Abstract
Coxiella burnetii and Legionella pneumophila are evolutionarily related pathogens with different intracellular infection strategies. C. burnetii persists within and is transmitted by mammalian hosts, whereas, L. pneumophila is found primarily in the environment associated with protozoan hosts. Although a type IV secretion system encoded by the defect in organelle trafficking (dot) and intracellular multiplication (icm) genes is a virulence determinant that remains highly conserved in both bacteria, the two pathogens encode a different array of effector proteins that are delivered into host cells by the Dot/Icm machinery. This difference suggests that adaptations to evolutionarily distinct hosts may be reflected in the effector protein repertoires displayed by these two pathogens. Here we provide evidence in support of this hypothesis. We show that a unique C. burnetii effector from the ankyrin repeat (Ank) family called AnkG interferes with the mammalian apoptosis pathway. AnkG was found to interact with the host protein gC1qR (p32). Either the addition of AnkG to the repertoire of L. pneumophila effector proteins or the silencing of p32 in mouse dendritic cells resulted in a gain of function that allowed intracellular replication of L. pneumophila in these normally restrictive mammalian host cells by preventing rapid pathogen-induced apoptosis. These data indicate that p32 regulates pathogen-induced apoptosis and that AnkG functions to block this pathway. Thus, emergence of an effector protein that interferes with a proapoptotic signaling pathway directed against intracellular bacteria correlates with adaptation of a pathogen to mammalian hosts.
Keywords: Legionella pneumophila, ankyrin repeat proteins, gC1qR (p32), type IV secretion, pyroptosis
The bacterial pathogen Legionella pneumophila replicates inside protozoan hosts in soil and freshwater ecosystems (1). A type IV secretion apparatus encoded by the defect in organelle trafficking (dot) and intracellular multiplication (icm) genes is an essential virulence determinant used by L. pneumophila to deliver bacterial effector proteins into the eukaryotic cell (2, 3). The Dot/Icm effectors delivered into host cells by L. pneumophila play an important role in modulating host vesicular transport processes to facilitate the biogenesis of a vacuole that enables intracellular replication (4).
Although human infections sometimes can result in a severe pneumonia known as “Legionnaires’ disease” (5), under most conditions aerosolized L. pneumophila that gain access to the lungs of a mammalian host are detected and cleared without signs of clinical disease, indicating that the mammalian immune system can recognize this pathogen efficiently and mount a response that limits replication (6). One mechanism used by mammalian hosts to limit replication of L. pneumophila is to initiate a cell-death response shortly after intracellular infection. Mouse dendritic cells have been shown to block intracellular replication of L. pneumophila by rapidly activating a mitochondrial-dependent apoptosis pathway (7). Mouse macrophages and dendritic cells can independently restrict intracellular replication of L. pneumophila by sensing flagellin through the host sensors nucleotide-binding domain, leucine-rich repeat containing protein (NLR) family, caspase recruitment domains (CARD) domain-containing 4 (NLRC4) and NLR family apoptosis inhibitory protein 5 (Naip5) (8), which activate caspase-1 and trigger a form of cell death called “pyroptosis” (9–11). Activation of pyroptosis and apoptosis by L. pneumophila is observed only when cells are infected with bacteria having a functional Dot/Icm system, indicating that mammalian cells respond to the activities controlled by this secretion apparatus.
Coxiella burnetii is an intracellular pathogen of mammalian hosts that causes an acute disease, Q-fever. Phylogenetic analysis of rRNA sequences revealed that C. burnetii is evolutionarily related to L. pneumophila (12), and the families Legionellaceae and Coxiellaceae now comprise the order Legionellales. The genome sequence of C. burnetii confirmed this phylogenetic relationship and revealed that C. burnetii encodes a Dot/Icm apparatus that is similar in sequence and function to the Dot/Icm apparatus in L. pneumophila (13). More than 150 genes have been identified in L. pneumophila that encode effector proteins translocated into host cells by the Dot/Icm system (2, 3). Although some of the predicted C. burnetii effectors contain sequence motifs that are found in L. pneumophila effectors, there does not appear to be significant overlap in the repertoires of the Dot/Icm effectors in the genomes of these two organisms (13–16). This lack of overlap suggests that different selective pressures applied by their natural hosts may have shaped the repertoire of effector proteins in each pathogen.
