Abstract
A cyclical control circuit composed of four master regulators drives the Caulobacter cell cycle. We report that SciP, a helix-turn-helix transcription factor, is an essential component of this circuit. SciP is cell cycle-controlled and co-conserved with the global cell cycle regulator CtrA in the α-proteobacteria. SciP is expressed late in the cell cycle and accumulates preferentially in the daughter swarmer cell. At least 58 genes, including many flagellar and chemotaxis genes, are regulated by a type 1 incoherent feedforward motif in which CtrA activates sciP, followed by SciP repression of ctrA and CtrA target genes. We demonstrate that SciP binds to DNA at a motif distinct from the CtrA binding motif that is present in the promoters of genes co-regulated by SciP and CtrA. SciP overexpression disrupts the balance between activation and repression of the CtrA-SciP coregulated genes yielding filamentous cells and loss of viability. The type 1 incoherent feedforward circuit motif enhances the pulse-like expression of the downstream genes, and the negative feedback to ctrA expression reduces peak CtrA accumulation. The presence of SciP in the control network enhances the robustness of the cell cycle to varying growth rates.
Keywords: CtrA, feedback, genetic circuit
The Caulobacter crescentus cell cycle is controlled by a cyclical genetic circuit comprising four proteins—DnaA, GcrA, CtrA, and CcrM—that pace the progress of the cell cycle and assure that the subsystems that implement the cell cycle are initiated in a tightly controlled order (1–8). DnaA, GcrA, and CtrA are global transcriptional regulators that control nearly 200 cell cycle-regulated genes (4, 5, 9). This cell cycle control system drives progression of the Caulobacter cell cycle in a near-mechanical step-like fashion. Simulation and analysis of the Caulobacter cell cycle control system has shown that the overall circuit design has been selected to be robust to the inevitable stochastic variations in intrinsic reaction rates and nutrient supplies (2). The Caulobacter cell cycle regulatory architecture is widely conserved, in whole or part, among the α-proteobacteria (10).
The CtrA response regulator plays a pivotal role in the control of polar morphogenesis and cell cycle progression. To control the timing of CtrA function, the cell regulates both CtrA activity, by controlling its phosphorylation state, and CtrA accumulation, by inhibiting ctrA transcription upon DNA remethylation of newly replicated chromosomes (11), and by the temporal regulation of CtrA proteolysis (12, 13). We report here that SciP (CC0903), also identified by Gora et al. (14), provides another layer of control. SciP represses the transcription of late predivisional cell and swarmer cell genes that were activated by CtrA. We show that SciP is an essential helix-turn-helix (HTH) transcription factor, and we identify its binding motif by DNase I footprinting. Multiple flagella and chemotaxis genes are activated by CtrA and subsequently repressed by SciP that accumulates preferentially in the swarmer cell. The promoters of most of these genes contain a SciP DNA binding motif upstream of a CtrA binding motif. The transcription of sciP is tightly cell cycle-controlled, and mis-timed expression of sciP results in cell death. CtrA activates the transcription of sciP and SciP negatively regulates CtrA accumulation by binding to the promoter of ctrA and repressing its transcription. Our coimmunoprecipitation results and reported yeast two-hybrid and gel retardation experiments (14) suggest that interaction between SciP and CtrA mediates SciP repression of CtrA activated genes. We propose that SciP's principal function in cell cycle regulation is to enhance the robustness of the core cell cycle control circuit.
Results
sciP Is Cell Cycle-Regulated.
