Abstract
Studies of long-lived Caenorhabditis elegans mutants have identified several genes that function to limit lifespan, i.e., loss-of-function mutations in these genes promote longevity. By contrast, little is known about genes that normally act to delay aging and that when mutated cause premature aging (progeria). To seek such genes, we performed a genetic screen for C. elegans mutants that age prematurely. We found that loss-of-function mutations of the ketoacyl thiolase gene kat-1 result in an increased accumulation of the lipofuscin-like fluorescent aging pigment, shortened lifespan, early behavioral decline, and other abnormalities characteristic of premature aging. These findings suggest that kat-1 acts to delay C. elegans aging. kat-1 encodes a conserved metabolic enzyme that catalyzes the last step of fatty acid oxidation and was previously shown to regulate fat accumulation in worms. We observed that kat-1 is required for the extension of lifespan and enhanced thermotolerance mediated by extra copies of the deacetylase gene sir-2.1. kat-1 acts independently of other known pathways that affect longevity. Our findings suggest that defects in fatty acid oxidation can limit lifespan and accelerate aging in C. elegans and that kat-1-mediated fatty acid oxidation is crucial for overexpressed sir-2.1 to delay aging.
Keywords: progeria, lipofuscin, fatty acid oxidation, sirtuins, protein deacetylation
Over the past few decades, experiments using Caenorhabditis elegans and other organisms have resulted in progress toward an understanding of molecular mechanisms of aging (1–3). Loss-of-function mutations in single C. elegans genes can substantially increase lifespan (4–7), indicating that lifespan determination of nematodes has a major genetic component. Research in the aging field has largely focused on lifespan-extending mutations in genes that normally function to reduce longevity. This focus has been primarily a consequence of the difficulties in distinguishing accelerated aging from sickness of short-lived mutants.
Though longevity remains the most commonly used measure of the rate of nematode aging, additional biomarkers of C. elegans aging have been characterized. These biomarkers include behavioral decline and tissue aging (8–11). Aging of C. elegans is characterized by a progressive decline in locomotion, both in speed and coordination (9, 10, 12), and decreased defecation and pharyngeal pumping rate (8, 9, 13). Old nematodes, like old humans, show progressive tissue and organ deterioration: muscles, gonad, intestine, and connective tissues degenerate in aged worms (9–11).
The intestinal cells of aging C. elegans accumulate an autofluorescent aging pigment similar to the aging pigment lipofuscin present in postmitotic mammalian cells (13, 14). Lipofuscin accumulates in the secondary lysosomes (14–16) and consists primarily of lipid peroxidation products and oxidized proteins that resist proteolytic degradation (17). The amount of lipofuscin gradually increases throughout C. elegans adulthood, and autofluorescent granules can be detected in aging adult nematodes by fluorescence microscopy.
Accumulation of lipofuscin, tissue degeneration, and behavioral decline characterize aged worms and can be used to seek progeroid mutants. Such mutants might define a new class of aging genes.
Here we report the identification and characterization of progeroid mutants of C. elegans defective in the gene kat-1 (3-ketoacyl thiolase). We show that kat-1 functions to delay C. elegans aging and that kat-1 is specifically required for the increased lifespan of worms bearing extra copies of the sir-2.1 protein deacetylase gene.
Results
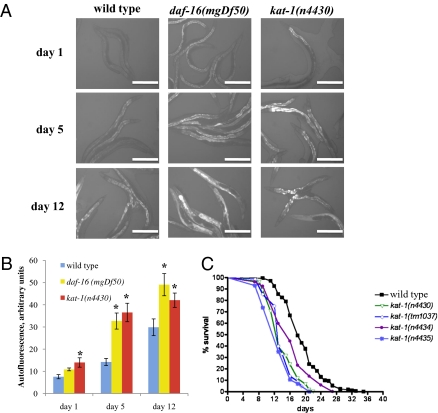
To identify genes that act to delay aging, we sought mutants that age prematurely, using accumulation of the autofluorescent pigment lipofuscin as our marker. We used a fluorescence-equipped dissecting microscope to screen for mutants that showed a premature increase of lipofuscin. Old wild-type adults and animals with a daf-16 loss-of-function mutation, which age prematurely, accumulated high levels of lipofuscin (13) (Fig. 1 A and B). We mutagenized wild-type hermaphrodites and screened F2 offspring for an increased accumulation of intestinal autofluorescence. Overall, we screened ≈20,000 mutagenized haploid genomes and obtained 11 mutant strains that reliably exhibited premature accumulation of intestinal autofluorescence (Fig. S1, and see SI Text for details).
