouped in this manner and are plotted in Figure 2. It can be seen that, for any given base pair combination, the reaction in which H is in the RNA template proceeds with greater catalytic efficiency. Alternatively, these results may be viewed as disfavoring reactions which require the binding and orienting of dITP at the polymerase active site. In the case of H·A pairs, insertion of dATP opposite template H is favored ∼20-fold relative to insertion of dITP opposite A. The differences in second order rate constants are primarily reflected in a nearly 40-fold difference in KM, while the kcat values differ by <2-fold.
Figure 2.
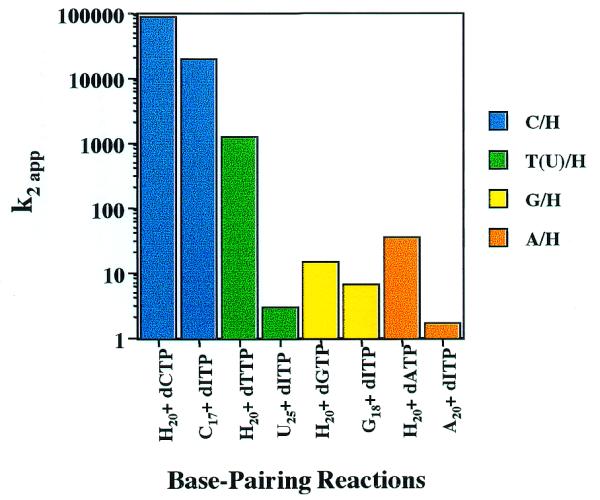
Apparent second order rate constants describing the formation of heteroduplex H·N base pairs and the corresponding N·H inversion pair. The k2app values for reactions rH + dNTP and rN + dITP for a given N are grouped by color.
In the case of H·G, the reaction with template H is only modestly favored by a 2-fold greater kcat, while the KM values are nearly equivalent. A larger ‘RNA template effect’ is observed for H·Pyr pairings. The H·C pairing reaction favors H in the template by nearly 5-fold, due to enhanced kcat and lowered KM. The much larger variation observed for the second order rate constants for the rH + dTTP and rU + dITP reactions (>400-fold) may reflect, in part, differences between thymine and uracil. It is difficult, however, to rationalize this large effect on the basis of the 5-methyl group alone. The KM for the favored reaction with H in the template is ∼200-fold lower, while the kcat advantage is small, only 2-fold (Table 2). Thus, for every possible pairing involving H catalyzed by HIV-RT, H in the RNA template provides a more favorable opportunity for non-standard pairing. The magnitude of the differences in kcat/KM values between rH + dNTP versus rN + dITP undoubtedly reflect, to some extent, the influence of nearest-neighbor bases. Nonetheless the values for reactions with H in the RNA template are greater in every case, regardless of the exact sequence environment. This suggests that the RNA template effect overrides insertion preferences due to sequence context.
Insertion of dITP on DNA and RNA templates within the same sequence context
The formation of base pairs involving hypoxanthine by HIV-RT was also examined on a DNA oligonucleotide identical in sequence to the RNA template except for replacement of U with T, and A for H at position 20 (Table 1). All possible mispairs on the DNA template involving H were examined using dITP (Table 3). These reactions could be compared directly to the corresponding insertion reactions of dITP on RNA templates (Table 2). The complete steady-state kinetic values measured for all reactions using the DNA template are presented in Table 3.
Table 3. HIV-RT steady-state kinetic data for DNA template.