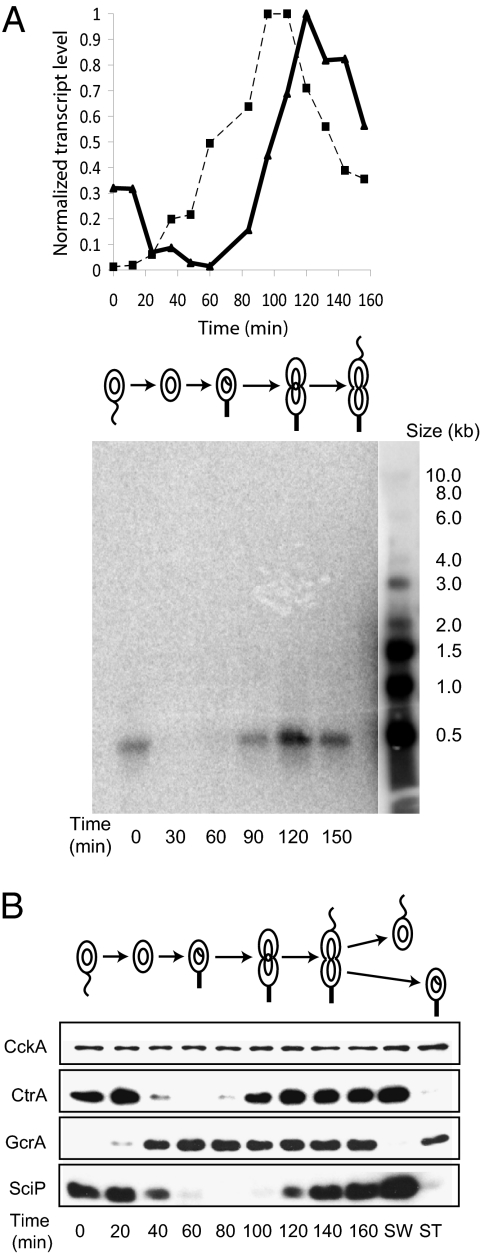
Microarray gene expression profiles (Fig. 1A, Upper) and Northern blot assays (Fig. 1A, Lower) showed that the sciP transcript peaks approximately 30 min after the peak of ctrA transcription. Immunoblots using antibodies to SciP showed that the level of SciP increased in the late predivisional cell and peaked at the time of cell division (Fig. 1B). Immunoblots of samples from the same synchrony using antibodies to the CtrA and GcrA global regulators and to the CckA histidine kinase were used as controls. The swarmer and stalked cell progeny were separated and immunoblots showed that SciP accumulated specifically in the daughter swarmer cell, as did CtrA, as previously reported (15). Also as previously reported, GcrA accumulated in the stalked daughter cell (5), and CckA was present throughout the cell cycle (16).
Fig. 1.
The expression of sciP is cell cycle-regulated. (A) Upper: The transcript levels of sciP (solid line) and ctrA (dotted line) over the course of the cell cycle, beginning with swarmer cells, as determined by microarray analysis (18). Lower: Northern blot analysis of sciP mRNA as a function of the cell cycle. RNA was extracted from a synchronized population of WT C. crescentus (LS101) at the indicted time intervals. sciP mRNA was detected using a probe against nucleotides 169 to 197 within the sciP coding region. A single transcript of approximately 500 bp peaked at the 120-min time point. (B) Immunoblots of samples from a synchronized population of WT cells (LS101) using antibodies to SciP and to three cell cycle regulators, GcrA, CtrA, and CckA. Upon cell division, swarmer and stalked cells were separated and immunoblot analysis showed that SciP and CtrA appeared specifically in the swarmer cell, whereas GcrA appeared specifically in the stalked cell.
To investigate the effects of constitutive expression of sciP, we placed the sciP gene on a high copy plasmid under the control of a vanillate-inducible promoter (PvanA) in the presence of a chromosomal copy of the native sciP gene. Within 6 h of induction with 0.5 mM vanillate, cell division was aberrant; after 11 h, the cells were very filamentous (Fig. S1A). Assays of colony forming units showed that survival dropped 100-fold over approximately 30 h (Fig. S1B). We generated a strain (MHT277) in which sciP was placed on a high copy plasmid under the control of its own promoter so that the levels of SciP remained cell cycle-regulated (Fig. S2A). Although DIC images showed some filamentation, this culture appeared similar to WT (Fig. S2A), suggesting that correct timing of SciP accumulation is critical for normal cell cycle progression.