Fig. 1.
kat-1 mutations cause increased fluorescence and short lifespan. (A) Intestinal fluorescence accumulation in young (1-d), middle-aged (5-d), and old (12-d) kat-1(n4430) adults compared with wild-type (N2) and progeroid daf-16(mgDf50) animals of the same ages. Intestinal autofluorescence was recorded using a DAPI filter (excitation 365–380 nm, emission 450 nm) with an exposure time of 1,000 ms. (Scale bar, 0.2 mm.) (B) Quantitation of the intestinal autofluorescence intensity in wild-type, kat-1(lf), and daf-16(lf) animals using ImageJ software. Fluorescence intensity was calculated by subtracting background fluorescence (ranging from 90 to 110 arbitrary units) from total intestinal fluorescence. Five animals per genotype per time point were analyzed. *P < 0.05 compared with wild type of the same age. (C) kat-1 mutants have shortened lifespans. Median lifespans in this experiment were: wild-type, 18 d, n = 79; n4430, 13 d, n = 105; n4434, 16 d, n = 95; n4435, 13 d, n = 122, tm1037, 13 d, n = 89. Lifespans af all four kat-1 mutants were significantly different from that of the wild type (P < 0.0001).
Age-dependent accumulation of fluorescence in the homozygous mutant isolates was comparable to that of animals bearing loss-of-function mutations in daf-16 (example shown in Fig. 1 A and B is n4430, an allele of the kat-1 gene identified below). We mapped n4430, n4434, n4435, and n4501 to linkage group II (LGII), and n4427, n4428, n4429, n4431, n4432, n4433, and n4436 to LGV. n4434 and n4435 failed to complement n4430, whereas n4501 complemented n4430 for increased autofluorescence accumulation. All LGV mutants were strongly semidominant and appeared to fail to complement each other. In short, the 11 isolates with high autofluorescence represent at least three complementation groups: two on LGII and one or more on LGV (Table 1), suggesting that at least three genes control the age-related accumulation of autofluorescence in C. elegans.
Table 1.
Genetic and phenotypic analyses of screen isolates
| Genotype | Phenotypea | LG | Comp. group | Behavioral decline | Median lifespan 20 °C (25 °C) |
| Wild type | — | — | — | Normal | 21 (12) |
| kat-1(n4430) | Weakly SD | II | 1 | Premature | 14* (11*) |
| kat-1(n4434) | Weakly SD | II | 1 | Premature | 14* (9*) |
| kat-1(n4435) | Weakly SD | II | 1 | Premature | 14* (10*) |
| n4501 | Recessive | II | 2 | Normal | 18* (11*) |
| n4427 | SD | V | 3b | Normal | 16* (11*) |
| n4428 | SD | V | 3b | Normal | 19* (11*) |
| n4429 | SD | V | 3b | Normal | 19* (11*) |
| n4431 | SD | V | 3b | Normal | 19* (11*) |
| n4432 | SD | V | 3b | Normal | 12* (10*) |
| n4433 | SD | V | 3b | Normal | 19* (11*) |
| n4436 | SD | V | 3b | Normal | 16* (11*) |
aRefers to autofluorescence accumulation.
bThese mutations might or might not be allelic; they cause a SD phenotype.
Assays were performed as described in Materials and Methods. SD, semidominant; LG, linkage group; Comp., complementation. *P < 0.05.
In their studies of aging C. elegans, Gerstbrein et al. (13) distinguished two distinct sources of autofluorescence: the aging pigment lipofuscin, which increases with age and has an emission range of 400–460 nm, and total protein fluorescence (mostly tryptophan fluorescence), which remains constant during adulthood and has an emission range of 330–380 nm. To elucidate which type of fluorescent pigment accumulated in our mutants, we extracted proteins from the mutant strains and tested fluorescence accumulation in worm lysates, comparing young to old animals of each strain as well as mutant to wild-type animals at different ages. Our lysis buffer was designed to extract proteins, so we were not expecting to detect lipofuscin components of lipid origin in lysates. Nonetheless, both wild-type animals and our mutants showed in whole-worm lysates an age-dependent increase in fluorescence with a spectrum similar to that of lipofuscin (420 nm emission peak), possibly because fluorescent moieties of nonprotein origin were also extracted during lysis. Both young and old adult LGV mutants showed levels of protein fluorescence substantially higher than those in the wild type, and LGII mutants also showed an age-dependent increase in protein fluorescence but had overall levels more similar to and possibly less than those of the wild type (Fig. S2). It is noteworthy that because wild-type animals showed an age-dependent increase in fluorescence accumulation by this method, and the LGII mutants did not show abnormally high levels in young adults, it is possible that not all features of aging are accelerated in the LGII mutants. To determine whether the isolated mutants show additional characteristics of premature aging, we performed tests to assess other aging parameters of the screen isolates, as described below.