| Template basea |
dNTP |
KM (µM) |
Vmax (pM/s) |
kcat (s–1) |
Second order rate
(s–1M–1) |
fins |
| A20 | H | 210.000 | 56 | 4.5 × 10–4 | 2.1 × 100 | 6.7 × 10–5 |
| C17 | H | 0.082 | 210 | 1.7 × 10–3 | 2.0 × 104 | 1.0 × 100 |
| G19 | H | 550.000 | 100 | 8.0 × 10–4 | 1.4 × 100 | 4.1 × 10–5 |
| T25 | H | 34.000 | 68 | 5.4 × 10–4 | 1.6 × 101 | 2.6 × 10–4 |
| A20 | A | 23.000 | 17 | 1.36 x 10–4 | 5.9 x 10–6 | 1.8 × 10–4 |
| A20 | C | 52.000 | 95 | 7.6 × 10–4 | 1.5 × 101 | 4.1 × 10–4 |
| A20 | G | 78.000 | 35 | 2.8 × 10–4 | 3.6 × 100 | 1.0 × 10–4 |
| C17 | A | 5.400 | 35 | 2.8 × 10–4 | 5.2 × 101 | 1.4 × 10–3 |
| G19 | A | 34.000 | 43 | 3.4 × 10–4 | 1.0 × 101 | 1.7 × 10–4 |
| C17 | G | 0.075 | 190 | 1.5 × 10–3 | 2.0 × 104 | |
| G19 | C | 0.026 | 104 | 8.3 × 10–4 | 3.2 × 104 | |
| A20 | T | 0.056 | 250 | 2.0 × 10–3 | 3.6 × 104 | |
| T25 | A | 0.027 | 205 | 1.6 × 10–3 | 6.1 × 104 |
aNumbers correspond to template base positions from Table 1.
For ease of comparison, fins for the reaction of dITP at specific positions on DNA and RNA templates are displayed graphically in Figure 3. The values of fins measured for insertion of dITP opposite template RNA bases are significantly greater than those determined opposite bases in the DNA template for the same sequence positions. For example, the fins for dITP insertion opposite rG19 catalyzed by HIV-RT is ∼20-fold greater than that for insertion opposite dG19. The fidelity for any ‘non-standard’ pairing event occurring is defined as 1/fins. Thus, insertion of dITP at rG19 can be expected to occur approximately once every 1000 replication events, whereas insertion of dITP at dG19 within the same sequence context would occur once in 22 000 cycles.
Figure 3.
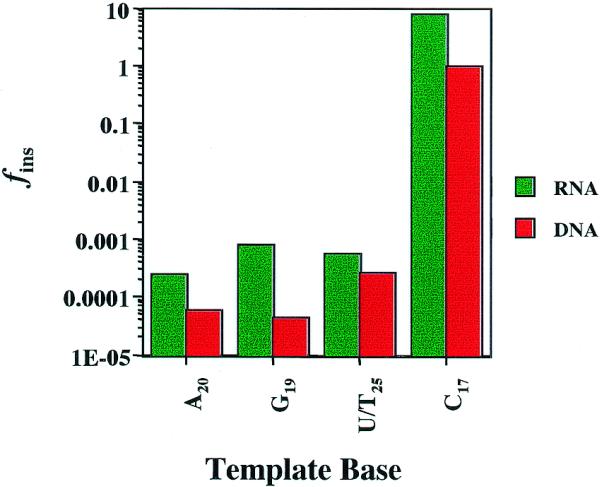
Incorporation of dITP into RNA and DNA templates of identical sequence by HIV-RT. The apparent second order rate constants for the reactions NDNA/RNA + dITP were divided by the second order rate constants for the corresponding Watson–Crick base pairs at the indicated positions to obtain fins. Values for RNA (green bars) and DNA (red bars) templates for all N were taken from Tables 2 and 3, respectively. Numbered bases in the ordinate refer to positions in the RNA template 5.
The base pairing reaction with dITP at rC17 was catalyzed 8-fold more readily than the reaction at dC17. In the case of the DNA template reaction, the insertion of dITP at dC17 was characterized by an apparent second order rate constant which was virtually identical to that measured for the insertion of dGTP at the same position.
The insertion of dITP at rA20 is preferred over insertion at dA20 by ∼4-fold. The least discrimination in favor of RNA templates was observed for the insertion of dITP at (U/T)25, where the reaction at U was favored by ∼2-fold. These comparisons reveal that the incorporation of dITP from nucleotide precursor pools by HIV-RT is more likely to occur during first-strand DNA synthesis from the retroviral RNA template rather than during copying of the cDNA (second-strand DNA synthesis).
Examination of the individual components of the fins values, the apparent second order rate constants (k2app), reveals that some of the enhanced values of fins for the reactions of dITP on the RNA template reflect lower values of k2app for the Watson–Crick base pairing reactions. For example, the reactions of dITP at C17 are characterized by identical k2app values on both DNA and RNA (2 × 104 s–1M–1, Tables 2 and 3). However, the k2app for the insertion of dGTP opposite C17 (the denominator component of fins for the dITP + C17 reaction) is nearly 10-fold lower on the RNA template. Thus, the lower efficiency of formation of C17·G pairs on RNA enables dITP to compete more efficiently for insertion with dGTP, increasing the probability of insertion. Watson–Crick base pair formation on the DNA template can more efficiently compete with dITP insertion from the dNTP precursor pool.