CtrA Activates Expression of sciP.
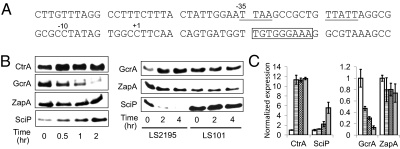
Because the sciP promoter contains a CtrA motif between its −10 and −35 regions (Fig. 2A), we monitored the level of expression of both the SciP protein and the sciP transcript when the level of active CtrA was perturbed.
Fig. 2.
CtrA activates the expression of sciP. (A) The nucleotide sequence of the sciP promoter is shown. A CtrA-binding motif, TTAA(n7)TTATT, is underlined, and a DnaA motif with a 7/9 match to the consensus sequence is boxed. Numbers are relative to the +1 transcription start site (24). (B) Left: Immunoblots show that 0.3% xylose-induced expression of a nondegradable phosphomimetic of ctrA, ctrAD51EΔ3Ω, on a high copy plasmid, increased the level of SciP accumulation. Serving as controls, the level of GcrA accumulation decreased, whereas the level of zapA, a gene whose expression is independent of a CtrA-binding site (9), remained unchanged. Right: Immunoblot showing that the SciP protein level decreased with time after LS2195 (ctrA401ts) cells were shifted from 28 °C to 37 °C, compared with no change for WT LS101 cells. As controls, 2 h after the shift to the restrictive temperature, ZapA levels remained unchanged and the level of GcrA, whose expression is inhibited by CtrA (5), increased. (C) Quantitative real-time PCR was used to measure the relative transcript levels of ctrA, sciP, gcrA, and zapA in strain LS4190 upon addition of 0.3% xylose to induce expression of ctrAD51EΔ3Ω. Left: The transcript level of ctrA increased approximately 11-fold, whereas that of sciP increased approximately sixfold. Right: The transcript level of gcrA decreased by approximately sevenfold, whereas that of zapA did not change significantly. In both panels, white bars indicate 0 min, striped bars indicate 15 min, stippled bars indicate 30 min, and gray bars indicate 60 min after addition of 0.3% xylose. The error bars indicate SDs from three technical replicates.
Using strain LS4190, we induced the expression of a CtrA phosphomimetic encoded by ctrAD51EΔ3Ω (15) on a high copy plasmid and determined the relative levels of the SciP and CtrA at the indicated times after induction by immunoblot analysis (Fig. 2B, Left). GcrA and ZapA served as controls. As expected, the level of GcrA dropped rapidly, as CtrA represses the transcription of gcrA (5). In contrast, the level of SciP increased, indicating that CtrA activates sciP expression. To confirm this observation, we examined the protein levels of GcrA and SciP in a mutant strain, LS2195, bearing a temperature sensitive allele, ctrA401ts (17). Upon a shift to restrictive temperature, the level of GcrA increased whereas the level of SciP decreased rapidly (Fig. 2B, Right). No significant changes were observed when we shifted the WT strain LS101 to 37 °C. The level of the sciP transcript was measured by quantitative real-time PCR upon induction of the stable CtrA variant CtrAD51EΔ3Ω (Fig. 2C). The transcript level of sciP increased approximately sixfold within 1 h after induction, whereas that of gcrA decreased. Cumulatively, these results indicate that CtrA is a positive regulator of sciP expression.
SciP Is an Essential Global Transcriptional Regulator.
