Abstract
Considerable progress has been made in understanding variations in gene sequence and expression level associated with phenotype, yet how genetic diversity translates into complex phenotypic differences remains poorly understood. Here, we examine the relationship between genetic background and spatial patterns of gene expression across seven strains of mice, providing the most extensive cellular-resolution comparative analysis of gene expression in the mammalian brain to date. Using comprehensive brainwide anatomic coverage (more than 200 brain regions), we applied in situ hybridization to analyze the spatial expression patterns of 49 genes encoding well-known pharmaceutical drug targets. Remarkably, over 50% of the genes examined showed interstrain expression variation. In addition, the variability was nonuniformly distributed across strain and neuroanatomic region, suggesting certain organizing principles. First, the degree of expression variance among strains mirrors genealogic relationships. Second, expression pattern differences were concentrated in higher-order brain regions such as the cortex and hippocampus. Divergence in gene expression patterns across the brain could contribute significantly to variations in behavior and responses to neuroactive drugs in laboratory mouse strains and may help to explain individual differences in human responsiveness to neuroactive drugs.
Keywords: brain evolution, gene regulation, genetic background, neuroanatomy, species difference
There are numerous and complex mechanisms by which genetic variation within or across species contributes to phenotypic diversity (1–3). The most appreciated and easily testable of these involve sequence modifications that alter the amino acid coding regions of genes and measurably affect protein function. Quantitative differences in transcript abundance, due to gene dosage or regulatory mutations have also been associated with phenotype (4–6). However, outside the realm of developmental biology and recent studies of copy number variation (7, 8), the spatial distribution of gene expression within defined tissues and cell types remains relatively unexplored as a means by which genetic differences confer phenotypic differences.
To investigate the relationship between spatial gene expression patterns and genetic background, we conducted a large-scale, systematic, brainwide survey of gene expression (9) in seven inbred strains of adult male mice. The genealogies of inbred mouse strains have been characterized previously (10), and the strains selected for comparison span a broad range of genealogical relationships. Specifically, we analyzed three widely used, closely related Mus musculus inbred laboratory strains [C57BL/6J (C57), 129S1/SvImJ (129), DBA/2J (DBA)]; three wild-derived inbred M. musculus subspecies {M. m. domesticus [WSB/EiJ (WSB)], M. m. castaneus [CAST/EiJ (CAST)], M. m. musculus [PWD/PhJ (PWD)]}; and the wild-derived inbred species M. spretus [SPRET/EiJ (SPRET)]. The three M. musculus subspecies represent the ancestral founder strains (11) for the common laboratory strains, with WSB being most genetically similar to the laboratory strains as suggested by single nucleotide polymorphism (SNP) analysis (12, 13). SPRET was chosen as the most divergent laboratory mouse strain used in research.
Because the mouse is an important animal model for human disease and drug development, and because mouse strains respond differently, like human individuals, to pharmaceutical agents, expression patterns were examined for 49 well-known genes that represent sites of action for a wide range of existing therapeutic agents (Dataset S1). In addition to potential clinical relevance, the gene panel was functionally diverse and included genes encoding proteins involved in neurotransmitter synthesis, transport and signaling, G protein-coupled receptors, ion channels, and other intracellular signaling molecules. Cellular-level gene expression was assessed in brain sections using a standardized, automated platform for colorimetric in situ hybridization (ISH) (9), with data for each gene collected simultaneously in all strains to allow cross-strain comparison. In contrast to typical transcriptional profiling studies, which tend to analyze homogenates of gross brain structures comprising multiple cell types (14, 15), this approach revealed expression pattern differences in specific subnuclei and cell types. All ISH data are freely available online (http://mousediversity.alleninstitute.org).
Results
Interstrain Gene Expression Pattern Differences Are Frequent and Localized.
To elucidate differences in gene expression between mouse strains, data from over 15,000 brain sections were collected and manually analyzed. Gene expression was scored relative to the C57 reference strain for each gene in each strain in 203 distinct brain regions (Dataset S2).
Overall, individual genes exhibited similar global expression patterns across strains, indicating an expected high degree of conservation in overall brain organization (Fig. 1, left column).