C. burnetii has a robust ability to interfere with apoptosis upon infection of mammalian cells (17, 18), distinguishing it from L. pneumophila. If changes in the type IV effector repertoire are shaped in part by pathogen adaptations to new hosts, it is possible that C. burnetii might encode effectors that play a specific role in preventing apoptosis in mammalian cells. The only C. burnetii proteins that have been demonstrated to be delivered into host cells by the Dot/Icm system are those belonging to the Ankyrin repeat (Ank) family of effectors containing ankyrin repeat homology domains (ARHDs) (19, 20). The role and function of these effectors during bacterial infection remain unknown. Thus, in an effort to determine whether C. burnetii has acquired effectors with a potent ability to prevent cell death, we decided to test whether any of the Ank proteins might have the capacity to block apoptosis.
Results
AnkG Is an Effector That Interferes with Apoptosis.
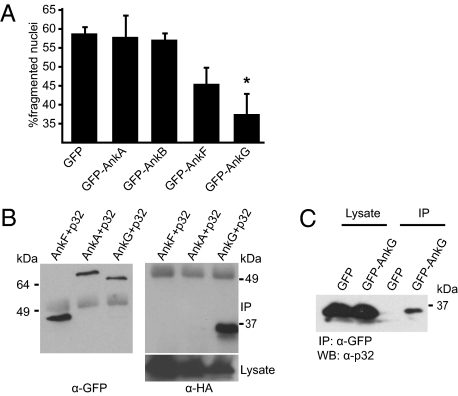
To determine if any of the C. burnetii Ank proteins shown previously to be translocated into host cells by the Dot/Icm system might be sufficient to interfere with host-cell apoptosis, we ectopically produced these proteins in CHO cells and measured apoptosis after stimulation with the proapoptotic agent staurosporine. Nuclear morphology visualized by DAPI staining revealed that the expression of GFP-AnkG protected cells from staurosporine-induced apoptosis (Fig. 1A). Conversely, expression of other C. burnetii Ank proteins was not sufficient to interfere with apoptosis when compared with the expression of GFP alone (Fig. 1A). These results suggest that AnkG is a C. burnetii effector protein that interferes with the apoptosis pathway.
Fig. 1.
AnkG interferes with apoptosis and binds the host protein p32. (A) CHO cells ectopically expressing GFP or the indicated GFP-tagged C. burnetii Anks were incubated with staurosporine. The nuclei were stained with DAPI, and nuclei of GFP-expressing cells were scored for fragmentation. One hundred nuclei from GFP-expressing cells per sample from four independent experiments were counted. *P < 0.001. (B) HEK 293 cells were cotransfected with plasmids encoding HA-tagged p32 and the indicated GFP-tagged C. burnetii ankyrin repeat-containing proteins. Proteins were precipitated from the cell lysates with an anti-GFP antibody and analyzed by immunoblot. The α-GFP blot shows Ank protein levels in the immunoprecipitate (IP), and the α-HA blot shows p32 levels in the immunoprecipitate (IP) and lysate. (C) HEK 293 cells were transfected with a plasmid encoding GFP-AnkG or the GFP control. Proteins were precipitated from the cell lysates with an anti-GFP antibody, and endogenous p32 was analyzed by immunoblotting with an anti-p32 antibody.
To understand how the AnkG protein prevents apoptosis, host proteins that interact with AnkG were identified by affinity chromatography (Fig. S1). The identity of host proteins that associated with AnkG, and not with GST alone, was determined by mass spectrometry and validated by coimmunoprecipitation studies. Testing interactions between GFP-tagged AnkG and candidate host proteins tagged with HA validated a complex containing AnkG and gC1qR (p32) (Fig. S1). The host protein p32 was found to interact with AnkG and not with other C. burnetii Ank proteins tested (Fig. 1B). An interaction between GFP-AnkG and endogenous p32 was also detected (Fig. 1C). Thus, the binding of p32 to AnkG is specific, and p32 binding is not a general property displayed by C. burnetii Ank proteins.