Three Isolates Prematurely Display Multiple Attributes of Old Age.
Progeroid mutants should have shorter lifespans than wild-type animals. We performed lifespan analyses of our mutants (Table 1). Six had 25–40% shorter lifespans than the wild type, and five had slightly shorter or normal lifespans (Table 1). Notably, all three allelic LGII mutants had short lifespans, with mean lifespans of 14–15 d, compared with 21 d for the wild type (Fig. 1C and Table 1). As will be described below, these three mutants carry alleles of the gene kat-1. The other LGII mutant, n4501, had only a slightly (15%) shorter lifespan, and of the seven LGV mutants, three had short lifespans, and the other four had nearly wild-type lifespans.
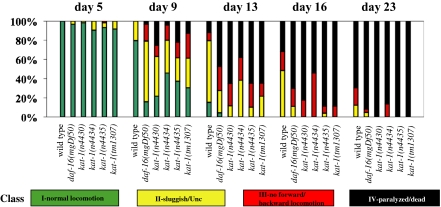
We also characterized the age-dependent behavioral decline of our mutants. The most obvious behavioral abnormality associated with C. elegans aging is a progressive slowing of the rates of feeding (pharyngeal pumping) and locomotion, as well as uncoordinated locomotion (11, 13). kat-1 mutants showed a premature onset of uncoordinated locomotion and paralysis, as did progeroid daf-16(lf) mutants (Fig. 2). The kat-1 mutants n4430, n4434, and n4435, but none of the other mutants, including those with short lifespans, showed a premature decline in both locomotion and pumping (Fig. 2 and Fig. S3).
Fig. 2.
Behavioral abnormalities of kat-1 mutants. Aging animals were classified into four groups based on locomotion: I, animals with robust, coordinated sinusoidal locomotion (green bars); II, uncoordinated and/or sluggish animals (yellow bars); III, animals that did not move forward or backward but did move their heads or shudder in response to prodding (red bars); and IV, paralyzed/dead animals (black bars). Four kat-1 mutants were tested: n4430, n4434, n4435, and tm1037. Wild-type animals (N2) and progeroid daf-16(mgDf50) mutants were used as controls. At least 60 animals per strain were tested for each time point. We used Student's t test to determine statistical significance of the differences between wild-type and mutant animals. For all mutants, differences in one or more classes were significant (P < 0.05).
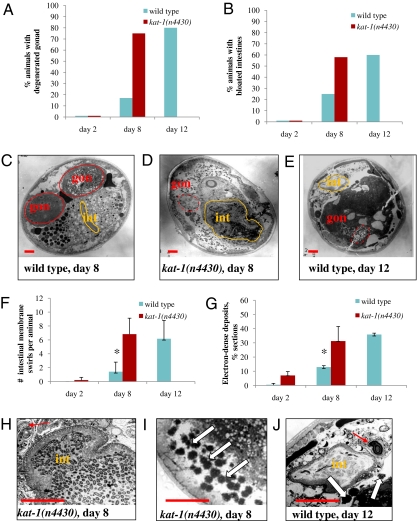
We performed electron microscopic analyses to examine tissue integrity in aging wild-type and kat-1(n4430) mutant animals. Old wild-type animals show a striking decline in the integrity of many organs, including the gonad, intestine, and muscle. Aging C. elegans undergo marked gonad degeneration, and old animals lack a detectable gonad, though yolk and lipofuscin accumulate within their intestines (10, 11). The apparent size of the intestinal lumen increases as old animals become bloated with bacteria, and the intestinal cells of old worms show membrane swirls typical of necrosis (10). Old adults also accumulate electron-dense material in the intestine and other tissues (11). kat-1(n4430) animals showed early degeneration of the gonad compared with the wild type (Fig. 3 A and C–E). The intestines of kat-1 animals showed multiple signs of premature decline: increased bloating of the intestinal lumens (Fig. 3 B, C–E, H, and J) and early appearance of membrane swirls (Fig. 3 F, H, and J). kat-1(n4430) animals also had greatly increased accumulation of electron-dense material (Fig. 3 G, I, and J) compared with the wild type.