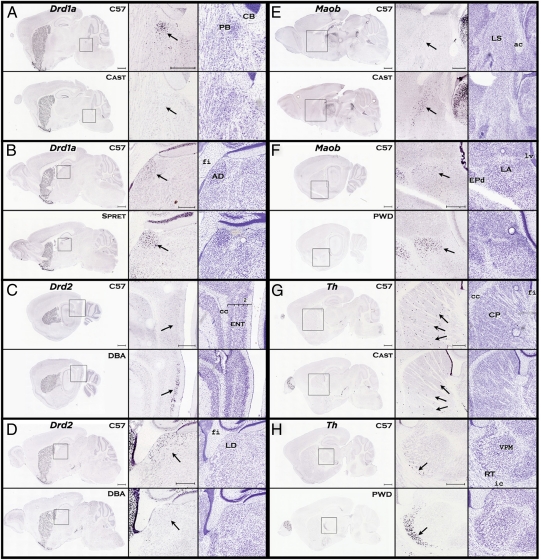
Fig. 1.
Strain differences in gene expression within members of the dopamine family. Each panel shows a comparison between the C57 reference strain and a strain that differed in expression pattern for a particular gene. The low magnification images at the Left of each panel provide anatomical context and show similar expression patterns in other brain areas. The Center column shows the expression pattern difference at high magnification, denoted by arrows. The column on the Right shows the nearest corresponding Nissl section for each strain. (A) The CAST strain shows lower density of Drd1a in the parabrachial (PB) nucleus. (B) The SPRET strain shows greater density of Drd1a expression in the anterodorsal (AD) nucleus of the thalamus. (C) The DBA strain shows greater density of Drd2 expression in the entorhinal area, lateral part (ENTl). (D) The DBA strain shows lower density of Drd2 expression in the lateral dorsal nucleus (LD) of the thalamus. (E) The CAST strain shows greater density of Maob expression in the lateral septal nucleus (LS). (F) The PWD strain shows greater density of Maob expression in the lateral amygdalar (LA) nucleus. (G) The CAST strain shows lower density of Th expression in the caudate-putamen (CP). (H) The PWD strain shows higher density of Th expression in the reticular nucleus (RT) of the thalamus. [Scale bars, (low magnification) 1,000 μm; (high magnification) 500 μm.] Drd1a, dopamine receptor 1a; Drd2, dopamine receptor 2; Th, tyrosine hydroxylase; Maob, monoamine oxidase b.
However, higher-resolution analysis reveals numerous striking, localized expression differences across all strains (Fig. 1 and Figs. S1 and S2). The interstrain differences were often robust and frequently found in small nuclei or sparse cell populations, such as the tyrosine hydroxylase (Th) positive striatal interneurons (Fig. 1G), that would likely be missed by traditional transcriptional analyses of gross brain regions. Variation in gene expression pattern was observed to differ across strain and structure and increased or decreased relative to the C57 reference strain, as demonstrated by nonuniform differences in several dopamine signaling pathway genes (Fig. 1 and Fig. S2). For example, the dopamine D2 receptor (Drd2) was clearly expressed in the entorhinal cortex of the DBA but not detected there in the C57 or 129 strains, whereas, the Drd2 transcript was expressed in the lateral dorsal thalamus of the C57 and 129 strains but was undetectable in the DBA strain (Fig. 1 C and D and Fig. S1).
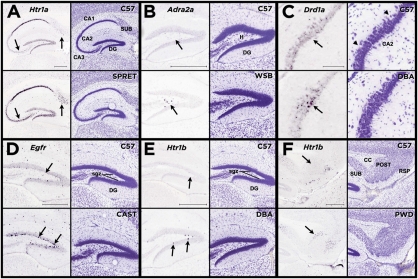
Interstrain differences in gene expression tended to occur at the level of specific cell classes, most clearly demonstrated by the stereotyped cellular architecture of the hippocampus (Fig. 2). For example, clear expression pattern differences were seen in pyramidal neurons in CA3 and CA2 subfields (Fig. 2 A and C), dentate gyrus hilar interneurons (Fig. 2B), and subgranular zone (Fig. 2 D and E) and the subiculum (Fig. 2 A and F) in a variety of distinct signaling pathways including the serotonergic, dopaminergic, and adrenergic systems.
Fig. 2.