Inhibition of Apoptosis Correlates with AnkG Binding p32.
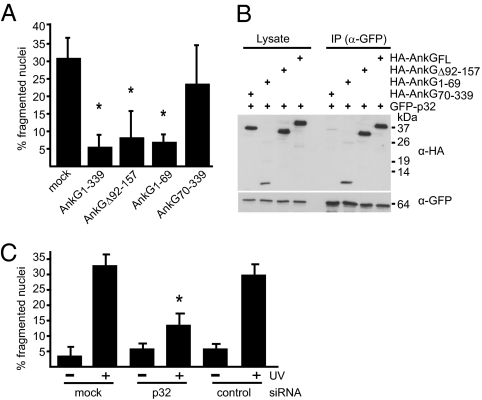
Truncation derivatives were used to identify the amino acid regions in AnkG required for inhibition of apoptosis. The production of HA-AnkG inhibited staurosporine-induced apoptosis (Fig. 2A), confirming the antiapoptotic activity observed for GFP-AnkG. The HA-AnkGΔ92–157 protein inhibited staurosporine-induced apoptosis, indicating that the ARHDs encoded in the deleted region were not essential for the antiapoptotic activity of AnkG (Fig. 2A). Production of HA-AnkG1–69 was sufficient to interfere with apoptosis, and production of HA-AnkG70–339 did not interfere with apoptosis (Fig. 2A). Similar deletion derivatives were used to identify the p32-interacting region in AnkG. Coprecipitation of HA-p32 was observed with GFP-AnkG1–339, GFP-AnkGΔ92–157, and GFP-AnkG1–69 but not with AnkG70–339 (Fig. 2B). Thus, the amino terminal AnkG1–69 region encodes a domain that is both necessary and sufficient for inhibition of apoptosis and p32 interaction.
Fig. 2.
The AnkG antiapoptotic domain mediates binding to the proapoptotic protein p32. (A) CHO cells ectopically expressing the indicated HA-tagged truncations of AnkG were incubated with staurosporine. The cells were stained with DAPI and HA-specific antibody. The nuclear morphology of HA-expressing cells was scored as in Fig.1A. *P < 0.003. (B) HEK 293 cells were cotransfected with plasmids encoding GFP-tagged p32 and the indicated HA-tagged truncations of AnkG. Proteins were precipitated from the cell lysates with an anti-GFP antibody. Immunoblot analysis was used to detect p32 (α-GFP) and AnkG (α-HA) in the lysates and immunoprecipitates (IP). (C) HeLa cells transfected with control siRNA (control), with p32 siRNA (p32), or untransfected (mock) were exposed to UV light as indicated. The cells were stained with DAPI, and nuclear morphology was scored. The percentage of cells with apoptotic nuclear morphology was calculated. One hundred nuclei per sample from three independent experiments were counted. *P < 0.006.
Reducing the levels of p32 in mammalian cells has been shown to make them more resistant to apoptosis (21, 22), suggesting that p32 has a proapoptotic activity. Consistent with these data, when p32 levels were reduced using siRNA (Fig. S2) and cells were exposed to UV light, apoptosis was diminished in the silenced cells (Fig. 2C). Taken together, these data suggest that AnkG may interfere with p32-dependent activities important for activating apoptosis.
Translocated AnkG Interferes with Pathogen-Induced Apoptosis.