A similar explanation appears to hold for the 4-fold increased probability in favor of insertion of dITP at rA20 relative to dA20. The k2app values are virtually identical for the respective reactions with dITP (∼2 s–1M–1), yet the reaction forming A20·T base pairs is substantially less efficient on the RNA template (Tables 2 and 3). Comparison of fins values shows that the insertion of dITP opposite rG19 relative to dG19 is favored in the former case by 19-fold. In this example, the k2app for the rG19 + dITP reaction is ∼5-fold greater than the value describing dITP insertion at dG19. The rG19 + dCTP reaction has a 4-fold lower value of k2app compared to the reaction on DNA, which contributes to the large increase in insertion fidelity. Interestingly, we observe in all cases for each position within this template sequence that the k2app values for Watson–Crick base pair formation catalyzed by HIV-RT are invariably lower (4–13-fold) on RNA compared to DNA templates.
Base substitution analyses of HIV sequences
In order to better analyze the base substitution trends in HIV-1 which could potentially arise via the intermediacy of hypoxanthine, a comprehensive analysis of env subtypes A and B was undertaken using the ‘Search and Align’ feature of the HIV Sequence Database site (http ://hi v-web.lanl.gov). Frequencies of base substitutions could be compared with biases predicted from our in vitro data. This database includes updated HIV-1 sequence information in addition to a number of tools for genome analysis. Analyses were carried out within distinct subtypes of env since these are normally considered to have evolved independently. Thus, in theory the base substitution patterns within different subtypes might be dissimilar. The TAR sequence was also analyzed for base substitutions since it is a known double-stranded element in the HIV genome, and has been shown to be a substrate for the dsRAD (11). Base substitution frequencies for all three sequences are provided in Table 4.
Examination of Table 4 reveals that the most frequent base substitution found among all sequences is A to G. In the TAR sequence, this accounts for as much as 30% of all base changes. The phenomenon of G to A hypermutation (30) is also observed in the analyses of these sequences, accounting for ∼20% of base changes in the env sequences. The extent of this substitution is lower in TAR sequences (15%). Other types of base substitutions are more represented in TAR than in the env subtypes. For example, G to T substitutions are substantially higher in TAR (∼11 versus ∼3%) than in env subtypes. Other substitutions, for example T to A, do not vary significantly among the analyzed sequences. It is interesting to note that the relative ratio of A to C and A to T substitutions in the TAR sequence (1:2) resembles the ratio of insertion frequencies for dGTP and dATP opposite H by HIV-RT. This is not the case for the env subtypes, where A to C changes are more numerous than A to T (1.5–2:1).
DISCUSSION
The overrepresentation of adenine in the HIV genome (∼60%; 31) suggests that even modest levels of deamination of this base could potentially contribute to hypermutagenesis. We have examined the steady-state kinetics of formation of base pairs involving the adenine deamination product, hypoxanthine, by HIV-RT during first-strand DNA synthesis on RNA templates. Kinetic analyses of base pairing events involving hypoxanthine have not previously been described. By substituting H for A at a specific site in an oligoribonucleotide template, the miscoding potential in RNA due to a single deamination event within a defined sequence context could be examined in detail. The frequencies with which dITP from nucleoside precursor pools could be incorporated at various positions during first-strand synthesis on RNA templates and on DNA templates were also examined within identical sequence contexts.
The hierarchy of kinetic insertion preferences for dNTPs opposite H in RNA by HIV-RT determined in our studies was found to be H·C >> H·T > H·A > H·G. The order of thermodynamic stabilities of base pairs containing H in DNA has been determined to be H·C > H·A >H·T ∼ H·G (1). According to thermodynamic melting studies H·C is less stable than an A·T base pair within the same sequence context of RNA. However, our data indicate that formation of an H·C base pair occurs 13-fold more frequently than an A·T base pair in RNA with identical flanking sequences. If the same stability trends found in DNA hold for the heteroduplex primer/template complexes, it would appear that thermodynamic considerations may not provide a reliable indicator of insertion frequencies. Thermodynamic data from Martin et al. (1) suggest that H·T base pairs should be the least stable among all four possible H-containing base pairs. However, we find that H·T base pair formation is characterized by relatively high insertion frequencies, surpassed only by H·C base pairs (Fig. 1). This is true not only for heteroduplex base pairs formed on RNA templates (rH20 + dTTP), but for reactions on DNA templates as well (dT25 + dITP; Fig. 3).
