Abstract
Bacterial survival requires adaptation to different environmental perturbations such as exposure to antibiotics, changes in temperature or oxygen levels, DNA damage, and alternative nutrient sources. During adaptation, bacteria often develop beneficial mutations that confer increased fitness in the new environment. Adaptation to the loss of a major non-essential gene product that cripples growth, however, has not been studied at the whole-genome level. We investigated the ability of Escherichia coli K-12 MG1655 to overcome the loss of phosphoglucose isomerase (pgi) by adaptively evolving ten replicates of E. coli lacking pgi for 50 days in glucose M9 minimal medium and by characterizing endpoint clones through whole-genome re-sequencing and phenotype profiling. We found that 1) the growth rates for all ten endpoint clones increased approximately 3-fold over the 50-day period; 2) two to five mutations arose during adaptation, most frequently in the NADH/NADPH transhydrogenases udhA and pntAB and in the stress-associated sigma factor rpoS; and 3) despite similar growth rates, at least three distinct endpoint phenotypes developed as defined by different rates of acetate and formate secretion. These results demonstrate that E. coli can adapt to the loss of a major metabolic gene product with only a handful of mutations and that adaptation can result in multiple, alternative phenotypes.
Author Summary
Bacteria must constantly adapt to many different environmental challenges, but how do they adapt when they lose a key gene product? We addressed this question using Escherichia coli lacking pgi, a major metabolic gene involved in sugar utilization, by serially passing replicates lacking the pgi gene for 50 days and resequencing the entire genome from endpoint clones isolated from nine of the replicates. We repeatedly found mutations in rpoS, a gene mostly active during stationary phase but one that regulates the expression of about fifty genes during exponential phase growth, and udhA and pntAB, three genes involved in maintaining redox balance in the cell. We also found multiple distinct endpoint phenotypes; the replicates could be stratified into three different groups after adaptation based on their rates of acetate and formate secretion. These results support the view that the metabolic network in E. coli is robust and can adjust to loss of a major metabolic gene in alternative ways.
Introduction
Recent advances in DNA sequencing technology enable bacterial genomes to be fully sequenced with a resolution high enough to find all differences relative to a reference sequence. These developments make possible the study, at the whole-genome level, of the genetic basis through which bacteria adapt to different perturbations. For example, E. coli adaptively evolved to achieve optimal growth on glycerol were found to have two to three mutations when endpoint clones were re-sequenced and compared to the parental wild-type strain [1]. Allelic replacement introducing the discovered mutations into the parental strain was then used to show the phenotypic causality of each mutation [1]. In another example, a long-term adaptive evolution study of E. coli revealed that genomic evolution did not decrease over time as expected. Instead, genomic evolution remained nearly constant over 20,000 generations and nearly all of the mutations that appeared were beneficial [2], [3]. Other adaptive evolution and resequencing-based studies have examined at the genome level how E. coli adapts to growth on other carbon sources besides glycerol [4]–[6], how Myxococcus xanthus transitions from cooperative behavior to cheating and back [7], and how different pathogens develop antibiotic resistance [8]–[11].
Another topic that has received intense investigation is that of compensatory mutations, especially with regard to antibiotic resistance [12]. Bacteria that develop antibiotic resistance through mutation of the target enzyme or through horizontal gene transfer (HGT) are often less fit than their drug-sensitive counterparts, but wild-type fitness levels can sometimes be restored if they acquire additional mutations that compensate for the lower fitness. Crucially, the former mechanism does not alter the structure of different networks within a bacterium since the same suite of genes remains. In contrast, the latter does change network structure: the organism gains new genes that must be assimilated into networks controlling transcriptional regulation, metabolism, and transcription/translation.
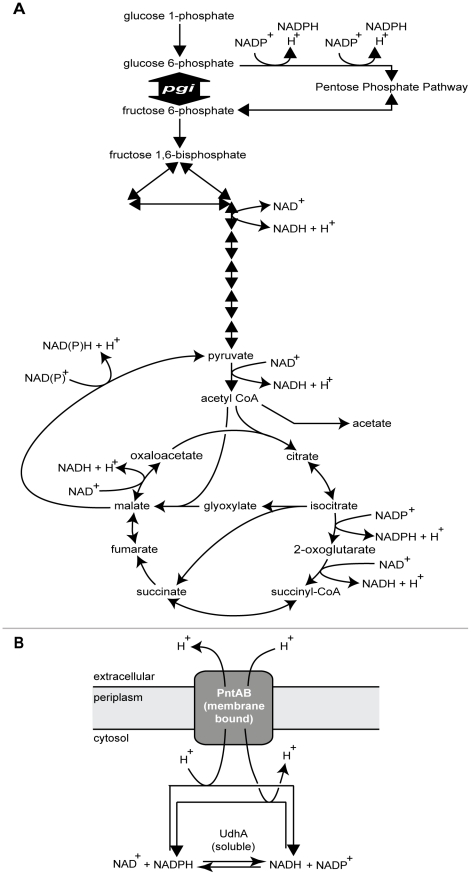
Here we address an important related question: how does a bacterium adjust to the complete loss of a major gene product, not just to mutations in the gene? The answer to this question would provide insight into the plasticity of both bacterial genomes and the networks that emerge from the proteins they encode. We selected the gene pgi for study as it plays a major role in central metabolism by transcribing the enzyme that catalyzes the second step in glycolysis (Figure 1A). In E. coli, loss of pgi significantly alters the structure of its metabolic network by disabling the use of upper glycolysis, a situation that cripples growth (growth rate <20% of wild-type levels in glucose minimal media [13]) but does not kill the organism. E. coli Δpgi mutants remain viable because glycolytic flux is rerouted through the pentose phosphate pathway (PPP) [14]–[15]; however, this introduces a redox imbalance problem since excess NADPH is produced which, in turn, perturbs a significant portion of the metabolic network [16]. Thus, pgi represents a good candidate gene to study mechanisms of compensation to gene loss.
Figure 1. Role of pgi, udhA, and pntAB in cellular metabolism.
A. The gene pgi catalyzes the isomerization of glucose 6-phosphate to fructose 6-phosphate in upper glycolysis. Removal of this gene forces glycolytic flux through the pentose phosphate pathway, creating a redox imbalance due to excess NADPH production. B. The genes udhA and pntAB catalyze the interconversion of NAD/NADH and NADP/NADPH. UdhA is a soluble protein whereas PntAB is membrane-bound.
To study how E. coli might overcome limitations imposed by the loss of pgi and consequent flux imbalances, we serially passed ten E. coli Δpgi replicates for 50 days in glucose minimal media and characterized all evolved, endpoint clones at the phenotypic level through measurements of growth rates and rates of substrate uptake and secretion. We also assessed genotypic changes in nine of the strains through whole-genome resequencing using Nimblegen tiling arrays (all nine strains) and Illumina technology (three strains) to identify possible adaptive mutations that arose during evolution.