Strain differences in gene expression within the hippocampal formation. Each panel shows a comparison between the C57 reference strain and a strain that differed in expression pattern for a particular gene. The Left column of each panel shows the difference in gene expression pattern, denoted by arrows, and the Right column shows the nearest corresponding Nissl section for each strain. (A) The SPRET strain shows greater density of Htr1a expression in the subiculum (Sub) and in cornu ammonis field 3, pyramidal layer (CA3). (B) The WSB strain shows greater density of Adra2a expression in the dentate gyrus, polymorph layer (DG). (C) The DBA strain shows greater density of Drd1a expression in the cornu ammonis field 2, pyramidal layer (CA2). (D) The CAST strain shows greater density of Egfr expression in the dentate gyrus, subgranular zone. (E) The DBA strain shows greater density of Htr1b expression in the dentate gyrus, subgranular zone. (F) The PWD strain shows greater density of Htr1b expression in the postsubiculum. [Scale bars, 500 μm; C (high magnification), 100 μm.] Htr1a, 5-hydroxytryptamine receptor 1A; Adra2a, adrenergic receptor, type 2a; Drd1a, dopamine receptor 1a, Egfr, epidermal growth factor receptor; Htr1b, 5hydroxytryptamine receptor 1B.
Remarkably, over 50% (26/49) of transcripts analyzed exhibited interstrain expression variations in one or more brain structures. In addition, variation occurred in many, but not all structures, with 60 of the 203 structures analyzed displaying at least one difference in expression pattern. Overall, these results may actually underrepresent the full magnitude of interstrain variance, as the results are strongly biased toward expression pattern rather than expression level differences, which are harder to detect reliably using colorimetric ISH. Additionally, stringent scoring criteria were used, counting only consistent, unambiguous differences, and accounting for cytoarchitectural differences between strains.
Extent of Interstrain Gene Expression Pattern Differences Reflects Genetic Divergence.
To elucidate potential organizing principles governing the relationship between genetic background and gene expression, we performed hierarchical clustering analysis on the matrix of expression differences across structure (203), strain (7), and gene (49). This analysis (Fig. 3 and Figs. S3 and S4) revealed clear strain relationships mirroring previously described genealogies (10, 12, 13). As expected, the 129 and DBA strains were the most similar in expression pattern compared with the closely related C57 reference strain. Also, M. m. domesticus (WSB) emerged as the subspecies most similar to C57, with M. m. musculus (PWD) and M. m. castaneus (CAST) as more distant subspecies, and M. spretus (SPRET) as the most divergent strain. These findings concur with prior work, which used SNP variation across the entire genome, identifying M. m. domesticus (WSB) as the primary ancestral contributor to the common laboratory strains (12, 13).
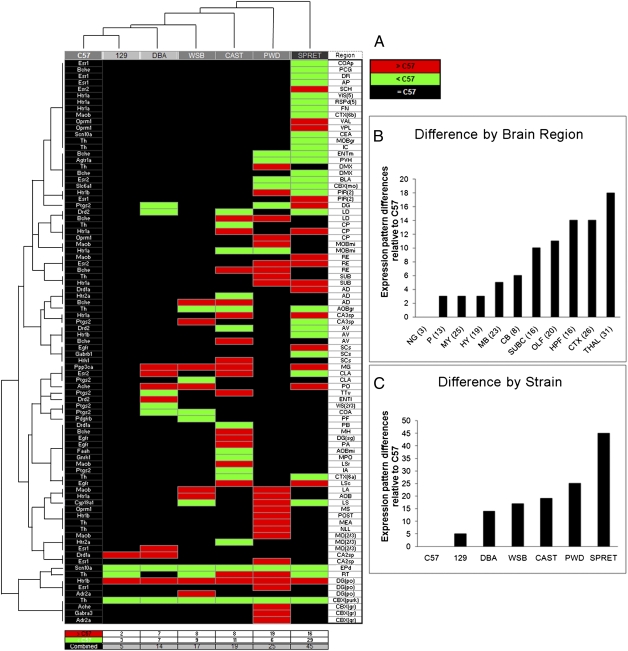
Fig. 3.