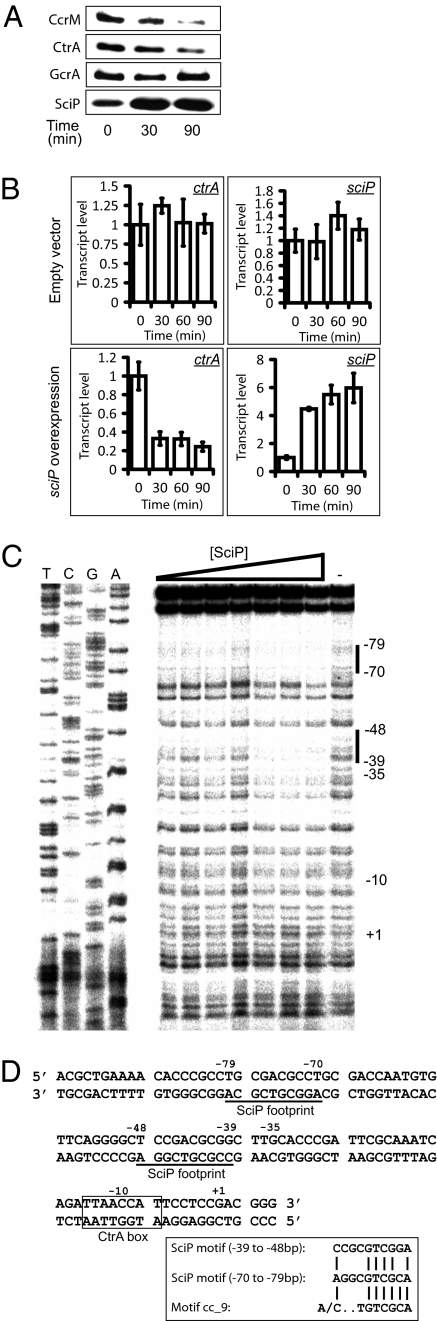
To determine if SciP functions to control the expression of specific cell cycle-regulated genes, we carried out immunoblots of strain MHT68, with the native copy of sciP on the chromosome and a copy of sciP under the control of a xylose-inducible promoter on a high copy plasmid (Fig. 3A). At 30 min after induction of sciP, SciP levels increased and both CcrM and CtrA exhibited a decrease in protein accumulation. By 90 min after induction of sciP, the levels of both CcrM and CtrA, but not GcrA, were significantly reduced. When we overproduced SciP in a strain containing the ctrAΔ3Ω allele, which encodes a version of CtrA that cannot be degraded, as the only copy of ctrA, the protein levels of CtrA were still found to decrease (Fig. S3), suggesting that increased proteolysis was not the reason for the decreased level of CtrA during sciP over production. We found, as did Gora et al. (14), that CtrA was stabilized during sciP overexpression (Fig. S4).
Fig. 3.
Overexpression of sciP represses transcription of the ctrA master regulator. (A) Immunoblots of SciP and three essential cell cycle regulators—CtrA, CcrM, and GcrA—in strain MHT68, in which sciP is under the control of the xylose-inducible promoter on a high copy plasmid, at 0, 30, and 90 min after the addition of 0.3% xylose. The protein levels of CtrA and CcrM, but not GcrA, decreased upon SciP overproduction. (B) Measurements of the relative transcript levels of sciP and ctrA by quantitative real-time PCR in strains bearing an empty plasmid (MHT338) or a plasmid-borne sciP expressed from a vanillate-inducible promoter (MHT171) in a background containing WT sciP. In both cases, cultures were grown in the presence of 0.5 mM vanillate for the times indicated. The transcript levels of ctrA decreased as sciP levels increased upon induction with vanillate. The error bars indicate the SDs from three technical replicates. (C) DNase I protection (footprinting) of the ctrA P1 promoter by purified His6-SciP. The antisense strand was radiolabeled at the 5′ end and the first four lanes contain the sequencing ladder of the promoter region. His6-SciP was added in increasing concentrations from 6.4 nM to 512 nM and the last lane contains no purified protein. Black vertical bars on the right indicate regions of His6-SciP protection against DNase I digestion. Numbers are relative to the +1 transcription start site (15). (D) Sequence of the region upstream of the transcription start site of the P1 promoter of ctrA. The two SciP-binding sites are underlined and the CtrA-binding site is boxed (25).