Echols and Goodman (32) have demonstrated that base pair thermodynamic stability does not usually predict base pairing preferences by DNA polymerases. Additional thermodynamic studies on H-containing heteroduplexes will be required to evaluate what role, if any, thermodynamic stability plays in base pairing preferences when RNA is the template. Echols and Goodman have also formulated a principle of geometric selection, dictated by base pair geometry, to explain the observed order of insertion frequencies by polymerases (32,33).
The greater importance of base pair geometry relative to simple thermodynamic considerations in rationalizing frequencies of mispair formation is further supported in our studies by examining homopurine pairing events. Somewhat surprisingly, homopurine base pairs may perturb overall helix geometry in a negligible manner. However, the overall contribution of such pairs to thermodynamic stability may be destabilizing. For example, A·G pairs in RNA produce large helical perturbations, whereas H·A pairs are accommodated with little deviation from the normal A helix dimensions (34). Yet A·G pairs contribute greater thermodynamic stability to duplexes than H·A pairs. We observe an ∼30-fold preference for H20·A over A20·G pairing by HIV-RT in our studies, in spite of the greater thermodynamic stability of the latter pair.
A similar rationale may hold for the observation of preferential A·A over A·G base pair formation in RNA templates. The rA20 + dATP reaction is favored over rA20 + dGTP by a factor of ∼4-fold (Table 2). The NMR structure of the A·A pair shows remarkably little deviation from the normal B configuration in DNA, and effective stacking within the duplex is observed (35). This is in spite of the fact that in the anti/anti configuration, only a single hydrogen bond (NH2·N1) may be formed. Although there is a greater thermodynamic contribution to duplex stability by A·G base pairs (36), geometric discrimination by HIV-RT may favor the A·A pairing arrangement. The A·A pair is also favored over A·G in DNA homoduplexes, although to a lesser extent.
For each H·N base pair, we have shown that reactions where H is in the RNA template invariably proceed with a greater efficiency than when participating as the incoming triphosphate. This suggests that in retroviral first-strand synthesis, deamination of rA in the template is potentially more mutagenic than deamination of dATP in the nucleotide triphosphate precursor pool. This variation may be due in part to differences in the polarity of stacking for a given base pair with respect to the helix, depending upon whether it is in the 3′ or 5′ strand. The geometrical and thermodynamic properties of each configuration are expected to be non-equivalent. Partitioning between the relative contributions of each of these effects will require NMR or crystallographic studies of H·N pairs in both possible orientations in heteroduplexes in conjunction with calorimetric studies.
The notion that hypoxanthine pairs like guanine in DNA is supported by the fact that the apparent second order rate constants for the dC17 + dITP and dC17 + dGTP reactions are the same (2 × 104 M–1s–1). The respective contributions of KM and Vmax to this value of k2app are nearly identical for both reactions. This is not the situation, however, when the template strand is RNA. Here, the rC17 + dITP reaction is characterized by an apparent second order rate constant which is nearly 10-fold greater than that for rC17 + dGTP. Thus, when RNA is the template, the condition of first-strand synthesis, HIV-RT appears to favor the formation of H·C even over G·C pairs within the same sequence context. This large preference for H·C formation in heteroduplexes can also be observed by comparing kcat/KM values for the rH20 + dCTP reaction with the formation of G·C pairs at independent sites (different sequence contexts). This reaction is 11- and 36-fold more efficient than the rG19 + dCTP and rC17 + dGTP reactions, respectively (Table 2).