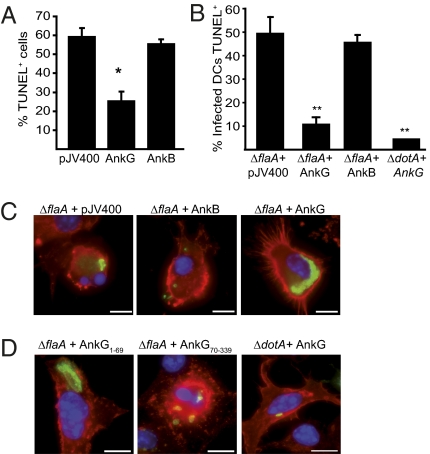
Although AnkG was able to interfere with apoptosis when overproduced in mammalian cell lines, it remained to be determined whether delivery of the native AnkG protein by the Dot/Icm system was sufficient to interfere with apoptosis during bacterial infection. Because the construction of an isogenic C. burnetii ankG mutant is not feasible, a gain-of-function analysis was conducted to determine if L. pneumophila producing AnkG could prevent apoptosis. Staurosporine-induced apoptosis was reduced significantly in CHO-FcR cells infected with L. pneumophila that produced the C. burnetii AnkG protein, whereas the majority of cells infected with L. pneumophila containing vector alone or containing vector producing the control C. burnetii AnkB protein showed signs of apoptosis, as determined by TUNEL staining (Fig. 3A). Thus, translocated AnkG is sufficient to block staurosporine-induced apoptosis in immortalized CHO FcR cells.
Fig. 3.
Translocation of AnkG prevents rapid pathogen-induced apoptosis, allowing L. pneumophila replication in DCs. (A) CHO-FcR cells infected with L. pneumophila harboring the vector pJV400, pJV400-AnkB (AnkB), or pJV400-AnkG (AnkG) were incubated with staurosporine. L. pneumophila was stained with a specific anti-Legionella antibody, and the nuclei were scored by the TUNEL assay. The nuclei of at least 100 infected cells per sample from three independent experiments were counted. *P < 0.01. (B) Bone marrow DCs derived from B6 mice were infected for 6 h with L. pneumophila ΔflaA expressing the pJV400 vector, AnkG, or AnkB or with L. pneumophila ΔdotA expressing AnkG. L. pneumophila was stained with a specific anti-Legionella antibody, and the nuclei of infected cells were scored by the TUNEL assay. Data shown are the mean ± SD of 300 cells counted per each coverslip in triplicate and are representative of two independent experiments. **P < 0.01. (C and D) Representative fluorescence micrographs of B6 DCs infected with (C) L. pneumophila ΔflaA + pJV400, ΔflaA + AnkB, or ΔflaA + AnkG and (D) L. pneumophila ΔflaA expressing AnkG1–69 or AnkG70–339 or L. pneumophila ΔdotA expressing full-length AnkG. DCs were fixed at 10 h postinfection and were stained with an antibody specific for MHC class II (red), DAPI (blue), and an anti-Legionella antibody (green).
L. pneumophila infection causes rapid apoptosis in mouse bone marrow-derived dendritic cells (DCs), and this pathogen-induced event limits the capacity of L. pneumophila to replicate in these cells (7). We reasoned that if the AnkG protein was a type IV effector with the ability to limit pathogen-induced apoptosis, then the addition of AnkG to the repertoire of L. pneumophila effector proteins might enable this pathogen to block apoptosis following infection of primary mouse DCs. A flagellin-deficient (ΔflaA) strain of L. pneumophila was used to avoid activating the caspase-1–dependent pyroptosis pathway of cell death regulated by the host sensors Naip5 and NLRC4. Approximately 50% of the DCs infected for 6 h with L. pneumophila ΔflaA containing vector alone (pJV400) had apoptotic nuclei as determined by TUNEL staining, whereas that percentage decreased significantly, to roughly 10%, in DCs infected with the isogenic L. pneumophila ΔflaA strain encoding AnkG (graphs in Fig. 3B and images in Fig. S3). Bacterial uptake was not affected by the addition of AnkG (Fig. S3), suggesting that the AnkG delivered by the L. pneumophila Dot/Icm system is able to disrupt pathogen-induced apoptosis in DCs.