To determine if SciP is a transcriptional regulator of ctrA, we measured the transcript level of ctrA during sciP overexpression by quantitative real-time PCR. In a strain carrying an empty vector control (MHT338), addition of inducer did not alter the transcript level of sciP or ctrA, whereas in the MHT171 strain carrying a high-copy plasmid with sciP under the control of PvanA, addition of the vanillate inducer resulted in an increase in the transcript level of sciP and a concomitant decrease in the transcript level of ctrA (Fig. 3B). Thus, the overproduction of SciP repressed, either directly or indirectly, the transcription of ctrA.
We used microarray analysis after chromatin immunoprecipitation (ChIP-on-chip) to determine the direct targets of SciP. Sixty-six distinct peaks were identified, 52 of which corresponded to the promoter regions of predicted ORFs. Twenty-one peaks corresponded to DNA positioned between divergently transcribed genes and thus could be binding to either or both promoters of two different annotated genes. Thus, the promoter regions of at least 73 genes were predicted by our ChIP-on-chip experiment to be bound by SciP. Many of these genes are the first gene in operons, so including downstream genes, SciP could affect transcription of approximately 130 genes. We could not delete SciP in either rich or minimal media, suggesting it is an essential gene.
SciP Directly Controls the Transcription of ctrA.
To determine if SciP binds directly to the CtrA promoter, we carried out DNase I protection experiments with purified His6-SciP and the ctrA P1 promoter. These in vitro footprinting experiments revealed that SciP protected two sites within the region upstream and on the opposite strand of the CtrA binding motif; −39 to −48 and −70 to −79 (Fig. 3C). The direct binding of SciP to the ctrA promoter was confirmed by ChIP experiments followed by quantitative real-time PCR (ChIP-PCR) using a strain expressing chromosomal m2-sciP under control of the native sciP promoter as the only copy of sciP in the cell (MHT267). Cumulatively, these results demonstrate that SciP binds to the ctrA promoter in vivo and in vitro. The sequences in the ctrA promoter protected by SciP are shown in Fig. 3D. Both SciP binding sites align with the previously identified cc_9 motif (18) that is found in the promoter regions of genes expressed late in the cell cycle and whose promoters have a CtrA binding motif between their +1 transcription start site and the SciP binding site (18). In a less common configuration, the ccrM promoter has the CtrA binding site directly upstream of the SciP binding motif.
SciP Regulon Includes Genes with both SciP and CtrA Binding Motifs in Their Promoters.
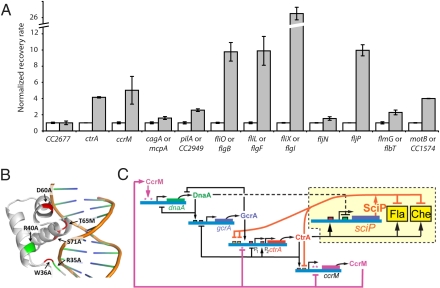
By combining experimental and genomic evidence, we identified the genes in the SciP regulon. A subset is shown in Fig. 4A, and the complete list is in Dataset S1. Fig. 5A shows results of ChIP followed by quantitative real-time PCR of a subset of these genes including ctrA and ccrM, as well as many flagella and chemotaxis genes. The genes in Fig. 4A have at least one CtrA and one SciP binding site on either the coding or noncoding strand, and their expression profile peaks in the predivisional cell after accumulation of CtrA. CtrA activates these genes. Then, after a delay for SciP accumulation, SciP represses their expression. The regulation of the ctrA P2 promoter fits this pattern, as do many flagella and chemotaxis genes. The SciP binding motif logo (Fig. 4B) is derived from the SciP regulon. Fig. 5C shows the Caulobacter cell cycle control circuit with the addition of SciP regulation of ctrA, flagella genes, and chemotaxis genes.
Fig. 4.
SciP is a global transcriptional regulator that controls the expression of late cell cycle genes. (A) A subset of genes in the SciP regulon. ChIP column: “x” indicates promoters detected. OE (overexpression) column: “x” indicates response detected in our overexpression experiment; “G” indicates response reported in ref. 14. (B) The consensus SciP binding motif.
Fig. 5.