Results
The growth rates and glucose uptake rates of 50-day evolved Δpgi mutants increased 3.6- and 2.6-fold on average, respectively, after adaptive evolution
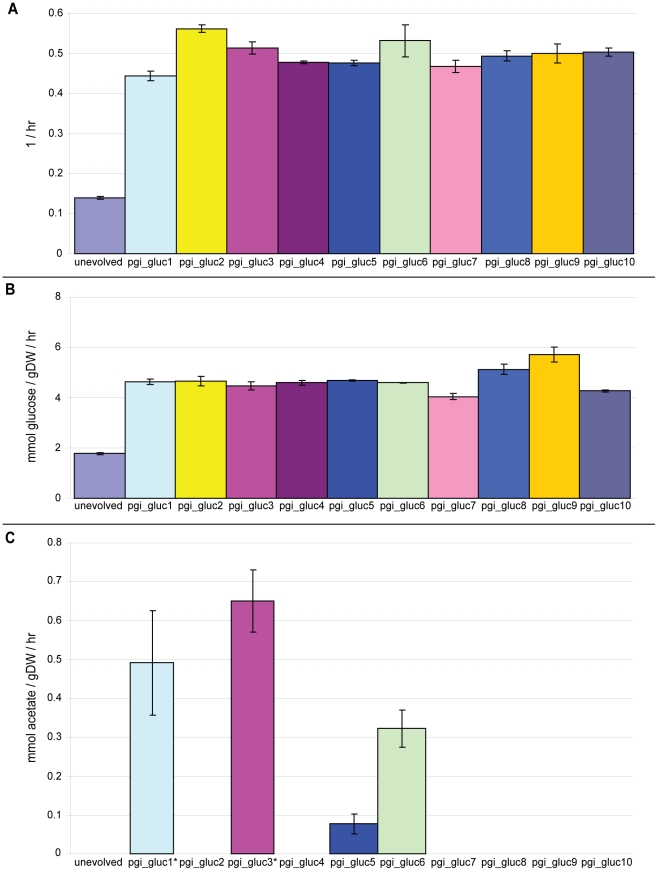
All ten replicates converged to similar endpoint phenotypes after fifty days of serial passage and adaptive evolution in glucose M9 minimal media when assessed for changes in growth rates (Figure 2A) and glucose uptake rates (Figure 2B). These ten strains, as well as all other strains used in this study, are summarized in Table 1. On average, the growth rates for the ten strains exhibited a 3.6-fold increase over the starting unevolved Δpgi strain to a final value of 0.50±0.03 hr−1. That growth rates of parallel replicates converge during adaptive evolution has been observed previously and thus seems to be a reproducible phenotypic outcome of such studies [17]. The glucose uptake rates for the evolved strains exhibited more variability; it increased 2.6-fold to a final average value of 4.68±0.46 mmol/gram dry weight/hour.
Figure 2. Growth rates, glucose uptake rates, and acetate secretion rates for unevolved and all evolved strains.
A. Growth rates for the starting unevolved Δpgi strain and all ten evolved strains after adaptive evolution. The growth rate of unevolved and 50-day evolved wild-type E. coli K12 MG1655 in the same medium was 0.69(0.0069 and 0.79(0.0092 hr-1. B. Glucose uptake rates for the unevolved (pgi strain and the ten evolved strains after adaptive evolution. Unevolved and 50-day evolved wild-type E. coli had a glucose uptake rate of 8.43(0.72 and 11.6(0.41 mmol/gDW/hour, respectively. C. Acetate secretion rates for the unevolved (pgi strain and the ten evolved strains after adaptive evolution. Unevolved and 50-day evolved wild-type E. coli had an acetate secretion rate of 12.8(8.87 and 5.61(0.24 mmol/gDW/hour, respectively. The symbol * indicates strains that also secrete formate (pgi_gluc1: 0.49(0.13 mmol/gDW/hour; pgi_gluc3: 0.22(0.02 mmol/gDW/hour). All error bars represent the standard deviation from three biological replicates. Abbreviations – gDW: gram dry weight; hr: hour.
Table 1. Strains used in this study.
| Strain | Characteristics | Source |
| Δpgi | Starting strain for adaptive evolutions | Fong et al. [45] |
| pgi_gluc1 | 50-day evolved Δpgi strain. Secretes acetate and formate | this study |
| pgi_gluc2 | 50-day evolved Δpgi strain. | this study |
| pgi_gluc3 | 50-day evolved Δpgi strain. Secretes acetate and formate | this study |
| pgi_gluc4 | 50-day evolved Δpgi strain. | this study |
| pgi_gluc5 | 50-day evolved Δpgi strain. Secretes acetate | this study |
| pgi_gluc6 | 50-day evolved Δpgi strain. Secretes acetate | this study |
| pgi_gluc7 | 50-day evolved Δpgi strain. | this study |
| pgi_gluc8 | 50-day evolved Δpgi strain. | this study |
| pgi_gluc9 | 50-day evolved Δpgi strain. | this study |
| pgi_gluc10 | 50-day evolved Δpgi strain. | this study |
| KI2_rpoS | Δpgi with in-frame duplication in rpoS at 595–603 | this study |
| KI4_rpoS | Δpgi with 1 base pair deletion in rpoS at position 841 | this study |
| KI5_rpoS | Δpgi with 1 base pair deletion in rpoS at position 850 | this study |
| KI6_rpoS | Δpgi with out-of-frame duplication in rpoS at 597–603 | this study |
| KI7_rpoS | Δpgi with c829t mutation in rpoS | this study |
| KI2_udhA | Δpgi with udhA g(-64)a mutation | this study |
| KI2_pntA | Δpgi with pntA c197a mutation | this study |
| KI2_RU | Δpgi double knock-in strain with rpoS 595–603 duplication and udhA g(-64)a SNP | this study |
| KI2_RP | Δpgi double knock-in strain with rpoS 595–603 duplication and pntA c197a SNP | this study |
| KI2_UP | Δpgi double knock-in strain with udhA g(-64)a and pntA c197a mutations | this study |
| KI2_3KI | Δpgi triple knock-in strain with rpoS 595–603 duplication and udhA g(-64)a and pntA c197a SNPs | this study |
Knock-in strains that were constructed by introducing mutations identified during adaptive evolution back into the starting unevolved Δpgi strain are designated “KI.” This designation is followed next by a number that corresponds to the evolved replicate on which the knock-in strain is based. The identity of the gene(s) containing the mutation follows last. Abbreviations – RU: rpoS + udhA; RP: rpoS + pntA; UP: udhA + pntA; 3KI: triple knock-in.
Three replicates of wild-type E. coli were also serially passed in glucose M9 for fifty days and assessed for changes in growth rate. The average initial growth rate was 0.69±0.0069 hr−1. The average growth rate after serial passage was 0.79±0.0092 hr−1, which constitutes a 1.1-fold increase. A separate study in which wild-type E. coli was evolved on glucose minimal media over 44 days reported slightly lower initial and final growth rates but a similar value for the fold increase over the evolutionary period [13].