Drug target gene expression pattern differences across brain structures and strains. (A) Organized by unsupervised two-way hierarchical clustering, the matrix depicts the incidence between the genes that varied in expression pattern (Left column) and the brain structures (Right column) where they differed, in each strain (column headings), relative to the C57 reference strain. Relative expression differences were qualitatively binned as either decreased (green cells) or increased (red cells), with black cells indicating no difference. Two major groupings were returned, composed of the classic laboratory strains (129 and DBA) with M. m. domesticus subspecies (WSB), and that of subspecies M. m. castaneus (CAST), M. m. musculus (PWD) with the outgroup species M. spretus (SPRET). (B) Total number of differences across brain structures ranked across major brain divisions. The number of brain structures contained within each division are listed in parentheses. Gene and structure abbreviations are defined in Datasets S1 and S2. (C) Total strain differences in expression organized by rank order. Total differences also showed the 129 strain most similar to the C57, and SPRET the most different from the C57 reference strain.
Importantly, further analysis of the full dataset revealed a nonuniform distribution of gene expression pattern differences across the brain. Brain areas exhibiting the highest degree of interstrain expression variance were biased toward more recently evolved forebrain regions including the cortex and hippocampus (Fig. 3B and Fig. S3). These regions are most commonly linked to higher order functions such as cognition, learning, and memory. Interestingly, the thalamus, which is functionally and anatomically connected to the cortex, also contained many pattern variations between strains. In contrast, brain structures involved in autonomic functions and that are structurally well-conserved across mammalian species, such as the hypothalamus, brainstem, and medulla, exhibited the least divergence in gene expression patterns.
Discussion
The observed bias toward higher-order brain areas, combined with the alignment of genealogical distance with the degree of expression variance, has interesting implications for understanding evolutionary relationships and determining the functional relevance of genetic differences between individuals and species. Although genomic studies of brain evolution have identified a plethora of cross-species genetic differences, links between such differences and changes to brain-related phenotypes are lacking (16). The results presented here represent a step toward filling that gap, complementing genomic analyses with molecular phenotypic detail. The abundance and nature of observed gene expression differences, generally reflecting changes in discrete cell populations, suggest that phenotypic differences may be driven combinatorially by many small differences in the spatial regulation of gene expression. Functional implications for variations of expression patterns within structures noted in this study are suggested in reports that indicate a role for dopamine D2 receptor acting in entorhinal cortex during associative visual learning in nonhuman primates (17), as well as for variations in creativity correlated with receptor binding in the human thalamus (18). Furthermore, a recent study of striatopallidal neurons and dopaminergic regulation of GABAergic synaptic transmission highlights the functional relevance of the complex interplay of receptor expression, cell type, and circuitry within highly localized brain regions (19).
Many of the genes examined in this study encode neuropsychiatric drug targets, and thus may have already been selected through drug screening methods to be particularly dynamic in terms of behavioral or other phenotypic consequences. Indeed the expression patterns of these genes varied considerably across strains. Therapeutics affecting the dopamine and serotonin systems are among the most commonly prescribed neuroactive drugs, covering a variety of indications ranging from depression and schizophrenia to movement disorders. The dopamine D2 receptor alone is a site of action for a number of therapeutics and has roles in locomotion, emotion, cognition, and drug abuse (17, 20, 21). Differences in individual responses to such drugs have been observed in humans (22) and previous studies have shown that C57 and DBA mice respond differently to drugs known to affect dopamine signaling (23). These findings imply that expression pattern variation may contribute to individual differences in response to drug treatments in terms of efficacy and side effect profiles. However, other classes of genes also exhibited a high degree of variance, suggesting that these data may be generalizable. For example, prostaglandin-endoperoxide synthase 2 (Ptgs2), which encodes the analgesic target cyclooxygenase-2 (COX-2), was one of the most variant genes observed and exhibited complex expression across strains within hippocampal areas involved in learning, memory, and neurogenesis (Fig. S1).
Some qualifications and limitations of the approach should be noted in regards to sample size and qualitative analysis. One caveat is that our sampling interval (200 μm) necessarily limits the size (and therefore the number of brain regions) available for analysis. This is due to our requirement for two or more sections per nucleus in order to assess expression variation. Throughout the study, observations of all available sections were examined to take into account possible differential gradients in structures such as the caudoputamen and hippocampus. Ordinal variations in pattern were noted by analysts in light of substantial experience with the region of interest. This is not uncommon in descriptive reports of pattern variation (24), which require rigorous training and consensus among observers to ensure that expression differences reported are unlikely to be confounded by observer artifact. Thus, differences reported should be further confirmed by other techniques.