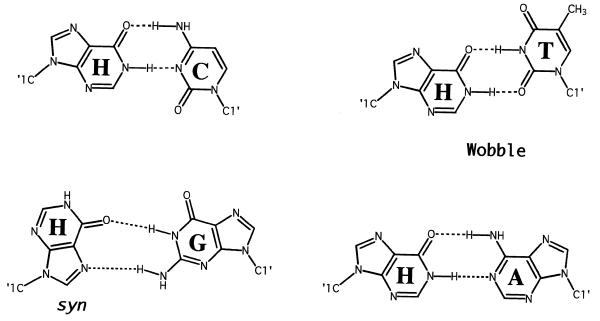
The structures of all possible base pairs involving hypoxanthine are shown in Figure 4. Crystal structures of H·C-containing duplexes in both B and Z DNA (36,37) reveal an overall geometry similar to that of G·C pairs. The two hydrogen-bond distances in the H·C base pair in DNA are nearly equivalent to the corresponding bonds in G·C pairs within the same sequence context (36). The local helix geometry of H·C pairs in B form DNA resembles that of a polyadenine tract in DNA, possessing large propeller twist angles and a narrow minor groove. Whether a similar perturbation exists in RNA–DNA heteroduplexes is currently unknown.
Figure 4.
Postulated hydrogen-bonding schemes for base pairs involving inosine. Redrawn from Martin et al. (1). The H(syn)·G(anti) configuration shown is proposed by analogy to G·G base pairs.
The duplexes involved in chain elongation by RT are either heteroduplexes (first-strand synthesis) or all DNA duplexes (second-strand synthesis). Since heteroduplexes have some structural features which distinguish them from DNA duplexes, it is possible that some of the kinetic differences we observe between first- and second-strand synthesis for a given pairing are due to these features. Heteroduplexes demonstrate a heteronomous configuration, meaning that the RNA strand maintains a C3′ endo (A form) sugar pucker while the DNA strand is found to be C2′ endo (38). However, under certain conditions, DNA homoduplexes may also display heteronomous configurations between strands (39). Thus, the extent to which this may contribute to the kinetic differences is unclear.
Heteroduplexes appear to differ significantly from RNA and DNA homoduplexes in the extent of hydration of the major groove. RNA homoduplexes are the least hydrated. Although more water is accessible to the major groove of DNA homoduplexes, there is more significant hydration in the heteroduplexes (40). This is facilitated by the 2′-OH group, which in the heteroduplex adopts a preferred orientation facing the complementary base, rather than towards C3′ (Φ = 0 to –30°) of the template base. Water molecules hydrogen bonded in this manner in an rA·dT base pair result in a significant amount of hydration at the C2 position of adenine (40). Analogously, one might anticipate a similar hydration environment at H in heteroduplexes. Water molecules oriented about the C2 position of H may facilitate the incorporation of dCTP by lowering the transition state energy for incorporation via hydrogen bonding interactions with O2 of dCTP (Fig. 4). This may facilitate the rapid adoption of an optimal orientation for polymerization within the RT active site, resulting in the unusually large k2app observed in our studies.
Alignment analyses of three HIV gene sequences revealed an unusual preponderance of A to G transitions. The largest percentage of such changes, nearly 30% were found in the double-stranded TAR sequence. Studies of base substitution patterns for other HIV genes, such as gag, have similarly revealed a high percentage of A to G transitions (41). The majority of these base substitutions are not lethal, suggesting that they may play a role in generating adaptive variants of the virus (42). It is also noteworthy that the base substitution patterns within the two env subtypes are remarkably similar, in spite of the fact that they must have evolved from different ancestors in non-identical environments. This suggests that the mechanistic forces acting to produce base substitutions are similar.
The steady-state kinetics of dITP insertion by HIV-RT on RNA templates were directly compared to insertion on DNA templates in order to evaluate the mutagenic potential of H introduced from the nucleoside precursor pool during first- and second-strand DNA synthesis. Base pairs are formed more readily when RNA is the template for all pairings with dITP. This suggests that during retroviral replication, incorporation of dITP from the nucleoside precursor pool is more likely to occur during first-strand rather than second-strand synthesis. This effect is largest for the G19 + dITP reaction, which is favored in RNA over DNA by a factor of 20. Second-strand DNA synthesis following incorporation of dITP opposite G would most likely pair dCTP with H in the initially synthesized DNA strand. This accomplishes an overall G to C transversion in the retroviral genomic sequence. A similar mechanism could account for some percentage of A to C transversions in the retroviral genome, due to mis-incorporation of dITP opposite A during first-strand synthesis.