Adaptive evolution produced multiple, different endpoint clones as defined by their rates of metabolic by-product secretion
All ten strains evolved toward similar endpoint growth rates and glucose uptake rates but, interestingly, different metabolic functional states. Specifically, the ten strains could be stratified into three distinct groups based on their rates of acetate and formate secretion: those that secrete both acetate and formate, those that secrete acetate only, and those that secrete neither acetate nor formate (Figure 2C). Those that do secrete acetate do so at a rate about one-tenth lower than the acetate secretion rate for both unevolved wild-type E. coli (12.8±8.87 mmol acetate/gram dry weight/hour) and E. coli strains adapted to grow in glucose M9 for 50 days (5.61±0.24 mmol acetate/gram dry weight/hour) (Figure 2C). The parental Δpgi clone did not secrete acetate.
Two to five mutations were detected during the course of adaptation in the nine evolved Δpgi strains that were sequenced
The mutations detected after the 50-day adaptive evolution period for the nine sequenced strains are summarized in Table 2. Mutations in rpoS were most common, appearing in six of nine endpoints. The rpoS mutation in pgi_gluc7 encodes a stop codon at that position; the rpoS mutations in pgi_gluc4, pgi_gluc5 and pgi_gluc6 likely result in truncated forms of the protein; the rpoS mutation in pgi_gluc3 is a SNP that results in a G279V change in the protein; and the rpoS mutation in pgi_gluc2 is an in-frame nine base pair duplication.
Table 2. Mutations detected in clones isolated from nine of the ten evolved Δpgi strains isolated after 50 days of adaptive evolution.
| pgi_gluc1 | pgi_gluc2 | pgi_gluc3 | pgi_gluc4 | pgi_gluc5 | pgi_gluc6 | pgi_gluc7 | pgi_gluc8 | pgi_gluc10 | |
| rpoA | c854t | Δ944..963 | t805c | ||||||
| rpoB | a3724c | ||||||||
| rpoC | c3520a | ||||||||
| rpoS | Dupl. (595–603) | g836t | Δ1bp (841) | Δ1bp (850) | Dupl. (597–603) | c829t | |||
| udhA | a949g | g(−64)a | g(−64)t | g(−64)a | g(−64)a; g58a | ||||
| pntA | c197a | Δ680..690 | |||||||
| pntB | Δ1bp (430) | Dupl. (698–703) | |||||||
| rep | t242g | ||||||||
| fabZ | c167t | ||||||||
| cpxR | g614a | Δ4bp, 585..588 | |||||||
| yfeH | Δ1bp (297) | ||||||||
| fruK | c886t | ||||||||
| rodA | c263a | ||||||||
| cyaA | a1175c | ||||||||
| bipA | 1bp Dupl. (960) | ||||||||
| ispU | t659g | ||||||||
| Large indel | e14 prophage deletion |
All nine were sequenced using Nimblegen tiling arrays. Strains pgi_gluc1, pgi_gluc7, and pgi_gluc10 were also sequenced using first-generation Solexa (Illumina) technology. Strain pgi_gluc9 was not sequenced. Sanger sequencing was used to validate all reported mutations. Additionally, for all nine strains we sequenced the rpoS, udhA, pntA and pntB genes in their entirety using Sanger sequencing, not just specific regions indicated by Nimblegen or Solexa technologies to contain a mutation. The full length of these four genes was similarly sequenced in the three evolved wild-type (rpoS +) replicates; no mutations were found at any position. No other genomic positions were sequenced in the evolved wild-type replicates. Abbreviations – bp: base pair; Dupl: duplication.
Mutations in the soluble transhydrogenase udhA [18] (six mutations in five strains) and membrane-bound transhydrogenase subunits pntA and pntB (two mutations each) were also common (Table 2). Loss of pgi directly perturbs both since they catalyze the oxidation and reduction of NAD(P)/NAD(P)H (Figure 1B). Interestingly, the same position, –64 base pairs (bp) upstream from the annotated udhA transcription start site, was mutated in four of the five strains with udhA mutations. This mutation was the only one that developed outside of a coding region. On the other hand, three of the four pntAB mutations likely result in truncated, nonfunctional proteins: the mutation in pgi_gluc2 pntA is a nonsense mutation while the pgi_gluc7 pntA and pgi_gluc4 pntB mutations truncate the proteins from 510 to 265 and 462 to 154 amino acids, respectively. The pgi_gluc10 pntB mutation extends the protein length by two amino acids: two additional alanines are added to a region where four alanines are already present, resulting in six consecutive alanine residues.
Whereas the evolved Δpgi replicates frequently developed mutations in rpoS, udhA, and pntAB, no mutations could be detected in these four genes in the three wild-type (pgi +) E. coli replicates evolved under the same growth conditions. This finding suggests that the mutations in rpoS, udhA and pntAB stemmed from adaptation to loss of pgi rather than other possible selection pressures such as the growth medium, a phenomenon that occurs when wild-type E. coli is cultured in minimal media containing glycerol or lactate as the sole carbon source [1], [4].
Another noteworthy observation is the frequency with which genes involved in global regulation were mutated – at least one in all nine strains. Besides rpoS, other genes that modulate transcription of multiple loci and that developed mutations included rpoA, rpoB, rpoC, cyaA and cpxR (Table 2). This result implies that adaptation required global, network-level changes to transcriptional regulation and metabolism, which is emerging as a general, recurring theme across multiple organisms. For example, E. coli strains adapted to grow on glycerol as the sole carbon source often develop causative mutations in RNAP [1]; a mutation in the transcription factor Spt15p confers greater ethanol tolerance in yeast [19]; and a mutation in the sensor kinase of a two-component signal transduction system enhanced transition to invasive infection and virulence in a mouse model of Group A Streptococcus pathogenesis [8].
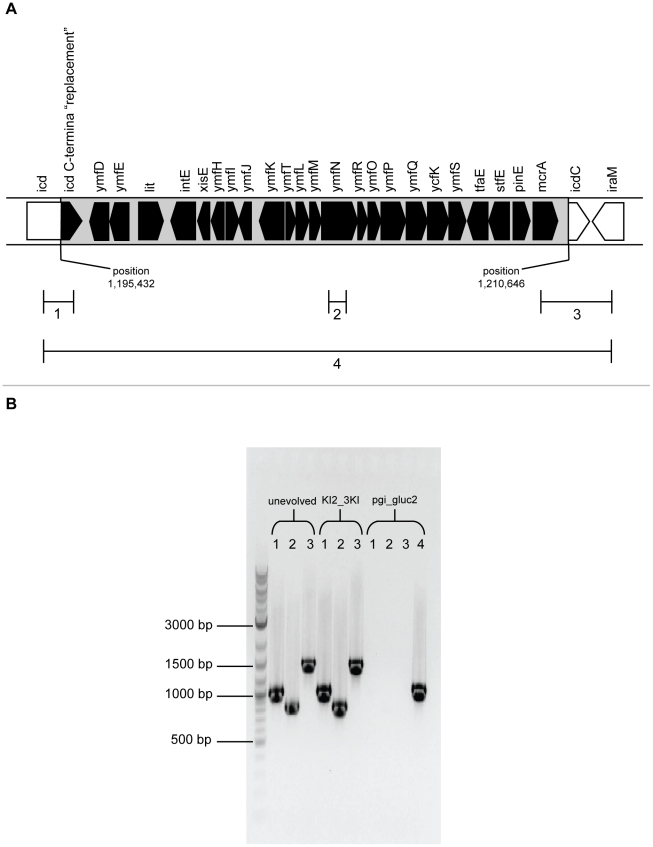
Lastly, we detected one large indel: loss of the 15.4 kbp e14 prophage in pgi_gluc2. Its deletion was initially suggested through both analysis of the resequencing data and an optical map for this strain. It was subsequently confirmed by PCR analysis of the ends of the prophage and flanking regions (Figure 3). We could not detect large indels in the resequencing data for any other strain; however, we cannot rule out the possibility that they might still be present, especially transposition of mobile elements such as insertion sequences (IS elements), because of limitations inherent in the two short-read resequencing technologies employed here. IS elements in particular have been shown to translocate frequently in E. coli when it undergoes adaptive evolution under a wide variety of conditions [20]–[23]. If the raw sequences do not assemble into contigs large enough to sufficiently span the mobile element and flanking regions, they can be difficult to map accurately to possible new locations in the genome.