To corroborate certain differences found using the ISH technique, we conducted rtPCR experiments comparing strains across small and large brain areas with qualitatively scored greater and lesser ISH expression differences. We compared expression values of three gene/strain pairs in which ISH data had shown gene expression to be higher in one brain area and lower in another brain area, relative to the C57 strain (Figs. S1 and S5). Results in all comparisons indicated that relative levels of gene expression in the non-C57 strains showed at minimum an approximately twofold difference in the predicted direction (Dataset S3 and Fig. S6).
Whereas the current study focused on a set of patterned, functionally relevant genes, these results likely represent a microcosm of expression variation across the genome. The notable abundance and diversity of interstrain gene expression differences observed suggest that divergent expression patterns, reflecting gene regulation at the level of discrete cell populations, may be a significant and underappreciated contributor to phenotypic variation. More detailed cellular-level data will provide increased resolution for understanding and interpreting genetic networks governing phenotype in both an evolutionary and functional context and for optimizing selection of model systems for preclinical studies. From an experimental standpoint, proving a direct link between differential region- or cell type-specific expression and behavioral phenotype will be informative but challenging, as the differential regulation may involve cis-acting factors (25), trans-acting factors, or both (26). In addition, it will be important to explore the extent to which genotype-dependent expression patterns are seen across other tissues (27), species, and individuals in human populations.
Materials and Methods
Animals.
All mice were purchased from The Jackson Laboratory West. Mice used were as follows: three closely related inbred laboratory strains [C57BL/6J (C57), 129S1/SvImJ (129), DBA/2J (DBA)], three wild-derived inbred M. musculus subspecies [M. domesticus (WSB/EiJ), M. castaneus (CAST/EiJ), and M. musculus (PWD/PhJ)], and a wild-derived inbred species M. spretus (SPRET/EiJ). Brain samples were collected between 8:00 AM and noon following at least 4 d of acclimation to the facilities. Mice were group housed by strain in microventilated cages and uniformly handled. A 12-h light/dark cycle was maintained, with free access to water and Purina Lab Diet 5001 mouse diet. All procedures were approved by the Allen Institute Institutional Animal Care and Use Committee.
In Situ Hybridization.
A semiautomated high-throughput platform was used to perform colorimetric ISH on serially sectioned fresh frozen brain tissue. The methods and supporting data on the sensitivity, reproducibility, and interpretation of signal have been previously described for this platform (9, 28). Briefly, brains from age matched (56 ± 5 d) male animals were serially sectioned (sagittal plane, 25 μm thick, 200-μm interval) beginning at the lateral border of the hippocampus and ending ∼1 mm past the midline of the brain (24 brain sections/gene), and then labeled using riboprobes specifically designed for each gene transcript. Reference sections every 100 μm were stained with Nissl. All Nissl reference and ISH sections were digitally imaged as previously described (9, 28) at 10× magnification (approximate resolution; 1 μm2/pixel). For this study, experimental variance was minimized by having all sections from all strains processed together for a given gene, whenever possible. A subset of probes (20) were run again and analyzed independently to confirm positive and negative findings. Female C57 mice (age and handling matched) were used for five of these replicates (noted in Dataset S1).
Gene and in Situ Hybridization Probe Selection.
A total of 49 “drug targets” (DTs) were selected to comprise the gene panel on the basis of their current use as clinical therapeutic targets (Dataset S1) and known expression in the brain during adulthood and during development. Specific sequences of the riboprobes used to elucidate expression of each gene are available within the Web application containing the ISH data at http://mousediversity.alleninstitute.org/.
Analysis and Comparison of Gene Expression Differences Across Strains.
Rapid, systematic, manual analysis across the large-scale dataset (∼15,000 brain sections) was enabled by advances in automated digitization, storage, and rendering of ISH data. A computer application designed for this project allowed simultaneous comparison of expression data across up to all seven strains within specific brain regions. To establish expertise with regular patterns necessary for understanding qualitative comparisons, a range of architectural and expression characteristics were noted and considered inherent for each nuclei and population of interest. ISH image data were first matched across strains (simultaneously) at the brain region of interest. To mitigate variance in plane of section, sections were then examined in procession across nuclei. Differences in expression were assessed for possible strain cytoarchitectural variance in corresponding Nissl sections and a heat mask (a scaled color rendering of the data, based on a densitometric algorithm to show cell expression intensity, ref. 9) for validation. Comparisons were made relative to C57 without regard to strain identification, blind to any predictions, and without prior assumption.