Fig. 3.
kat-1 tissues age prematurely. Electron microscopy (EM) was used to examine tissues of kat-1(n4430) mutants. All animals of a given age were grown identically and fixed on the same day. EM images were analyzed for the accumulation of electron-dense deposits and abnormalities in the gonad and intestine. Wild-type animals were analyzed on day 2, 8, and 12 of adulthood; kat-1 mutants were analyzed on day 2 and 8 of adulthood; 12-d kat-1 mutants were too sick to analyze by EM. (A) Gonad degeneration in wild-type animals (blue bars) and kat-1(n4430) mutants (red bars). Note that 8-d kat-1 adults had a similar proportion of animals with degenerated gonads as did 12-d wild-type animals. (B) Intestinal bloating in wild-type animals and kat-1 mutants (blue bars and red bars, respectively). On day 8 of adulthood, only a few wild-type animals showed marked signs of bloated/bacteria-packed intestinal lumens, whereas more than half of the kat-1(n4430) animals had bloated intestines. C–E and H–J show midbody cross-sections of wild-type worms and kat-1(n4430) mutants of the ages indicated, illustrating the abnormalities reported in A, B, F, and G. Intestinal lumens are outlined in orange; gonads are outlined in red; dashed red lines indicate degenerated gonads. int, intestinal lumen; gon, gonad. (Scale bars, 10 μm.) (C) An 8-d wild-type adult showed no significant signs of degeneration. (D) An 8-d kat-1 mutant adult showed degeneration of the intestine and the gonad, including a lack of membrane surrounding the gonad, as did the 12-d wild-type adult (E). (F) Necrotic membrane swirls that accumulate with age in intestinal cells were counted in EM micrographs of wild-type and kat-1(n4430) animals. Here and in G, error bars are SDs of the mean between animals. (F, H, and J) Membrane swirls were present in 8-d kat-1 mutants (H) and 12-d wild-type animals (J), red arrows, but not 8-d wild-type animals. (G, I, and J) Old wild-type and kat-1 mutants both accumulate electron-dense deposits. Note the increased accumulation of electron-dense material (white arrows) in 8-d kat-1 mutants (I) and 12-d wild-type animals (J). The numbers of animals analyzed by EM were as follows: day-2 wild-type, 5; day-8 wild-type, 12; day-12 wild-type, 5; day-2 kat-1(n4430), 8; day-8 kat-1(n4430), 14. Eight to 10 sections per animal were analyzed. *P < 0.05 compared with the wild type.
We conclude that kat-1 mutants have short lifespans and also display precocious onset and progression of many but not all features of C. elegans aging: we observed prematurely increased accumulation of lipofuscin-like pigment, early behavioral decline, and tissue degeneration. We failed to observe increased fluorescence in kat-1 mutant lysates but did observe increased fluorescence in lysates of old wild-type animals.
n4430, n4434, and n4435 Are Alleles of kat-1, Which Encodes a Conserved Thiolase Involved in Fatty Acid Metabolism.
We molecularly identified the gene mutated in n4430 animals using a combination of mapping with visible markers and polymorphisms, transgene rescue, and RNAi (see SI Text for details). We found that n4430 and the allelic mutants n4434 and n4435 all carry mutations in the gene T02G5.8 (Fig. S4). T02G5.8 was previously identified in a RNAi-based screen for animals with abnormal fat metabolism and was named kat-1 for ketoacyl thiolase-1 (18). kat-1 T02G5.8 is highly conserved from worms to humans (Fig. S5) and is predicted to encode a protein with 52% identity and 70% similarity to the mammalian enzyme acetoacetyl CoA thiolase (EC 2.3.1.9), which is involved in the β-oxidation of fatty acids in mitochondria and peroxisomes (Kegg Pathway Database, http://www.genome.ad.jp/kegg/pathway.html). The three kat-1 alleles identified in our screen are predicted to result in the substitutions of conserved amino acids (Fig. S5).
To analyze the null phenotype of kat-1, we obtained a deletion allele, tm1037, from the C. elegans Japanese knockout consortium (National Bioresource Project for the Experimental Animal Nematode C. elegans). kat-1(tm1037Δ) animals showed increased lipofuscin fluorescence, a shortened lifespan, and early behavioral decline comparable to those of the kat-1 mutants n4430, n4434, and n4435 (Figs. 1 and 2 and Fig. S6). Mortality rates of kat-1(tm1037) and kat-1(n4430) mutants were similar, and both increased prematurely compared with the wild-type animals, similarly to daf-16(lf) mutants (Fig. S6A). Notably, the lifespan abnormalities of kat-1(tm1037) and kat-1(n4430) mutants were also apparent when worms were grown without FUdR (Fig. S6B), a drug normally used in lifespan analyses and that we used in most of our studies; under some conditions, FUdR can mildly affect C. elegans lifespan (19). The injection of a PCR product containing multiple copies of the kat-1 gene into kat-1(n4430) mutant animals rescued the lifespan defect of the kat-1 mutants, resulting in normal (wild-type) lifespan (Fig. S6C). None of the transgenic lines overexpressing kat-1 in the kat-1(lf) background had extended lifespan.
kat-1 Is Required for the Lifespan Extension Mediated by the Protein Deacetylase Gene sir-2.1.