Figure 3. Loss of e14 prophage in pgi_gluc2.
A. Structure and location of the e14 prophage. It is integrated within the icd gene in the E. coli chromosome at position 1,195,432 to position 1,210,646. B. PCR analysis of the unevolved (pgi, pgi_gluc2 triple knock-in (KI2_3KI) and evolved pgi_gluc2 strains confirms loss of the e14 prophage in pgi_gluc2. Numbers 1 through 4 correspond to PCR amplification regions as indicated in the top panel. Regions 1 and 3 both span terminal segments of the integrated prophage and adjacent chromosomal DNA. Region 2 spans a segment wholly within the prophage. Region 4 spans the integration site and is amplified using the left primer from Region 1 and the right primer from Region 3.
Most mutations in rpoS likely result in nonfunctional proteins and none confer increased growth rate when present alone
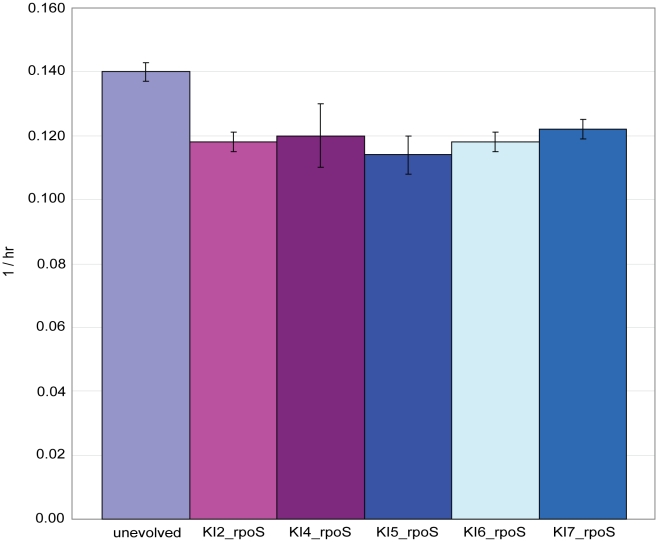
The high frequency of rpoS mutations prompted us to investigate whether this set of mutations had any impact on the growth rate increases seen in the evolved Δpgi strains. When five of the six rpoS mutations were introduced into the chromosome of the starting unevolved Δpgi clone, none of the five knock-in strains displayed an increase in growth rate (Figure 4). Surprisingly, all had growth rates slightly lower than that of the unevolved clone, indicating that they are neutral to slightly deleterious (Student's t-test, P<0.01 for all five). We then investigated indirectly whether the rpoS mutants still encoded functional proteins by flooding single colonies of each of the five knock-in strains with hydrogen peroxide. Vigorous bubbling occurs if rpoS is functional due to RpoS control of katE expression [24]. For comparison, we also performed this assay on the ten evolved strains, the starting unevolved Δpgi strain and a ΔrpoS mutant obtained from the Keio collection [25]. The five knock-in strains, the six evolved strains harboring rpoS mutations and, interestingly, one of the evolved strains that did not have an rpoS mutation (pgi_gluc1) all exhibited reduced bubbling upon contact with hydrogen peroxide (Table S1). The pgi_gluc1 result indicates that other genes besides rpoS can control katE expression. As expected, the ΔrpoS mutant also exhibited reduced bubbling. In contrast, bubbling remained vigorous for two of the three evolved strains (pgi_gluc8 and pgi_gluc10) that did not harbor an rpoS mutation. It therefore appears that the majority of rpoS mutations result in non-functional proteins and that they do not have a significant impact on growth rates when present alone.
Figure 4. Growth rates for five rpoS knock-in strains.
The strains were constructed by introducing five of the six rpoS mutations detected after adaptive evolution back into the starting unevolved Δpgi strain through site-directed mutagenesis. A similar knock-in strain containing the pgi_gluc3 rpoS mutation was not constructed. Growth rate data for the starting unevolved Δpgi strain is also shown for comparison. Error bars represent the standard deviation from three biological replicates. A full description for the strain abbreviations can be found in Table 1. Abbreviations – hr: hour.
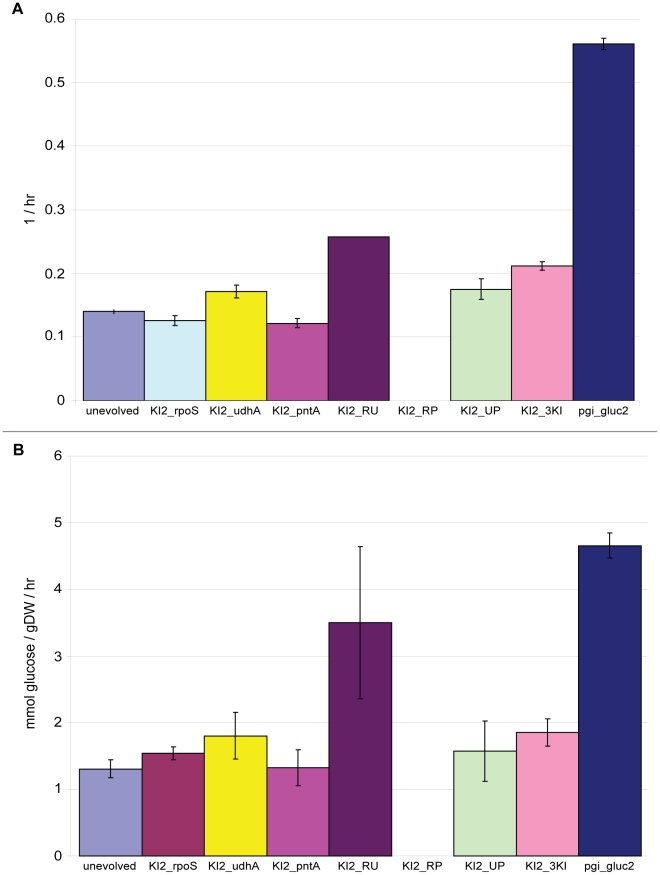
The three mutations in pgi_gluc2 display both positive and negative epistatic interactions
We next investigated how the rpoS, udhA and pntA mutations in pgi_gluc2 influence its growth rate by constructing all three single knock-ins, all three double knock-ins, and the triple knock-in from the starting unevolved Δpgi strain. Like rpoS, the pntA single knock-in strain grew slightly more slowly than the unevolved Δpgi strain (Figure 5). This slight growth rate reduction is perhaps expected since the c197a mutation in pntA changes the codon triplet at that position from a serine to a stop codon, truncating the protein from 510 to 65 amino acids and likely rendering it non-functional. The udhA mutation, on the other hand, does impact the growth rate; the udhA single knock-in strain grew at a rate 1.4 times faster than the rpoS and pntA single knock-in strains (P<0.001).