SciP is an integral component of the core circuit that controls cell cycle progression in Caulobacter. (A) ChIP followed by quantitative real-time PCR showed that SciP bound directly to the promoter regions of genes encoding CtrA, the CcrM DNA methyltransferase, and the PilA pilus subunit, as well as to the promoter regions of a subset of flagellar and chemotaxis genes. The endogenous sciP coding region on the chromosome was replaced by FLAG-sciP to create strain MHT267. Immunoprecipitation was performed on WT cells expressing untagged sciP (strain LS101; white bars) and on cells that expressed FLAG-sciP (strain MHT267; gray bars) using antibodies to the FLAG tag. The error bars indicate SDs from two technical replicates. The promoter of CC2677, which served as a control, does not contain a SciP motif (18). (B) Ribbon diagram depicting a homologous protein of SciP (Protein Data Bank ID code 1PDN) docked to DNA. Structural homology and sequence alignment was established by FFAS (20). Residues mutated in this study are red and residues mutated by Gora et al. (14) are green. All mutations were found to be in the HTH DNA-binding domain of SciP. (C) Topology of the core cell cycle circuit showing how sciP is connected to the other four previously known components, namely dnaA, gcrA, ctrA, and ccrM. CtrA activates the transcription of multiple flagellar and chemotaxis genes. It also activates the production of SciP in the late predivisional cell. SciP then directly represses the transcription of ctrA as well as many late predivisional cell and swarmer cell specific genes that had been activated by CtrA. CtrA represses the ctrA P1 promoter and autoactivates the P2 promoter.
SciP Binds DNA at Its HTH Motif.
We used the fold and function assignment system (FFAS; http://ffas.burnham.org/) that has been demonstrated to detect homologies beyond the reach of other methods to search for protein structures in the Protein Data Bank that are homologous to SciP (19, 20). Among the results was the structure of a SciP homologue bound to a cognate DNA segment. The predicted structure of the SciP:DNA complex is shown in Fig. 5B and Fig. S5. SciP has the HTH motif found in virtually all prokaryotic transcription factors, and the FFAS analysis showed this to be the SciP DNA-binding domain. Six strains with individual SciP amino acid substitutions in the predicted DNA recognition domain eliminated the filamentation phenotype when overexpressed (Fig. 5B), strongly suggesting that the regulatory action of DNA-bound SciP causes the phenotype. Two of these substitutions (R35A and R40A) were also identified by Gora et al. (14) and interpreted as being in the site involved in a CtrA-SciP interaction.
Interaction of CtrA and SciP.
To detect interactions between SciP and CtrA, we performed a coimmunoprecipitation experiment with strain MHT267 containing chromosomal m2-sciP under control of the native sciP promoter as the only copy of sciP in the cell. We divided the MHT267 cell lysate and added anti-FLAG M2 agarose to one sample and mouse IgG agarose to the other. We observed enrichment of CtrA when anti-FLAG M2 agarose was used compared with mouse IgG agarose, but there was no enrichment of the CckA control (Fig. S2C). These results suggest that SciP and CtrA interact directly or indirectly in vivo.
Discussion
In this study, we report that SciP is an essential global transcription factor with a major role in control of the Caulobacter cell cycle. The evidence reported in this article and results of Gora et al. (14) show that SciP acts as a repressor of genes activated by CtrA. Gora et al. (14) surmised that the mechanism of this repression involved SciP binding solely to CtrA and thus preventing CtrA from recruiting RNA polymerase. We have discovered that SciP, a winged HTH protein, binds directly to a specific motif present in the promoters of genes activated by CtrA and repressed by SciP. These genes have both SciP and CtrA binding motifs in their promoter regions, leading to a new model for SciP control in which SciP-DNA interaction is a component of SciP-mediated repression of CtrA-activated genes.