The rC17 + dITP reaction is 8-fold more efficient than the rC17 + dGTP reaction. However, since second-strand synthesis by HIV-RT would likely pair the resulting H with dCTP, no mutation results. Incorporation of dITP opposite rC17, rG19, rA20 and U25 occurs, respectively, 8-, 20-, 4- and 2-fold more frequently than for the corresponding DNA template bases. Thus, we would predict that there is a greater probability for incorporation of H from nucleotide precursor pools during first-strand synthesis, since dITP can compete more effectively with unmodified dNTPs when RNA is the template.
The relatively high efficiency of the rH + dTTP reaction implies that the standard method of assaying for the extent of adenine deamination using cDNA transcript analysis may actually underestimate the level of H in RNA. This efficiency may depend to some extent on the exact polymerase used to construct the cDNA. Footprinting methods for the direct detection of H in RNA have recently been developed (43), and should now make possible a more definitive quantitation of the occurrence of H in various RNA genomes.
Independent of the involvement of H, HIV-RT-catalyzed formation of A·C mispairs in RNA can also lead to A to G transitions. It is interesting to note that the rA20 + dCTP reaction has a 15-fold higher fins value than the dA20 + dCTP reaction. In fact, relative to mispairs formed on RNA with unmodified bases, the A·C mispair occurs with the highest frequency. However, the value of fins for the H·C mispair in RNA is greater than that of A·C by more than three orders of magnitude, thus, one would still predict a greater influence of H in the A to G transition phenomenon.
Tables 2 and 3 contain additional kinetic information on ‘normal’ mispairs with mutation-inducing potential for both DNA and RNA templates. Comparison of fins values for A·A and A·G mispairs reveals a 2- and 3-fold greater propensity for formation on RNA templates, respectively. It remains to be determined whether mispairs in general occur with greater frequency on RNA templates when HIV-RT is the replicative polymerase. Since most of the HIV-RT fidelity studies have relied on DNA templates in order to evaluate mismatch potential, an expanded RNA database of mispairs in different sequence contexts will be required to assess this possibility. This would suggest, as we have illustrated for replication involving H, that HIV-RT copying of heteroduplexes is inherently more error prone, and would imply that the majority of base substitutions likely occur during first-strand synthesis.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Gerald E.Wuenschell for helpful discussions and advice. The assistance of Toni Martinez in the preparation of this manuscript is gratefully appreciated. This work was supported by PHS Grant GM53692 to J.T.
References
- 1.Martin F.H., Castro,M., Aboul-ela,F. and Tinoco,I. (1985) Base pairing involving deoxyinosine: implications for probe design. Nucleic Acids Res., 13, 8927–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohtsuka E., Masuki,S., Ikehara,M., Takahashi,Y. and Matsubara,K. (1985) An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions J. Biol. Chem., 260, 2605–2608. [PubMed] [Google Scholar]
- 3.Bass B.L. (1997) RNA editing and hypermutation by adenosine deamination. Trends. Biochem. Sci., 22, 157–162. [DOI] [PubMed] [Google Scholar]
- 4.Haijar A.M. and Linial,M.L. (1995) Modification of retroviral RNA by double-stranded RNA adenosine deaminase. J. Virol., 69, 5878–5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Felder M.P., Laughier,D., Yatsula,B., Dezelee,P., Calothy,G. and Marx,M. (1994) Functional and biological properties of an avian variant long terminal repeat containing multiple A to G conversions in the U3 sequence. J. Virol., 68, 4759–4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass B.L., Weintraub,H., Cattaneo,R. and Billeter,M.A. (1989) Biased double-stranded RNA. Cell, 56, 331. [DOI] [PubMed] [Google Scholar]