Figure 5. Growth rates and glucose uptake rates for pgi_gluc2 single, double, and triple knock-in strains.
A. Growth rates for the three single, three double and triple knock-in strains constructed based on the three mutations that appeared in pgi_gluc2. The growth rates for the starting unevolved Δpgi strain and the evolved pgi_gluc2 strain are also shown for comparison. B. Corresponding glucose uptake rates for the seven knock-in strains. Data for the unevolved and evolved strains are again shown for comparison. Error bars for both represent the standard deviation from three biological replicates. A full description for the strain abbreviations can be found in Table 1. Abbreviations – gDW: gram dry weight; hr: hour.
Construction of the three double knock-in strains revealed both positive and negative epistasis among the three mutations. There is positive epistasis between the rpoS and udhA mutations. Since the rpoS single knock-in strain has a growth rate very similar to that of the unevolved Δpgi clone, one would expect the growth rate of the rpoS + udhA double knock-in strain to closely mimic that of the single udhA knock-in strain if the two mutations were independent. Instead, the double knock-in had a growth rate 1.8 and 1.5 times greater than that of the unevolved and the single udhA knock-in strains, respectively (Figure 5). We conclude from these data that, in addition to udhA, the rpoS mutation is causal. In contrast, there is negative epistasis between the rpoS and pntA mutations. This double knock-in strain grew well overnight as a pre-culture in LB medium, but failed to grow after washing twice and transferring to glucose M9 media, even after five days incubation. Lastly, the udhA and pntA mutations do not show any apparent epistasis since the growth rate for this double knock-in strain was essentially identical to that of the udhA single knock-in strain.
The growth rate for the triple knock-in strain was only 0.21 hr−1, less than half that of the evolved pgi_gluc2 strain (Figure 5). This finding implies that deletion of the e14 prophage likely contributes significantly to adaptation to loss of pgi in this genetic background.
Discussion
We have investigated how E. coli overcomes the loss of pgi, a challenge that forces glycolytic flux through the pentose phosphate pathway and creates a redox imbalance in the cell. The data presented here indicate that adapted Δpgi mutants accomplish this task through mutations in key genes, in particular rpoS and the transhydrogenases udhA and pntAB, that suppress the bacterial stress response and likely ameliorate the redox imbalance, respectively. Multiple alternative phenotypes arise from these mutations as defined by their rates of acetate and formate secretion.
One explanation for the high frequency of rpoS mutations is subordination of the rpoS-controlled stress response in favor of rpoD-controlled maximization of nutrient uptake and utilization. The rpoS gene encodes a sigma factor most active during stationary phase but which also affects the expression of about fifty genes during log phase [26]. It controls general stress response in E. coli and related bacteria [27]–[28] but does so at the considerable expense of reduced expression of rpoD-controlled housekeeping genes [29]. Perhaps most importantly, rpoD also controls expression of genes involved in nutrient scavenging in nutrient-limited environments [29]. This trade-off, designated stress protection and nutritional competence (SPANC) [30]–[31], consequently creates a conflict between the hunger and stress responses in E. coli since both sigma factors cannot simultaneously bind RNAP. Since the growth media for the ten Δpgi and three wild-type E. coli evolutions all contained the same initial glucose concentration (2 g/L), the greatly reduced growth rate of the unevolved Δpgi clone compared to wild-type E. coli demonstrates clearly that the former does not convert nutrients into biomass optimally. This growth rate reduction, which imposes a stress, is likely sufficient to induce high levels of RpoS in the unevolved Δpgi strain and shift the SPANC balance from metabolism to stress. The rpoS mutations that emerged in six of the evolved strains, all of which reduce functionality of the protein as indicated by the peroxidase assay (Table S1), would shift the balance from stress back to metabolism in these six to allow for greater rpoD-controlled nutrient acquisition and consequent faster growth.
There are several other notable mutations besides those in rpoS. As mentioned, the –64 position upstream of the udhA transcription start site was mutated in four out of five strains. Because udhA plays a major role in oxidizing NADPH with NAD during conditions of excess NADPH and its overexpression increases the growth rate of unevolved Δpgi mutants [32], we speculate that this mutation upregulates udhA expression. In contrast, three of the four pntAB mutations likely reduce or abolish PntAB function since they produce peptides that are severely truncated. Decreasing PntAB function prevents the redox imbalance condition caused by loss of pgi from worsening: 13C-flux measurements show that PntAB produces 35–45% of the NADPH in E. coli during standard batch growth on glucose [32]. The mutations in udhA and pntAB detected here thus support previous findings [32] that the two transhydrogenases have divergent functions despite their shared ability to catalyze the interconversion of NAD/NADH and NADP/NADPH reversibly. Whereas UdhA plays a major role in oxidizing NADPH with NAD, the membrane-bound PntAB plays a major role in reoxidizing NADH with NADP. The pgi_gluc1 fabZ mutation (missense; P167L) is noteworthy because fabZ is involved in fatty acid biosynthesis, a process that utilizes NADPH. This raises the possibility that NADPH overabundance in Δpgi mutants drives excessive fatty acids biosynthesis, translating the redox imbalance into an imbalance in fatty acid production. The fabZ mutation in pgi_gluc1 might help to alleviate this imbalance by decreasing flux through this pathway.
The observation that the pgi_gluc2 triple knock-in strain could not reproduce the same growth rate as the evolved strain implies that loss of the e14 prophage plays a role in adaptation. This prophage lies at position 1,195,432 bp to 1,210,646 bp [33] on the chromosome and contains 24 putative ORFs, many of which have unknown function [34]. Exposure to UV radiation consistently activates excision of this element [35]–[36], probably through induction of the SOS response [37]. This observation implies that the SOS response was probably induced at some point during adaptation to loss of pgi in pgi_gluc2. Deletion of e14 might contribute to adaptation to loss of pgi in several ways. First, e14 contains several genes whose presence/absence likely affects metabolism, for example a toxin encoded by kil that kills the cell in the absence of its repressor, which is also encoded by e14 [34], [38]. Second, deletion of e14 introduces ten synonymous and two non-synonymous mutations into the amino acid sequence of isocitrate dehydrogenase (icd), a key metabolic enzyme [39]. Although none of these mutations alters the specific activity of the protein in cell-free experiments [39], they could exert an effect in vivo through their potential impact on translation efficiency.