DCs obtained from mice that have genetic alterations that interfere with induction of apoptosis have been shown to support the intracellular replication of L. pneumophila (7). If translocated AnkG were sufficient to limit pathogen-induced apoptosis, then L. pneumophila producing AnkG should gain the ability to replicate in DCs from wild-type mice. Consistent with this hypothesis, large vacuoles harboring replicating bacteria were abundant in DCs infected with L. pneumophila ΔflaA producing AnkG but not in DCs infected with L. pneumophila ΔflaA with vector alone or with a vector encoding AnkB from C. burnetii (images in Fig. 3C and graphs in Fig. S3). Bcl-2–associated X (Bax) and BCL2-antagonist/killer (Bak) are host pore-forming proteins that regulate cytochrome c release from mitochondria and are required for induction of pathogen-induced apoptosis (7, 23). Replication of L. pneumophila producing AnkG was not enhanced in DCs deficient in Bax and Bak compared with control DCs (Fig. S3), suggesting that these proteins are in the same pathway. This epistatic relationship supports a genetic interaction between AnkG and host proapoptotic regulatory factors.
Truncation derivatives of AnkG were used to map the region required for inhibition of pathogen-induced apoptosis. Using adenylate cyclase toxin fusion proteins to measure protein delivery into the cytosol, we determined that both truncation derivatives AnkG1–69 and AnkG70–339 were translocated into mammalian cells by the L. pneumophila Dot/Icm apparatus (Fig. S4). Replication in DCs was observed for L. pneumophila producing AnkG1–69 but not for L. pneumophila producing AnkG70–339 (images in Fig. 3D and graphs in Fig. S3). Thus, the p32-interacting region of AnkG is both necessary and sufficient for inhibition of pathogen-induced apoptosis.
Pathogen-Induced Apoptosis Is Regulated by p32.
Our results suggest that AnkG prevents pathogen-induced apoptosis by interfering with p32 function and thus predict that p32 would be required for restriction of L. pneumophila replication in mouse DCs. To test this hypothesis, p32 protein levels were reduced in mouse DCs using siRNA (Fig. S5). Pathogen-induced apoptosis was reduced in p32-silenced DCs infected with L. pneumophila ΔflaA to levels similar to those observed for L. pneumophila ΔflaA producing AnkG and the Dot/Icm-deficient mutant (ΔdotA) (Fig. 4A). Bacterial uptake was unaffected by DCs after silencing of p32 (Fig. S5). Importantly, the blocking of pathogen-induced apoptosis resulting from p32 silencing was sufficient to allow replication of L. pneumophila ΔflaA in mouse DCs (graphs in Fig. 4B and images in Fig. S5). The ability of L. pneumophila ΔflaA to replicate in DCs and the associated defects in pathogen-induced apoptosis that resulted from silencing of p32 with a siRNA pool also were observed using individual siRNAs directed against nonoverlapping regions of the transcript encoding p32 (Fig. S5), indicating that these effects are specific to the silencing of p32. Last, it was determined that wild-type L. pneumophila expressing flagellin were unable to replicate in DCs after p32 silencing, indicating that p32 is not important for the pyroptotic cell death pathway controlled by Naip5 and NLRC4 (Fig. 4C). Thus, p32 plays a critical role in the pathogen-induced apoptosis pathway resulting from L. pneumophila infection.
Fig. 4.
Rapid pathogen-induced apoptosis in DCs requires p32 function. (A) DCs untreated, treated with siRNA against p32, or mock treated were infected with L. pneumophila ΔflaA, ΔflaA expressing AnkG, or ΔdotA. L. pneumophila were stained with a specific anti-Legionella antibody, and the nuclei of infected cells were scored using DAPI staining. Data shown are from one experiment representative of five independent experiments that yielded similar results. n = 200 MHC class II-positive DCs. (B) DCs untreated (black bars), treated with siRNA against p32 (white bars), or mock treated (gray bars) were infected with L. pneumophila ΔflaA, ΔflaA expressing AnkG, or ΔdotA. Vacuoles containing replicating bacteria at 10 h postinfection were counted. Data shown are from one representative experiment representative of five independent experiments that yielded similar results. n = 200 MHC class II-positive DCs. N.D., vacuoles containing replicating bacteria were not detectable; R.V., vacuoles containing replicating bacteria. (C) DCs treated with p32 siRNA (Right) or mock treated (Left) were infected with L. pneumophila ΔflaA or with L. pneumophila WT. Vacuoles containing replicating bacteria at 10 h postinfection were counted. Data are shown from one experiment representative of two independent experiments that yielded similar results. n = 200 MHC class II-positive DCs.