Our experiments, as well as those by Gora et al. (14), suggest that the two proteins interact. Gora et al. also reported gel shift experiments that support a direct interaction between SciP and CtrA (14). It remains an open question whether the mechanism of SciP repression is indirect, through disruption of CtrA∼P's ability to recruit RNAP, or through more direct interference in the activation of transcription by CtrA. The spacing on the DNA between the 5′ end of the CtrA binding site and the start of the SciP motif is quite variable, with most between 35 and 50 bp. A CtrA-SciP interaction on the DNA may provide incremental binding energy to SciP binding and thus lead to a more rapid onset of repression of CtrA-activated genes.
Function of SciP in Caulobacter Cell Cycle Regulation.
We have previously shown that Caulobacter cell cycle progression is driven by a cyclical genetic circuit (1, 2, 8) that is robust with respect to variations in environmental nutrients and to inevitable stochastic variations in reaction rates (2). SciP adds multiple feedback pathways that further modulate operation of this control circuit (Fig. 5C). ctrA-positive autoregulation favors rapid initial accumulation of CtrA following its initial activation by GcrA, and thus definitive regulation of the many downstream CtrA-regulated genes, but it also could lead to more CtrA in the cell than needed. The delayed negative regulatory feedback via the SciP pathway would allow the rapid initial CtrA accumulation followed by a tempering of peak CtrA concentration. We tested this by comparing simulated CtrA expression profiles with and without SciP at two different growth rates by using our Caulobacter cell cycle control simulation (2) (Table S1). The results predict that CtrA would grow to much higher peak levels under slower growth conditions without the SciP negative feedback, whereas with SciP, the peak CtrA level was much less sensitive to growth rate. The pulsed temporal pattern of CtrA in the cell was also much less sensitive to growth rate with SciP. As Caulobacter clears CtrA from the cell once per cell cycle by regulated proteolysis, production of excess CtrA is both energetically costly and deleterious to proper functioning of the cell cycle.
A DnaA binding motif is present downsteam of the SciP +1 site (Fig. 2A) and SciP levels increase in a DnaA deletion (Fig. S2B), suggesting that DnaA represses sciP. This repressive link would be useful in situations in which DnaA levels are unusually high (e.g., a stochastic high excursion of DnaA). In that case, SciP accumulation would be repressed, reducing SciP repression of ctrA, providing higher levels of CtrA to prevent reinitiation of DNA replication by the anomalously high DnaA level. This is another example of a cell cycle control circuit design feature that enhances robustness by preventing timing glitches (2).
The regulatory configuration of CtrA and SciP and their coregulated genes fits the type-1 incoherent feedforward loop circuit motif that is found in many regulatory circuits, often in arrangements in which multiple functionally related genes are coordinately regulated (21–23). The type-1 incoherent feedforward loop motif is a regulatory configuration in which an activator (i.e., CtrA) controls a target gene (e.g., a flagellar gene) and also activates a repressor of that target gene (i.e., SciP). Simulation of the dynamics of a downstream gene activated by CtrA with and without the SciP pathway again suggests greatly reduced sensitivity of both peak protein levels and of the pulse-like temporal pattern of expression to growth rate with the SciP pathway present. It thus appears that SciP's primary function is to enhance the robustness and reliability of Caulobacter cell cycle control in response to both intrinsic stochastic variations in reaction rates and environmental variations affecting growth rates.
Materials and Methods
Caulobacter strains used are listed in Table S2. His6-SciP protein was purified using a Nickel column and an ion exchange column. RNA was isolated using TRIzol-chloroform and separated in a glyoxal-agarose gel for Northern blotting. For ChIP, cells were fixed in formaldehyde before sonication and for coimmunoprecipitation, cells were cross-linked with dithiobis(succinimidyl propionate). All microarray experiments were performed using the custom-designed CauloHI Affymetrix chip. Additional details are provided in SI Materials and Methods and Table S3.