The threshold for difference in expression was defined as a clear, qualitative difference unlikely due to any variance of the experiment, cytoarchitecture, or observer. Specifically, a higher or lower perceived ratio of cells expressing relative to the number of cells available to express (Nissl) in a given region was scored as difference in density. A perceived factor of ≈2 was considered the minimum difference. Additional weight in a decision was given to differences in cell staining intensity when corroborated by the heat mask. Structures were required to span at least two tissue sections to qualify for a difference call and in replicates if available. Because neural structures across the brain exhibited either presence or absence of expression, all experiments provided an internal control for consistency (Figs. S2 and S5). The primary reference used in the analysis was Allen Reference Atlas (ARA) (29) and structural names are reported following the ARA ontology (Dataset S2).
Scoring.
Differences were recorded (Fig. 3 and Fig. S1) as exhibiting either higher or lower density and intensity of expression relative to the C57 reference strain, with subsequent verification by a second observer, and final verification by an expert neuroanatomist (J.A.M.). Putative differences not observed in available replicates were excluded. Following analysis completion, a literature search showed no confound of sex for any strain differences noted in the replicates conducted in C57 females for five genes. Validated differences were then converted to binary scores (= or ≠ C57) and compiled in a strain–structure–gene (dimension 7 × 203 × 49) matrix for informatics analysis (Fig. 3). The results for hierarchical clustering patterns by strain as well as brain region were analyzed using R software.
Laser Capture Microdissection (LCM) and rtPCR Methods.
LCM.
Successive sections (14 μm thick) were collected throughout the extent of the nuclei in one hemisphere. Sections were frozen at −80, until light staining with Cresyl Violet (2-min protocol). Tissue availability limited the number of brains and structures available (n = 3 C57; n = 2 for other strains). Genes selected for assay showed weak and strong ISH expression (Th, Htr2a, Esr2, and Maob), across brain regions of widely varying sizes (CP, LD, AD, 6b, MO 2/3, MO 5, MO6, ENT, and SCH), in males of four strains (C57, DBA, CAST, and SPRET).
rtPCR.
RNA was isolated and purified from homogenized tissues using the MELT Total Nucleic Acid Isolation System (Ambion) on a MagMAX-96 (Thermo) instrument. RNA concentration was normalized to 5 ng·μL−1. Equal amounts (∼10 ng) of total RNA were used in each reverse transcription reaction using a Qiagen Sensiscript kit (Invitrogen). Each reaction was run in duplicate with reverse transcriptase (RT+) or without (RT−) to control for any potential genomic DNA contamination. Real-time PCR was conducted using cDNA (from 1/20th of the reverse transcription reactions) with gene-specific primer pairs (designed against the ISH probe used, intron spanning, without known SNPs) as well as a positive control primer pair for Gapdh, using SYBR Green PCR Master Mix (Roche). Each sample was run in four replicates on a Roche LightCycler real-time PCR system. On completion of a real-time quantitative PCR experiment, the thermal denaturation profile of the resulting amplicon was determined to make sure that the same specific amplicon was detected in different samples. Difference in number of cycles needed to reach threshold fluorescence with gene-specific primers as compared with Gapdh primers (ΔCp) was used as a measure of relative mRNA abundance (Dataset S3 and Fig. S6).
Supplementary Material
Acknowledgments
The authors thank the Allen Institute for Brain Science founders, Paul G. Allen and Jody Allen, for their vision, encouragement, and support. We are grateful to the Allen Institute Scientific Advisory Board and to Paul G. Allen for proposing the strain comparison project, and to Robert Williams and Daniel Ciobanu for providing generous advice and feedback on the project. J.S.T. is an investigator in the Howard Hughes Medical Institute. Research was supported by the Allen Institute for Brain Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003732107/-/DCSupplemental.