Several pathways are known to control C. elegans aging, including insulin signaling, germ-line signaling, chemosensory signaling, mitochondrial function, dietary restriction (DR), and a pathway activated by the gene sir-2.1 (20). To see whether kat-1 acts to delay aging via one of these known longevity pathways, we crossed a kat-1 mutation into backgrounds of long-lived C. elegans mutants and tested whether kat-1 is required for the extended longevity of these mutants. We found that kat-1 is not necessary for lifespan extension by low insulin signaling, chemosensory and ETC abnormalities, and DR (SI Text and Table S1). A kat-1 mutation partially suppressed the long lifespan of mes-1 mutants that lacked germ lines (see SI Text for details), suggesting that the effect of germ-line signaling on lifespan is partially kat-1-dependent.
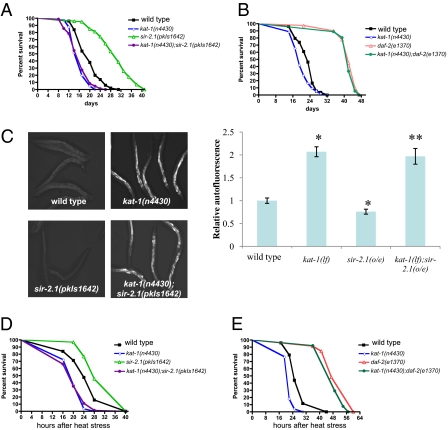
By contrast, kat-1 loss-of-function fully suppressed the lifespan extension caused by overexpression of the sir-2.1 deacetylase. sir-2.1 extends lifespan in response to stress by activating the forkhead transcription factor DAF-16 (21). Introducing a kat-1 mutation into a strain in which sir-2.1 is overexpressed eliminated the lifespan extension caused by sir-2.1 and resulted in a shortened lifespan similar to that of kat-1 mutants (Table S1 and Fig. 4A). As mentioned previously, kat-1 was not required for the extension of lifespan by a loss-of-function mutation in the insulin receptor gene daf-2, and kat-1;daf-2 double mutants had long lifespans similar to these of the single daf-2(lf) mutants (Fig. 4B), indicating that kat-1 is specifically required for the lifespan extension mediated by sir-2.1 overexpression.
Fig. 4.
kat-1 is required for the increased longevity and thermotolerance caused by extra copies of sir-2.1. (A) The increased longevity of sir-2.1-overexpressing animals was suppressed by loss of kat-1 function. Median lifespans were as follows: wild type (N2), 20 d, n = 50, pkIs1642[sir-2.1], 31 d, n = 101, kat-1(n4430), 17 d, n = 41, kat-1(n4430);pkIs1642 [sir-2.1], 17 d, n = 84. (B) The increased longevity of daf-2(lf) mutants was not suppressed by the loss of kat-1 function. Median lifespans were as follows: wild type (N2), 23 d, n = 76, kat-1(n4430), 18 d, n = 67, daf-2(e1370), 42 d, n = 92, kat-1(n4430);daf-2(e1370), 41 d, n = 98. (C) Intestinal lipofuscin accumulation in young (2-d-old) wild-type animals (N2) compared with long-lived sir-2.1-overexpressing animals, with and without the kat-1(n4430) mutation. Note that sir-2.1 animals accumulated less lipofuscin than the wild type, and the sir-2.1(o/e);kat-1(lf) animals accumulated more lipofuscin than did the wild type. Photographs were taken as described in Fig. 1A. *P < 0.05 vs. wild type, **P < 0.05 vs. sir-2.1[pkIs1642]. (D) kat-1 is required for increased resistance to heat stress caused by sir-2.1 overexpression. Median survival: wild type, 24 h, n = 63, pkIs1642[sir-2.1], 28 h, n = 99, kat-1(n4430), 20 h, n = 84, kat-1(n4430);pkIs1642 [sir-2.1], 20 h, n = 71. (E) kat-1 is dispensable for the increased resistance to heat stress of the insulin-like receptor mutant daf-2(e1370). Median survival: wild type, 25 h, n = 77, daf-2(e1370), 49 h, n = 65, kat-1(n4430), 22 h, n = 78, kat-1(n4430);daf-2(e1370), 47 h, n = 58.
sir-2.1-overexpressing animals accumulated less intestinal lipofuscin than did the wild type (Fig. 4C), consistent with the slower rate of aging and extended lifespan of sir-2.1 overexpressors. Loss of kat-1 function suppressed the reduced lipofuscin accumulation of sir-2.1 overexpressors, as sir-2.1-overexpressing animals homozygous for a kat-1 loss-of-function mutation accumulated more lipofuscin than did the wild type, just like kat-1 mutants (Fig. 4C).