Acetate secretion rates are zero to approximately ten times lower in the ten evolved Δpgi replicates compared to unadapted and glucose-adapted wild-type E. coli (Figure 2C), which implicates a connection between acetate metabolism and loss of pgi. We speculate that this connection stems from a need to maximize flux through the TCA cycle in E. coli Δpgi mutants. It is well known that wild-type E. coli secretes acetate during aerobic growth on glucose [40]–[41], a phenomenon known as overflow metabolism [42], when acetyl-CoA converts to acetate rather than enter the TCA cycle. On the other hand, the loss of pgi forces flux normally occurring in upper glycolysis through the pentose phosphate pathway, reducing flux through lower glycolysis [32] and presumably reducing the amount of acetyl-CoA that enters the TCA cycle relative to that in wild-type E. coli. Minimizing by-product secretion due to overflow metabolism would therefore serve to maximize the reduced amount of flux flowing through lower glycolysis that enters the TCA cycle. The regulation of acetate metabolism, however, is a complex process [43]–[44], and other factors could be involved.
Besides contrasting acetate secretion profiles, we hypothesize that evolved Δpgi strains utilize the glyoxylate shunt whereas their evolved wild-type (pgi +) counterparts do not since this shunt is already active in unevolved E. coli lacking pgi [32]. Routing flux partly through the glyoxylate shunt rather than the full TCA cycle would avoid exacerbating the redox imbalance problem because NAD(P)H biosynthesis normally arising from the conversion of isocitrate to α-ketoglutarate (NADPH) and α-ketoglutarate to succinyl-CoA (NADH) would not occur.
In conclusion, we have examined the robustness of the E. coli metabolic network and the genetic basis for adaptation to the loss of pgi through adaptive evolution, resequencing and phenotypic assays. Two to five mutations were detected after adaptation and were most frequently located in the alternative sigma factor rpoS, the soluble transhydrogenase udhA, and the membrane-bound transhydrogenase pntAB. The genetic and biochemical data collected here paints a picture in which one general mechanism through which E. coli adapts to loss of pgi and manages the redox imbalance problem occurs via 1) favoring rpoD-controlled hunger response over rpoS-controlled stress response, and 2) curtailing (and possibly eliminating) PntAB function to lessen PntAB-catalyzed NADPH biosynthesis. We also found evidence showing that adaptive evolution can lead to multiple, alternative phenotypes within the evolutionary landscape, defined in this study as a difference in byproduct secretion rates. Looking forward, adaptation to loss of pgi, and perhaps to loss of other metabolic genes, might constitute a model system to investigate the link between genotype and phenotype through creation of additional knock-in mutants and examining the effect of each mutation on metabolic flux. Such studies would further highlight the plasticity of the bacterial genome and cellular networks as well as delineate mechanisms through which bacteria adapt to genetic perturbations.
Methods
Strains
The strains used in this study are summarized in Table 1. The starting strain for all adaptively evolved replicates was an E. coli Δpgi strain constructed from wild-type E. coli K12 MG1655 (ATCC, Manassas, VA) as described previously [45]. Whole-genome resequencing identified three base pair changes in the MG1655 Δpgi strain utilized here that were not present in the sequenced MG1655 strain: a c→g mutation in ybeB (genomic position 667965), an a→g mutation in ylbE_1 (genomic position 547694) and a c→a mutation in nupC (genomic position 2511373). Sanger sequencing was used to confirm these three mutations. Strains pgi_gluc1 through pgi_gluc10 were all generated during this study through adaptive evolution of the starting E. coli Δpgi strain. Knock-in strains in which mutations detected after adaptive evolution were introduced into the starting unevolved Δpgi strain were created using the method of Tischer et al. [46] with the following modifications: 1) pKD46, a temperature-sensitive plasmid that carries bacteriophage λ red genes (γ, β, and exo) under the control of the arabinose-inducible ParaBAD promoter [47], was first electroporated into cells to be transformed and subsequently used for the first Red recombination step, 2) pKD13 [47] was used to amplify the kanamycin resistance gene instead of pACYC177, and 3) pACBSR, which contains a chloramphenicol-resistance selection marker and encodes I-SceI and the λ red system both under the control of an arabinose-dependent promoter [48], was used for the second Red recombination step. The primers used to construct the knock-in strains can be found in Table S2.
Growth media
The starting Δpgi clone was maintained and propagated in Luria-Bertrani (LB) broth (EMD Chemicals, Gibbstown, NJ). It was not exposed to glucose M9 minimal medium prior to this study. The composition of the glucose M9 was: dextrose (2 g/L), CaCl2 (100 µM), MgSO4 (200 mM), Na2HPO4 (13.6 g/L), KH2PO4 (6 g/L), NaCl (1 g/L), NH4Cl (2 g/L) and trace elements (500 µL). The trace element solution consisted of (per liter): FeCl3•6H2O (16.67 g), ZnSO4•7H2O (0.18 g), CuCl2•2H2O (0.12 g), MnSO4•H2O (0.12 g), CoCl2•6H2O (0.18 g) and Na2EDTA•2H2O (22.25 g).
Adaptive evolution protocol
The adaptive evolution protocol used here has been described previously [45]. To summarize, all ten Δpgi replicates and the three wild-type replicates were serially passed daily in 250 mL glucose M9 minimal medium in 500 mL Erlenmeyer flasks for 50 days at 37°C using magnetic stir bars for aeration. The volume transferred was adjusted each day to account for changes in growth rate in order to maintain cultures in prolonged exponential phase growth and thereby avoid entry into stationary phase. On Day 50, an aliquot of each replicate was streaked out on LB plates containing 20 g/L bacteriological agar (Sigma-Aldrich, St. Louis, MO) and incubated at 37°C for 24 hours. A well-isolated colony was then selected and suspended in LB broth containing 25% glycerol and stored long-term at −80°C.
Whole-genome resequencing
We isolated genomic DNA using a Qiagen DNeasy kit (QIAGEN, Valencia, CA) from the same single-colony sample collected for long-term storage. Whole-genome sequencing was then carried out using two platforms, Nimblegen hybridization-based tiling arrays [49] and Illumina technology. Nimblegen also provided data analysis capabilities [49] for identification of possible mutations, while Solexa results were analyzed using a software program developed in-house that counts the total number of each base for a given position and compares the consensus call to the reference E. coli genome. All reported mutations were confirmed by PCR amplification of the surrounding DNA region and Sanger sequencing. Additionally, we also used Sanger sequencing to sequence the full-length rpoS, udhA, pntA, and pntB genes for the nine resequenced, evolved Δpgi strains and the three evolved wild-type E. coli replicates, not just regions immediately surrounding mutations reported by Nimblegen and/or Illumina. For the evolved wild-type replicates, no other genomic positions were sequenced besides the full-length genes for these four. The list of primers used for both PCR amplification and Sanger sequencing is given in Table S3. The optical map for pgi_gluc2 was provided as a service by OpGen, Inc. (Gaithersburg, MD) using NcoI as the restriction enzyme.