Discussion
In addition to having a well-defined and critical role in development, apoptosis is emerging as an important innate immune response that enables cells damaged by either infectious agents or noninfectious pathological conditions to be cleared from the body. Although many intracellular pathogens inhibit apoptosis to enhance their intracellular survival and replication, the mechanisms that are used to interfere with apoptosis are not well understood. C. burnetii, the etiological agent of the human disease Q fever, was found to inhibit induction of apoptosis by targeting pathways upstream of cytochrome c release (17, 18). Although bacterial protein synthesis was shown to be required for inhibition of apoptosis, C. burnetii protein(s) involved in this process were not identified, and the mechanism by which they might function remained unknown. Here, we show that AnkG, a C. burnetii protein demonstrated to be a substrate of the Dot/Icm type IV secretion system (19, 20), had an activity that interfered with the induction of apoptosis.
The AnkG protein has a calculated mass of 39 kDa and is predicted to contain two ARHDs. Deletion derivatives revealed that the amino terminal 69 amino acids of AnkG, which did not contain either of the predicted ARHDs, were both necessary and sufficient for the antiapoptotic activity of this C. burnetii protein. This region of the AnkG protein was found to bind specifically to the mammalian protein p32. Thus, both the antiapoptotic activity and the p32-binding activity were localized to this amino terminal region in AnkG. The specificity of AnkG binding to p32 and the correlation between the region involved in p32 binding and inhibition of apoptosis strongly suggested that AnkG targeting of p32 could explain the mechanism by which AnkG interferes with host-cell apoptosis.
It has been speculated that p32 may participate in the regulation of apoptosis (24). Previous studies have shown that silencing of p32 reduced cisplatin-, harakiri-, and p14ARF-p16INK4a locus-induced apoptosis (21, 22, 25). Similarly, we found that p32 silencing conferred protection against UV-induced apoptosis. Our data also indicate that p32 may play a physiologically important role in regulating apoptosis in response to microbial infection. The bacterial replication and cell-death phenotypes observed when AnkG was added to the repertoire of L. pneumophila effector proteins were duplicated when p32 was silenced in mouse DCs. This result provides additional evidence that interfering with p32 function would be a possible mechanism to prevent pathogen-induced apoptosis from occurring during infection and that AnkG has evolved to function in this capacity.
Analysis of the genomes sequenced for L. pneumophila and C. burnetii revealed extensive plasticity in the type IV effector repertoires and conservation in components of the Dot/Icm apparatus (13–16). The versatility of the Dot/Icm apparatus in mediating the translocation of a large number of structurally diverse effector proteins into evolutionarily distinct eukaryotic host cells probably has contributed to the retention of the apparatus as members of the order Legionellales diverged. By contrast, effector plasticity probably is driven by the ease with which new genes obtained by horizontal transfer can be incorporated into the type IV effector repertoire and the ease with which effectors can be lost because of functional redundancy. As a result, L. pneumophila and C. burnetii now are remarkably different organisms, as reflected by an extensive nonoverlapping repertoire of effectors. Work presented in this study shows how the horizontal transfer of a single effector protein can result in a phenotypic change that enables L. pneumophila to replicate in a new eukaryotic host cell. In the example shown here, the protein AnkG was found to be an effector that inhibits apoptosis. Induction of apoptosis is an important host-cell defense mechanism of mammalian cells, the natural host for C. burnetii, but not of the protozoan host of L. pneumophila. Thus, AnkG illustrates how infection strategies for a specific host can be reflected in the repertoire of type IV effector proteins.
Materials and Methods
Reagents, Cell Lines, and Bacterial Strains.