References
- 1.Koentges G. Evolution of anatomy and gene control. Nature. 2008;451:658–663. doi: 10.1038/451658a. [DOI] [PubMed] [Google Scholar]
- 2.Wu C, et al. Gene set enrichment in eQTL data identifies novel annotations and pathway regulators. PLoS Genet. 2008;4:e1000070. doi: 10.1371/journal.pgen.1000070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20:1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 4.Hovatta I, et al. DNA variation and brain region-specific expression profiles exhibit different relationships between inbred mouse strains: Implications for eQTL mapping studies. Genome Biol. 2007;8:R25. doi: 10.1186/gb-2007-8-2-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McClurg P, et al. Genomewide association analysis in diverse inbred mice: Power and population structure. Genetics. 2007;176:675–683. doi: 10.1534/genetics.106.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakobsson M, et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 7.Henrichsen CN, et al. Segmental copy number variation shapes tissue transcriptomes. Nat Genet. 2009;41:424–429. doi: 10.1038/ng.345. [DOI] [PubMed] [Google Scholar]
- 8.Cahan P, Li Y, Izumi M, Graubert TA. The impact of copy number variation on local gene expression in mouse hematopoietic stem and progenitor cells. Nat Genet. 2009;41:430–437. doi: 10.1038/ng.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 10.Beck JA, et al. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 11.Guénet JL, Bonhomme F. Wild mice: An ever-increasing contribution to a popular mammalian model. Trends Genet. 2003;19:24–31. doi: 10.1016/s0168-9525(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 12.Frazer KA, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- 14.Chesler EJ, et al. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–242. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 15.Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci USA. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vallender EJ, Mekel-Bobrov N, Lahn BT. Genetic basis of human brain evolution. Trends Neurosci. 2008;31:636–644. doi: 10.1016/j.tins.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Z, et al. DNA targeting of rhinal cortex D2 receptor protein reversibly blocks learning of cues that predict reward. Proc Natl Acad Sci USA. 2004;101:12336–12341. doi: 10.1073/pnas.0403639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Manzano O, Cervenka S, Karabanov A, Farde L, Ullén F. Thinking outside a less intact box: thalamic dopamine D2 receptor densities are negatively related to psychometric creativity in healthy individuals. PLoS ONE. 2010;5:e10670. doi: 10.1371/journal.pone.0010670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tecuapetla F, Koós T, Tepper JM, Kabbani N, Yeckel MF. Differential dopaminergic modulation of neostriatal synaptic connections of striatopallidal axon collaterals. J Neurosci. 2009;29:8977–8990. doi: 10.1523/JNEUROSCI.6145-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper JR, Bloom FE, Roth RH. The Biochemical Basis of Neuropharmacology. 8th Ed. New York: Oxford Univ Press; 2002. [Google Scholar]
- 21.Zhang Y, et al. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci USA. 2007;104:20552–20557. doi: 10.1073/pnas.0707106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nelson JC. A review of the efficacy of serotonergic and noradrenergic reuptake inhibitors for treatment of major depression. Biol Psychiatry. 1999;46:1301–1308. doi: 10.1016/s0006-3223(99)00173-0. [DOI] [PubMed] [Google Scholar]
- 23.Puglisi-Allegra S, Cabib S. Psychopharmacology of dopamine: The contribution of comparative studies in inbred strains of mice. Prog Neurobiol. 1997;51:637–661. doi: 10.1016/s0301-0082(97)00008-7. [DOI] [PubMed] [Google Scholar]
- 24.Campbell P, Reep RL, Stoll ML, Ophir AG, Phelps SM. Conservation and diversity of Foxp2 expression in muroid rodents: Functional implications. J Comp Neurol. 2009;512:84–100. doi: 10.1002/cne.21881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.GuhaThakurta D, et al. Cis-regulatory variations: A study of SNPs around genes showing cis-linkage in segregating mouse populations. BMC Genomics. 2006;7:235. doi: 10.1186/1471-2164-7-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stranger BE, et al. Population genomics of human gene expression. Nat Genet. 2007;39:1217–1224. doi: 10.1038/ng2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu X, Su Z. Tissue-driven hypothesis of genomic evolution and sequence-expression correlations. Proc Natl Acad Sci USA. 2007;104:2779–2784. doi: 10.1073/pnas.0610797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Visel A, Thaller C, Eichele G. GenePaint.org: An atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 2004;32(Database issue):D552–D556. doi: 10.1093/nar/gkh029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong HW. Allen Reference Atlas: A Digital Color Brain Atlas of the C57Bl/6J Male Mouse. Hoboken, New Jersey: John Wiley and Sons; 2008. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.