Aging mutants often show altered resistance to stress: sir-2.1 and daf-16 loss-of-function mutants are hypersensitive to heat stress, whereas daf-2 loss-of-function mutants and sir-2.1 overexpressors are stress resistant (6, 21) (Fig. 4 D and E). We tested the thermotolerance of kat-1 mutants, alone and in daf-2 loss-of-function and sir-2.1 overexpression backgrounds. kat-1 mutants showed an increased sensitivity to heat, and kat-1 was required for the stress resistance mediated by sir-2.1 overexpression (Fig. 4D). By contrast, kat-1 was not required for the increased thermotolerance of daf-2 mutants, as kat-1;daf-2 double mutants had increased stress tolerance similar to that of daf-2 single mutants (Fig. 4E). Our findings suggest that kat-1 function is specifically required for the effect of sir-2.1 overexpression on aging, stress resistance/thermotolerance, and lifespan.
Discussion
We isolated loss-of-function (lf) mutations in the C. elegans ketoacyl thiolase gene kat-1 in a genetic screen for animals that age prematurely. The kat-1 premature aging phenotype is reminiscent of that of the well-characterized C. elegans progeroid daf-16(lf) mutant: both kat-1 and daf-16 mutants precociously accumulate high levels of autofluorescence associated with the aging pigment lipofuscin, show early behavioral decline typical of old age, have premature tissue aging, and are short-lived.
kat-1 is dispensable for the regulation of lifespan by most known longevity pathways, including the insulin-like pathway and that involving mitochondrial function, but is required for the lifespan extension caused by the overexpression of sir-2.1. We previously showed that SIR-2.1 can bind to and activate the transcription factor DAF-16 in a manner dependent on stress and the 14-3-3 gene ftt-2 and acting in parallel to the insulin-like pathway (21). Like ftt-2, kat-1 is not required for the longevity and heat stress resistance mediated by low insulin-like signaling but is required for the longevity mediated by sir-2.1 overexpression. Recently, Gerhart–Hines et al. (22) demonstrated that mammalian SIRT1, the closest homolog of worm SIR-2.1, acts to increase fatty acid oxidation in mouse myocytes. Under conditions of nutrient deprivation, metabolism switches from glucose to fatty acid oxidation (23). It is plausible that sir-2.1, which is activated by a high NAD/NADH ratio (24), acts to induce and maintain fatty acid oxidation in response to limited food. For example, sir-2.1 might specifically activate a subset of daf-16 target genes that induce fatty acid oxidation. In this case, blocking fatty acid oxidation by reducing kat-1 function might preclude this metabolic switch, render the animals less resistant to metabolic stress, and accelerate aging. Alternative models are also possible. For example, kat-1 might function to promote sir-2.1 activity. Either way, our findings suggest that the fatty acid oxidation enzyme KAT-1 acts in the SIR-2.1 pathway to delay aging and provide resistance to stress.
Efficient Fatty Acid β-Oxidation Can Slow Aging.
Loss of function of the fatty acid oxidation thiolase gene kat-1 causes accelerated aging in C. elegans. This effect of kat-1 might be a consequence of an effect on fatty acid oxidation or might result from a kat-1 function distinct from fatty acid oxidation, such as a direct role in lipofuscin accumulation.
Mutations in kat-1 were previously identified in a screen for genes that regulate fat accumulation (18): whereas tub-1 mutants display a modest increase in fat storage, kat-1 tub-1 double mutants have significantly elevated fat accumulation. tub-1 encodes the C. elegans homolog of mammalian Tubby, which is involved in glucose and fat metabolism. Mice homozygous for a Tubby loss-of-function mutation fail to activate carbohydrate metabolism and instead rely on increased fat metabolism for their energy needs (25). It is possible that C. elegans tub-1 mutants have an increased rate of fat metabolism, which causes kat-1;tub-1 double mutants to have higher levels of incompletely-processed fatty acids than do kat-1 single mutants. That a loss of tub-1 function results in an extension of lifespan (26) is consistent with the hypothesis that increased fat oxidation delays C. elegans aging. Interestingly, a recent study by Wang et al. (27) shows that in C. elegans fat hydrolysis modulated by a specific fat lipase is important for the lifespan extension mediated by a lack of germ-line signaling. A kat-1 loss-of-function mutation partially suppressed the lifespan extension induced by a lack of the germ line in mes-1 mutants. These findings further support the hypothesis that efficient fat metabolism can delay C. elegans aging.