Growth rate and substrate uptake/secretion rate measurements
Overnight pre-cultures of each strain grown in LB medium were spun down and washed twice with glucose M9 minimal medium, after which they were used to inoculate 500 mL Erlenmeyer flasks containing 250 mL glucose M9 in triplicate. These flasks were incubated overnight in an air incubator maintained at 37°C using magnetic stir bars for aeration, conditions which were identical to those used during the adaptive evolutions. The next day an inoculum of each replicate was transferred to a new 500 mL Erlenmeyer flask again containing 250 mL of glucose M9 such that the optical density at 600 nm (OD600) had been reduced to 0.005. These flasks were then placed in a water bath maintained at 37°C. Magnetic stir bars were again used for aeration.
Once the OD600 reached 0.05 and then continuing periodically thereafter, the optical density of each sample was recorded and culture media were collected and filtered through 0.22 µm membranes. HPLC analysis (Waters, Milford, MA) was then carried out on the filtered media using a Bio-Rad Aminex HPX-87H ion exclusion column (300 mm×7.8 mm) with 5 mM H2SO4 as the mobile phase at a flow rate of 0.5 mL/min and temperature of 45°C.
Supporting Information
Indirect assessment of RpoS activity utilizing the peroxidase assay [19]. Colonies with functional RpoS exhibit vigorous bubbling when they come into contact with hydrogen peroxide via a mechanism based on rpoS control of katE expression. We defined vigorous bubbling (v) as bubble formation occurring within five seconds after contact with hydrogen peroxide; medium bubbling (m) as bubble formation occurring between five and ten seconds after contact; and slight bubbling (s) as bubble formation occurring ten seconds after contact. This assay was performed 24 and 48 hours after colonies had been inoculated onto LB plates. Abbreviations: repl - replicate.
(0.06 MB DOC)
Primers used to introduce mutations detected after adaptive evolution back into the starting unevolved Δpgi strain according to the method of Tischer et al [46].
(0.04 MB DOC)
Primers used for Sanger sequencing to confirm mutations reported by Nimblegen and Illumina sequencing technologies. The rpoS, udhA, pntA, and pntB genes were sequenced in their entirety in all nine evolved strains using Sanger sequencing, not just regions immediately surrounding reported mutations. Primers were designed for this purpose by dividing the four genes into regions of approximately 800–900 base pairs with an overlap between regions of approximately 100 base pairs.
(0.06 MB DOC)
Acknowledgments
We thank Markus Herrgard and Byung-Kwok Cho for helpful discussions and Jaqueline Miller, Elisa Abate, Courtney Chew, and Howard Li for technical assistance.
Footnotes
The authors have declared that no competing interests exist.
We acknowledge funding from NIH R01GM062791. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 2.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 3.Barrick JE, Lenski RE. Genome-wide Mutational Diversity in an Evolving Population of Escherichia coli. Cold Spring Harb Symp Quant Biol. 2009. [DOI] [PMC free article] [PubMed]
- 4.Conrad TM, Joyce AR, Applebee MK, Barrett CL, Xie B, et al. Whole-genome resequencing of Escherichia coli K-12 MG1655 undergoing short-term laboratory evolution in lactate minimal media reveals flexible selection of adaptive mutations. Genome Biol. 2009;10:R118. doi: 10.1186/gb-2009-10-10-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee DH, Palsson BO. Adaptive Evolution Of Escherichia coli K-12 MG1655 On A Non-Native Carbon Source, L-1,2-Propanediol. Appl Environ Microbiol. [DOI] [PMC free article] [PubMed]
- 6.Maharjan R, Zhou Z, Ren Y, Li Y, Gaffe J, et al. Genomic identification of a novel mutation in hfq that provides multiple benefits in evolving glucose-limited populations of Escherichia coli. J Bacteriol. [DOI] [PMC free article] [PubMed]
- 7.Velicer GJ, Raddatz G, Keller H, Deiss S, Lanz C, et al. Comprehensive mutation identification in an evolved bacterial cooperator and its cheating ancestor. Proc Natl Acad Sci U S A. 2006;103:8107–8112. doi: 10.1073/pnas.0510740103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sumby P, Whitney AR, Graviss EA, DeLeo FR, Musser JM. Genome-wide analysis of group a streptococci reveals a mutation that modulates global phenotype and disease specificity. PLoS Pathog. 2006;2:e5. doi: 10.1371/journal.ppat.0020005. doi: 10.1371/journal.ppat.0020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mwangi MM, Wu SW, Zhou Y, Sieradzki K, de Lencastre H, et al. Tracking the in vivo evolution of multidrug resistance in Staphylococcus aureus by whole-genome sequencing. Proc Natl Acad Sci U S A. 2007;104:9451–9456. doi: 10.1073/pnas.0609839104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman L, Alder JD, Silverman JA. Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:2137–2145. doi: 10.1128/AAC.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, et al. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science. 2005;307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 12.Andersson DI, Hughes D. Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol. 2010;8:260–271. doi: 10.1038/nrmicro2319. [DOI] [PubMed] [Google Scholar]
- 13.Fong SS, Palsson BO. Metabolic gene-deletion strains of Escherichia coli evolve to computationally predicted growth phenotypes. Nat Genet. 2004;36:1056–1058. doi: 10.1038/ng1432. [DOI] [PubMed] [Google Scholar]
- 14.Hua Q, Yang C, Baba T, Mori H, Shimizu K. Responses of the central metabolism in Escherichia coli to phosphoglucose isomerase and glucose-6-phosphate dehydrogenase knockouts. J Bacteriol. 2003;185:7053–7067. doi: 10.1128/JB.185.24.7053-7067.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Canonaco F, Hess TA, Heri S, Wang T, Szyperski T, et al. Metabolic flux response to phosphoglucose isomerase knock-out in Escherichia coli and impact of overexpression of the soluble transhydrogenase UdhA. FEMS Microbiol Lett. 2001;204:247–252. doi: 10.1111/j.1574-6968.2001.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 16.Feist AM, Henry CS, Reed JL, Krummenacker M, Joyce AR, et al. A genome-scale metabolic reconstruction for Escherichia coli K-12 MG1655 that accounts for 1260 ORFs and thermodynamic information. Mol Syst Biol. 2007;3:121. doi: 10.1038/msb4100155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fong SS, Joyce AR, Palsson BO. Parallel adaptive evolution cultures of Escherichia coli lead to convergent growth phenotypes with different gene expression states. Genome Res. 2005;15:1365–1372. doi: 10.1101/gr.3832305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boonstra B, French CE, Wainwright I, Bruce NC. The udhA gene of Escherichia coli encodes a soluble pyridine nucleotide transhydrogenase. J Bacteriol. 1999;181:1030–1034. doi: 10.1128/jb.181.3.1030-1034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alper H, Moxley J, Nevoigt E, Fink GR, Stephanopoulos G. Engineering yeast transcription machinery for improved ethanol tolerance and production. Science. 2006;314:1565–1568. doi: 10.1126/science.1131969. [DOI] [PubMed] [Google Scholar]