- 7.Rueda P., Garcia-Barreno,B. and Melero,J.A. (1994) Loss of conserved cysteine residues in the attachment (G) glycoprotein of two human respiratory syncytial virus escape mutants that contain multiple A-G substitutions (hypermutations). Virology, 198, 653–662. [DOI] [PubMed] [Google Scholar]
- 8.Murphy D.G., Dimock,K. and Kang,C.Y. (1991) Numerous transitions in human parainfluenza virus 3 RNA recovered from persistently infected cells. Virology, 181, 760–763. [DOI] [PubMed] [Google Scholar]
- 9.O’Hara P.J., Nichol,S.T., Horodyski,F.M. and Holland,J.J. (1984) Vesicular stomatitis virus defective interfering particles can contain extensive genomic sequence rearrangements and base substitutions. Cell, 36, 915–924. [DOI] [PubMed] [Google Scholar]
- 10.Kumar M. and Carmichael,G.G. (1997) Nuclear antisense RNA induces extensive adenosine modifications and nuclear retention of target transcripts. Proc. Natl Acad. Sci. USA, 94, 3542–3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharmeen L., Bass,B., Sonenberg,N., Weintraub,H. and Groudine,M. (1991) Tat-dependent adenosine-to-inosine modification of wild-type transactivation response RNA. Proc. Natl Acad. Sci. USA, 88, 8096–8100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Verdier C.H., Niklasson,F., van Waeg,G., Ericson,A. and Högman,C. (1987) Purine metabolism in normal and high-ITP human erythrocytes. Attempts to evaluate the ability to store the cells. Biomed. Biochim. Acta, 46, S263–S267. [PubMed] [Google Scholar]
- 13.Frederico L.A., Kunkel,T.A. and Shaw,B.R. (1990) A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochemistry, 29, 2532–2537. [DOI] [PubMed] [Google Scholar]
- 14.Shen J.-C., Rideout,W.M. and Jones,P.A. (1994) The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res., 22, 972–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindahl T. (1967) Irreversible heat inactivation of transfer ribonucleic acids. J. Biol. Chem., 242, 1970–1973. [PubMed] [Google Scholar]
- 16.Shapiro R. and Pohl,S.H. (1968) The reaction of ribonucleosides with nitrous acid. Side products and kinetics. Biochemistry, 7, 448–455. [DOI] [PubMed] [Google Scholar]
- 17.Wink D.A., Kasprzak,K.S., Maragos,C.M., Elespuru,R.K., Misra,M., Dunams,T., Cebula,T.A., Koch,W.H., Andrews,A.W., Allen,J.S. and Keefer,L.K. (1991) DNA deaminating ability and genotoxicity of nitric oxide and its progenitors. Science, 254, 1001–1003. [DOI] [PubMed] [Google Scholar]
- 18.Nguyen T., Brunson,D., Crespi,C.L., Penman,B.W., Wishnok,J.S. and Tannenbaum,S.R. (1992) DNA damage and mutation in human cells exposed to nitric oxide in vitro. Proc. Natl Acad. Sci. USA, 89, 3030–3034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caulfield J.L., Wishnok,J.S. and Tannenbaum,S.R. (1998) Nitric oxide-induced deamination of cytosine and guanine in deoxynucleosides and oligonucleotides. J. Biol. Chem., 273, 12689–12695. [DOI] [PubMed] [Google Scholar]
- 20.Orgel L.E. (1965) The chemical basis of mutation. Adv. Enzymol., 27, 289–346. [DOI] [PubMed] [Google Scholar]
- 21.Schwarz K.B. (1996) Oxidative stress during viral infection: a review. Free Radic. Biol. Med., 21, 641–649. [DOI] [PubMed] [Google Scholar]
- 22.Myrnes B., Guddal,P.-H. and Krokan,H. (1982) Metabolism of dITP in HeLa cell extracts, incorporation into DNA by isolated nuclei and release of hypoxanthine from DNA by a hypoxanthine–DNA glycosylase activity. Nucleic Acids Res., 10, 3693–3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas K.R., Manlapaz-Ramos,P., Lundquist,R. and Olivera,B.M. (1979) Formation of Okazaki pieces at the Escherichia coli replication fork in vitro. Cold Spring Harbor Symp. Quant. Biol., 43, 231–237. [DOI] [PubMed] [Google Scholar]
- 24.Scaringe S.A., Wincott,F.E. and Caruthers,M.H. (1998) Novel RNA synthesis method using 5′-O-silyl-2′-O-orthoester protecting groups. J. Am. Chem. Soc., 120, 11820–11821. [Google Scholar]