Numerous studies over the past 30 y have revealed an association between aging and decreased fatty acid oxidation. Triglycerides and various products of incomplete lipid oxidation accumulate during old age, leading to so-called lipotoxicity, and this accumulation is associated with age-related disorders, such as type 2 diabetes, metabolic syndrome, and Alzheimer's disease (28–30). That loss of function of KAT-1, an enzyme involved in fatty acid breakdown, leads to accelerated aging suggests a causal link between the decline in fatty acid β-oxidation and aging.
Materials and Methods
Strains.
We provide a thorough description of the C. elegans strains used in the SI Text.
Isolation of Mutants with Increased Lipofuscin Autofluorescence.
We mutagenized N2 hermaphrodites with ethyl methanesulphonate (EMS) as described by Brenner (31). We screened F2 animals equivalent to ≈20,000 mutagenized haploid genomes for high intestinal autofluorescence at day 2 or 3 of adulthood, using a dissecting microscope equipped with fluorescence optics; the filter set (Leica) was designed to detect low levels of lipofuscin autofluorescence: the excitation filter wavelength was 350–365 nm, and the emission filter range was 440–460 nm. For further details, see SI Text.
Mapping and Cloning.
We cloned the gene mutated in n4430 animals using a combination of visual mapping, polymorphism mapping, and RNAi. We injected a PCR product containing the entire T02G5.8 ORF and 2 kb of the upstream promoter region into n4430 animals and observed that transgenes containing the single gene T02G5.8 rescued the phenotype of fluorescence accumulation of n4430 mutants. For a detailed description of mapping and cloning, see SI Text.
Analyses of Lifespan, Stress Resistance, and RNAi.
Unless stated otherwise, lifespan assays were performed by standard methods at 20 °C using plates containing 10 μM FUdR. We used Prism 4 GraphPad software, which uses nonparametric log-rank test that compares strain distributions, to perform statistical analyses of survival curves. RNAi lifespan assays were performed according to the standard RNAi feeding protocol (32). See SI Text for details.
Heat-shock assays were performed at 32 °C using 1-d adults. For each genotype, 50–70 animals were transferred to 60-mm Petri plates seeded with OP50 bacteria, and viability was scored every 2–8 h after the shift to 32 °C. Statistical analysis was performed using Prism 4 software as for the lifespan assays.
Behavioral Assays.
Classification of aging animals into behavioral classes was done as described by Herndon et al. (11). At least 80 animals per genotype were assayed. Student's t test was applied to determine statistical significance of the differences.
Electron Microscopy.
Adult hermaphrodites of various ages were fixed in a mixture of 0.8% gluteraldehyde and 0.8% OsO4 in 0.1 M cacodylate buffer (pH 7.2) for 1 h at 4 °C in the dark. Worms were cut in this fixative using a razor blade; head regions were discarded, and animals were postfixed in 2% OsO4 in 0.1 M cacodylate buffer overnight at 4 °C. Four posterior regions were then aligned in each agar block, and the agar blocks were dehydrated and infiltrated into an Epon-Araldite mixture. Sections of the midbody region about 30-nM thick were collected on Pioloform-coated slotted grids. Sections were poststained with 1% uranyl acetate and Reynolds lead citrate. Sections were viewed using a JEOL 1200EX electron microscope at 80 kV.
Supplementary Material
Acknowledgments
We thank current and former members of the H.R.H. and L.G. laboratories for helpful discussions; Erika Hartwieg and Tori Hatch for performing electron microscopic studies; and David Harris, Brendan Galvin, and Hillel Schwartz for comments concerning the manuscript. Support for this work was provided by the Ellison Medical Foundation (A.B., S.N., K.B., and H.R.H.), and the National Institutes of Health and the Paul F. Glenn Foundation (L.G.). H.R.H is an investigator at the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013854107/-/DCSupplemental.