- 20.Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156:477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stoebel DM, Hokamp K, Last MS, Dorman CJ. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 2009;5:e1000671. doi: 10.1371/journal.pgen.1000671. doi: 10.1371/journal.pgen.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naas T, Blot M, Fitch WM, Arber W. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics. 1994;136:721–730. doi: 10.1093/genetics/136.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riehle MM, Bennett AF, Long AD. Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci U S A. 2001;98:525–530. doi: 10.1073/pnas.021448998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zambrano MM, Siegele DA, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- 25.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2006. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong T, Schellhorn HE. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol Genet Genomics. 2009;281:19–33. doi: 10.1007/s00438-008-0389-3. [DOI] [PubMed] [Google Scholar]
- 27.Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the sigma(S) (RpoS) subunit of RNA polymerase. Microbiol Mol Biol Rev. 2002;66:373–395, table of contents. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hengge-Aronis R. Recent insights into the general stress response regulatory network in Escherichia coli. J Mol Microbiol Biotechnol. 2002;4:341–346. [PubMed] [Google Scholar]
- 29.Notley-McRobb L, King T, Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol. 2002;184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.King T, Ishihama A, Kori A, Ferenci T. A regulatory trade-off as a source of strain variation in the species Escherichia coli. J Bacteriol. 2004;186:5614–5620. doi: 10.1128/JB.186.17.5614-5620.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol. 2005;57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- 32.Sauer U, Canonaco F, Heri S, Perrenoud A, Fischer E. The soluble and membrane-bound transhydrogenases UdhA and PntAB have divergent functions in NADPH metabolism of Escherichia coli. J Biol Chem. 2004;279:6613–6619. doi: 10.1074/jbc.M311657200. [DOI] [PubMed] [Google Scholar]
- 33.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 34.Mehta P, Casjens S, Krishnaswamy S. Analysis of the lambdoid prophage element e14 in the E. coli K-12 genome. BMC Microbiol. 2004;4:4. doi: 10.1186/1471-2180-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aertsen A, Van Houdt R, Vanoirbeek K, Michiels CW. An SOS response induced by high pressure in Escherichia coli. J Bacteriol. 2004;186:6133–6141. doi: 10.1128/JB.186.18.6133-6141.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harris DR, Pollock SV, Wood EA, Goiffon RJ, Klingele AJ, et al. Directed evolution of ionizing radiation resistance in Escherichia coli. J Bacteriol. 2009;191:5240–5252. doi: 10.1128/JB.00502-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greener A, Hill CW. Identification of a novel genetic element in Escherichia coli K-12. J Bacteriol. 1980;144:312–321. doi: 10.1128/jb.144.1.312-321.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plasterk RH, van de Putte P. The invertible P-DNA segment in the chromosome of Escherichia coli. Embo J. 1985;4:237–242. doi: 10.1002/j.1460-2075.1985.tb02341.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hill CW, Gray JA, Brody H. Use of the isocitrate dehydrogenase structural gene for attachment of e14 in Escherichia coli K-12. J Bacteriol. 1989;171:4083–4084. doi: 10.1128/jb.171.7.4083-4084.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andersen KB, von Meyenburg K. Are growth rates of Escherichia coli in batch cultures limited by respiration? J Bacteriol. 1980;144:114–123. doi: 10.1128/jb.144.1.114-123.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holms WH, Bennett PM. Regulation of isocitrate dehydrogenase activity in Escherichia coli on adaptation to acetate. J Gen Microbiol. 1971;65:57–68. doi: 10.1099/00221287-65-1-57. [DOI] [PubMed] [Google Scholar]
- 42.Vemuri GN, Altman E, Sangurdekar DP, Khodursky AB, Eiteman MA. Overflow metabolism in Escherichia coli during steady-state growth: transcriptional regulation and effect of the redox ratio. Appl Environ Microbiol. 2006;72:3653–3661. doi: 10.1128/AEM.72.5.3653-3661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahman M, Shimizu K. Altered acetate metabolism and biomass production in several Escherichia coli mutants lacking rpoS-dependent metabolic pathway genes. Mol Biosyst. 2008;4:160–169. doi: 10.1039/b712023k. [DOI] [PubMed] [Google Scholar]
- 44.Veit A, Polen T, Wendisch VF. Global gene expression analysis of glucose overflow metabolism in Escherichia coli and reduction of aerobic acetate formation. Appl Microbiol Biotechnol. 2007;74:406–421. doi: 10.1007/s00253-006-0680-3. [DOI] [PubMed] [Google Scholar]
- 45.Fong SS, Nanchen A, Palsson BO, Sauer U. Latent pathway activation and increased pathway capacity enable Escherichia coli adaptation to loss of key metabolic enzymes. J Biol Chem. 2006;281:8024–8033. doi: 10.1074/jbc.M510016200. [DOI] [PubMed] [Google Scholar]
- 46.Tischer BK, von Einem J, Kaufer B, Osterrieder N. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques. 2006;40:191–197. doi: 10.2144/000112096. [DOI] [PubMed] [Google Scholar]
- 47.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Herring CD, Glasner JD, Blattner FR. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene. 2003;311:153–163. doi: 10.1016/s0378-1119(03)00585-7. [DOI] [PubMed] [Google Scholar]
- 49.Albert TJ, Dailidiene D, Dailide G, Norton JE, Kalia A, et al. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat Methods. 2005;2:951–953. doi: 10.1038/nmeth805. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Indirect assessment of RpoS activity utilizing the peroxidase assay [19]. Colonies with functional RpoS exhibit vigorous bubbling when they come into contact with hydrogen peroxide via a mechanism based on rpoS control of katE expression. We defined vigorous bubbling (v) as bubble formation occurring within five seconds after contact with hydrogen peroxide; medium bubbling (m) as bubble formation occurring between five and ten seconds after contact; and slight bubbling (s) as bubble formation occurring ten seconds after contact. This assay was performed 24 and 48 hours after colonies had been inoculated onto LB plates. Abbreviations: repl - replicate.
(0.06 MB DOC)
Primers used to introduce mutations detected after adaptive evolution back into the starting unevolved Δpgi strain according to the method of Tischer et al [46].
(0.04 MB DOC)
Primers used for Sanger sequencing to confirm mutations reported by Nimblegen and Illumina sequencing technologies. The rpoS, udhA, pntA, and pntB genes were sequenced in their entirety in all nine evolved strains using Sanger sequencing, not just regions immediately surrounding reported mutations. Primers were designed for this purpose by dividing the four genes into regions of approximately 800–900 base pairs with an overlap between regions of approximately 100 base pairs.
(0.06 MB DOC)