- 25.Starcich B.R., Hahn,B.H., Shaw,G.M., McNeely,P.D., Modrow,S., Wolf,H., Parks,E.S., Parks,W.P., Josephs,S.F., Gallo,R.C. and Wong-Staal,F. (1986) Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell, 45, 637–648. [DOI] [PubMed] [Google Scholar]
- 26.LeGrice S.F.J. and Grüninger-Leitch,F. (1990) Rapid purification of homodimer and heterodimer HIV-1 reverse transcriptase by metal chelate affinity chromatography. Eur. J. Biochem., 187, 307–314. [DOI] [PubMed] [Google Scholar]
- 27.Einolf H.J., Schnetz-Boutaud,N. and Guengerich,F.P. (1998) Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry, 37, 13300–13312. [DOI] [PubMed] [Google Scholar]
- 28.Boosalis M.S., Petruska,J. and Goodman,M.F. (1987) DNA polymerase insertion fidelity. Gel assay for site-specific kinetics. J. Biol. Chem., 262, 14689–14686. [PubMed] [Google Scholar]
- 29.Eisenthal R. (1974) The direct linear plot. A new graphical procedure for estimating enzyme kinetic parameters. Biochem. J., 139, 715–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martinez M.A., Vartanian,J.P. and Wain-Hobson,S. (1994) Hypermutagenesis of RNA using human immunodeficiency virus type 1 reverse transcriptase and biased dNTP concentrations. Proc. Natl Acad. Sci. USA, 91, 11787–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berkhout B. and van Hemert,J. (1994) The unusual nucleotide content of the HIV RNA genome results in a biased amino acid composition of HIV proteins. Nucleic Acids Res., 22, 1705–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Echols H. and Goodman,M. (1991) Fidelity mechanisms in DNA replication. Annu. Rev. Biochem., 60, 477–511. [DOI] [PubMed] [Google Scholar]
- 33.Goodman M.F. (1997) Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl Acad. Sci. USA, 94, 10493–10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carter R.J., Baeyens,K.J., SantaLucia,J., Turner,D.H. and Holbrook,S.R. (1997) The crystal structure of an RNA oligomer incorporating tandem adenosine–inosine mismatches. Nucleic Acids Res., 25, 4117–4122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gervais V., Cognet,J.A.H., Le Bret,M., Sowers,L.C. and Fazakerley,V. (1995) Solution structure of two mismatches A.A and T.T in the K-ras gene context by nuclear magnetic resonance and molecular dynamics. Eur. J. Biochem., 228, 279–290. [PubMed] [Google Scholar]
- 36.Xuan J.-C. and Weber,I.T. (1992) Crystal structure of a B-DNA dodecamer containing inosine, d(CGCIAATTCGCG), at 2.4 Å resolution and its comparison with other B-DNA dodecamers. Nucleic Acids Res., 20, 5457–5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kumar K.D., Harrison,R.W., Andrews,L.C. and Weber,I.T. (1992) Crystal structure at 1.5-Å resolution of d(CGCICICG), an octanucleotide containing inosine, and its comparison with d(CGCG) and d(CGCGCG) structures. Biochemistry, 31, 1541–1550. [DOI] [PubMed] [Google Scholar]
- 38.Arnott S., Chandrasekaran,R., Millane,R.P. and Park,H.-S. (1986) DNA–RNA hybrid secondary structures. J. Mol. Biol., 188, 631–640. [DOI] [PubMed] [Google Scholar]
- 39.Arnott S., Chandrasekaran,R., Hall,I.H. and Puigjaner,L.C. (1983) Heteronomous DNA. Nucleic Acids Res., 11, 4141–4155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gyi J.I., Lane,A.N., Conn,G.L. and Brown,T. (1998) The orientation and dynamics of the C2′-OH and hydration of RNA and DNA.RNA hybrids. Nucleic Acids Res., 26, 3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yoshimura F.K., Diem,K., Learn,G.H.,Jr, Riddell,S. and Corey,L. (1996) Intrapatient sequence variation of the gag gene of human immunodeficiency virus type 1 plasma virions. J. Virol., 70, 8879–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balfe P., Simmonds,P., Ludlam,C.A., Bishop,J.O. and Leigh Brown,A.J. (1990) Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J. Virol., 64, 6221–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morse D.P. and Bass,B.L. (1997) Detection of inosine in messenger RNA by inosine-specific cleavage. Biochemistry, 36, 8429–8434. [DOI] [PubMed] [Google Scholar]