References
- 1.Guarente L, Kenyon C. Genetic pathways that regulate ageing in model organisms. Nature. 2000;408:255–262. doi: 10.1038/35041700. [DOI] [PubMed] [Google Scholar]
- 2.Braeckman BP, Houthoofd K, Vanfleteren JR. Insulin-like signaling, metabolism, stress resistance and aging in Caenorhabditis elegans. Mech Ageing Dev. 2001;122:673–693. doi: 10.1016/s0047-6374(01)00222-6. [DOI] [PubMed] [Google Scholar]
- 3.Gershon H, Gershon D. Caenorhabditis elegans—a paradigm for aging research: Advantages and limitations. Mech Ageing Dev. 2002;123:261–274. doi: 10.1016/s0047-6374(01)00401-8. [DOI] [PubMed] [Google Scholar]
- 4.Klass MR. A method for the isolation of longevity mutants in the nematode Caenorhabditis elegans and initial results. Mech Ageing Dev. 1983;22:279–286. doi: 10.1016/0047-6374(83)90082-9. [DOI] [PubMed] [Google Scholar]
- 5.Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- 7.Arantes-Oliveira N, Berman JR, Kenyon C. Healthy animals with extreme longevity. Science. 2003;302:611. doi: 10.1126/science.1089169. [DOI] [PubMed] [Google Scholar]
- 8.Klass MR. Aging in the nematode Caenorhabditis elegans: Major biological and environmental factors influencing life span. Mech Ageing Dev. 1977;6:413–429. doi: 10.1016/0047-6374(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 9.Collins JJ, Huang C, Hughes S, Kornfeld K. The measurement and analysis of age-related changes in Caenorhabditis elegans. WormBook. 2008:1–21. doi: 10.1895/wormbook.1.137.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garigan D, et al. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herndon LA, et al. Stochastic and genetic factors influence tissue-specific decline in ageing C. elegans. Nature. 2002;419:808–814. doi: 10.1038/nature01135. [DOI] [PubMed] [Google Scholar]
- 12.Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerstbrein B, Stamatas G, Kollias N, Driscoll M. In vivo spectrofluorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans. Aging Cell. 2005;4:127–137. doi: 10.1111/j.1474-9726.2005.00153.x. [DOI] [PubMed] [Google Scholar]
- 14.Clokey GV, Jacobson LA. The autofluorescent “lipofuscin granules” in the intestinal cells of Caenorhabditis elegans are secondary lysosomes. Mech Ageing Dev. 1986;35:79–94. doi: 10.1016/0047-6374(86)90068-0. [DOI] [PubMed] [Google Scholar]
- 15.Koenig H. Neuronal lipofuscin in disease. Its relation to lysosomes. Trans Am Neurol Assoc. 1964;89:212–213. [PubMed] [Google Scholar]
- 16.Brunk UT, Terman A. Lipofuscin: Mechanisms of age-related accumulation and influence on cell function. Free Radic Biol Med. 2002;33:611–619. doi: 10.1016/s0891-5849(02)00959-0. [DOI] [PubMed] [Google Scholar]
- 17.Yin D. Biochemical basis of lipofuscin, ceroid, and age pigment-like fluorophores. Free Radic Biol Med. 1996;21:871–888. doi: 10.1016/0891-5849(96)00175-x. [DOI] [PubMed] [Google Scholar]
- 18.Mak HY, Nelson LS, Basson M, Johnson CD, Ruvkun G. Polygenic control of Caenorhabditis elegans fat storage. Nat Genet. 2006;38:363–368. doi: 10.1038/ng1739. [DOI] [PubMed] [Google Scholar]
- 19.Aitlhadj L, Stürzenbaum SR. The use of FUdR can cause prolonged longevity in mutant nematodes. Mech Ageing Dev. 2010;131:364–365. doi: 10.1016/j.mad.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 21.Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- 22.Gerhart–Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ockner RK. Integration of Metabolism, Energetics, and Signal Transduction. New York: Kluwer/Plenum; 2004. [Google Scholar]
- 24.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 25.Wang Y, et al. Defective carbohydrate metabolism in mice homozygous for the tubby mutation. Physiol Genomics. 2006;27:131–140. doi: 10.1152/physiolgenomics.00239.2005. [DOI] [PubMed] [Google Scholar]
- 26.Mukhopadhyay A, Pan X, Lambright DG, Tissenbaum HA. An endocytic pathway as a target of tubby for regulation of fat storage. EMBO Rep. 2007;8:931–938. doi: 10.1038/sj.embor.7401055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang MC, O'Rourke EJ, Ruvkun G. Fat metabolism links germline stem cells and longevity in C. elegans. Science. 2008;322:957–960. doi: 10.1126/science.1162011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Montine TJ, Morrow JD. Fatty acid oxidation in the pathogenesis of Alzheimer's disease. Am J Pathol. 2005;166:1283–1289. doi: 10.1016/S0002-9440(10)62347-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy JK, Rao MS. Lipid metabolism and liver inflammation. II. Fatty liver disease and fatty acid oxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290:G852–G858. doi: 10.1152/ajpgi.00521.2005. [DOI] [PubMed] [Google Scholar]
- 30.Weinberg JM. Lipotoxicity. Kidney Int. 2006;70:1560–1566. doi: 10.1038/sj.ki.5001834. [DOI] [PubMed] [Google Scholar]
- 31.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamath RS, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.