Supplementary Material
Acknowledgments
We thank Mohammed AlQuraishi for valuable suggestions regarding the SciP-DNA binding interactions and for Fig. 5B and Fig. S5. This work was supported by US Department of Energy Grant DE-FG02-05ER64136 (to H.H.M.), National Science Foundation Grant MCB-0923679 (to H.H.M.), and National Institutes of Health Grant GM032506-27 (to L.S.). M.H.T. was supported by a National Science Scholarship from the Singapore Agency for Science, Technology, and Research (A*Star).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014395107/-/DCSupplemental.
References
- 1.McAdams HH, Shapiro L. System-level design of bacterial cell cycle control. FEBS Lett. 2009;583:3984–3991. doi: 10.1016/j.febslet.2009.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shen X, et al. Architecture and inherent robustness of a bacterial cell-cycle control system. Proc Natl Acad Sci USA. 2008;105:11340–11345. doi: 10.1073/pnas.0805258105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laub MT, Shapiro L, McAdams HH. Systems biology of Caulobacter. Annu Rev Genet. 2007;41:429–441. doi: 10.1146/annurev.genet.41.110306.130346. [DOI] [PubMed] [Google Scholar]
- 4.Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58:1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- 5.Holtzendorff J, et al. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science. 2004;304:983–987. doi: 10.1126/science.1095191. [DOI] [PubMed] [Google Scholar]
- 6.McAdams HH, Shapiro L. A bacterial cell-cycle regulatory network operating in time and space. Science. 2003;301:1874–1877. doi: 10.1126/science.1087694. [DOI] [PubMed] [Google Scholar]
- 7.Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci USA. 2007;104:17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collier J, Shapiro L. Spatial complexity and control of a bacterial cell cycle. Curr Opin Biotechnol. 2007;18:333–340. doi: 10.1016/j.copbio.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA. 2002;99:4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brilli M, et al. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: A comparative genomic analysis. BMC Syst Biol. 2010;4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reisenauer A, Shapiro L. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 2002;21:4969–4977. doi: 10.1093/emboj/cdf490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGrath PT, Iniesta AA, Ryan KR, Shapiro L, McAdams HH. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell. 2006;124:535–547. doi: 10.1016/j.cell.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 13.Iniesta AA, McGrath PT, Reisenauer A, McAdams HH, Shapiro L. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc Natl Acad Sci USA. 2006;103:10935–19040. doi: 10.1073/pnas.0604554103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gora KG, et al. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell. 2010;39:455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domian IJ, Quon KC, Shapiro L. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell. 1997;90:415–424. doi: 10.1016/s0092-8674(00)80502-4. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs C, Hung D, Shapiro L. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc Natl Acad Sci USA. 2001;98:4095–4100. doi: 10.1073/pnas.051609998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quon KC, Marczynski GT, Shapiro L. Cell cycle control by an essential bacterial two-component signal transduction protein. Cell. 1996;84:83–93. doi: 10.1016/s0092-8674(00)80995-2. [DOI] [PubMed] [Google Scholar]
- 18.McGrath PT, et al. High-throughput identification of transcription start sites, conserved promoter motifs and predicted regulons. Nat Biotechnol. 2007;25:584–592. doi: 10.1038/nbt1294. [DOI] [PubMed] [Google Scholar]
- 19.Rychlewski L, Jaroszewski L, Li W, Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: A server for profile—profile sequence alignments. Nucleic Acids Res. 2005;33(web server issue):W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci USA. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eichenberger P, et al. The program of gene transcription for a single differentiating cell type during sporulation in Bacillus subtilis. PLoS Biol. 2004;2:e328. doi: 10.1371/journal.pbio.0020328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mangan S, Itzkovitz S, Zaslaver A, Alon U. The incoherent feed-forward loop accelerates the response-time of the gal system of Escherichia coli. J Mol Biol. 2006;356:1073–1081. doi: 10.1016/j.jmb.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 24.Chen LS, Mullin D, Newton A. Identification, nucleotide sequence, and control of developmentally regulated promoters in the hook operon region of Caulobacter crescentus. Proc Natl Acad Sci USA. 1986;83:2860–2864. doi: 10.1073/pnas.83.9.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci USA. 1999;96:6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.