Unless otherwise noted, chemicals were purchased from Sigma. Complete protease inhibitor and Fugene 6 were from Roche. Protein A and Protein G Sepharose were from Pharmacia. CHO and CHO-FcR fibroblasts were grown in alpha-MEM (Gibco) containing 10% heat-inactivated FBS. HeLa and HEK293 cells were maintained in DMEM containing 10% FBS. Bone marrow derived-DCs were prepared as described in ref. 7. Ted Hackstadt (NIH Rocky Mountain Laboratories, Hamilton, MT) generously provided a plaque-purified isolate of the C. burnetii phase II Nine Mile strain. Cultivation and host-cell infection by C. burnetii were conducted as described (17). Escherichia coli strains DH5α and BL21 (DE3) were cultivated in LB medium. L. pneumophila serogroup 1 strains were grown on charcoal yeast extract plates as described (19).
Mice.
C57BL/6 mice were purchased from Jackson Laboratories. All animals were maintained in accordance with the guidelines of the Yale Institutional Animal Use and Care Committee.
Plasmids and Primers.
Plasmids and primers used in this study are described in Tables S1 and S2.
Apoptosis Assays.
Details of the assays used to analyze apoptosis in the cells transfected with the indicated plasmids or infected with Coxiella burnetii or Legionella pneumophila are provided in SI Materials and Methods.
Assays to Measure Replication and Vacuole Formation in DCs.
Single-cell assays to measure uptake and formation of vacuoles that support L. pneumophila replication and cfu-based assays to measure intracellular replication of L. pneumophila in mouse DCs were conducted as described previously (7).
Coimmunoprecipitation, Immunoblotting, and siRNA Silencing.
Protein A Sepharose was used for all immunoprecipitations studies. ON-TARGETplus SMARTpools and the corresponding set of four individual siRNAs for siRNA silencing of p32 in human and mouse cells were purchased from Thermo Scientific. HeLa cell transfection of siRNA was conducted following the manufacturer's recommended protocol. DCs were electroporated with p32 siRNA as described previously (26). Details are provided in SI Materials and Methods.
Protein Purification.
AnkG-GST and GST were expressed in E. coli BL21 (DE3) and were purified by using glutathione–Sepharose columns (GE Healthcare). Proteins were coupled to Affi-Gel 15 (BioRad) and were incubated with CHO cell lysate. Bound proteins were eluted, separated by SDS/PAGE, and analyzed by MALDI-TOF. Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Eva Campodonico for editorial assistance, the Roy laboratory for helpful discussions, Jan Schulze-Lührmann for assistance with the RT-PCR analysis, and Claudia Giessler and Rita Eckart for technical assistance. This work was supported by a Fellowship from the Deutsche Forschungsgemeinschaft (to A.L.), by Ruth L. Kirschstein National Research Service Award Individual Postdoctoral Fellowship AI066547 (to K.L.C.), by Fundação para a Ciência e Tecnologia Grant BD/11758/2003 (to C.V.N.), and by National Institutes of Health Grants R01-AI064559 and R01-AI048770, and Northeast Biodefense Center Grant U54-AI057158-Lipkin (to C.R.R).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004380107/-/DCSupplemental.
References
- 1.Fields BS. The molecular ecology of Legionellae. Trends Microbiol. 1996;4:286–290. doi: 10.1016/0966-842x(96)10041-x. [DOI] [PubMed] [Google Scholar]
- 2.Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: Strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Ensminger AW, Isberg RR. Legionella pneumophila Dot/Icm translocated substrates: A sum of parts. Curr Opin Microbiol. 2009;12:67–73. doi: 10.1016/j.mib.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isberg RR, O'Connor TJ, Heidtman M. The Legionella pneumophila replication vacuole: Making a cosy niche inside host cells. Nat Rev Microbiol. 2009;7:13–24. doi: 10.1038/nrmicro1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fraser DW, et al. Legionnaires’ disease: Description of an epidemic of pneumonia. N Engl J Med. 1977;297:1189–1197. doi: 10.1056/NEJM197712012972201. [DOI] [PubMed] [Google Scholar]
- 6.Shin S, Roy CR. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 2008;10:1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- 7.Nogueira CV, et al. Rapid pathogen-induced apoptosis: A mechanism used by dendritic cells to limit intracellular replication of Legionella pneumophila. PLoS Pathog. 2009;5:e1000478. doi: 10.1371/journal.ppat.1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lightfield KL, et al. Critical function for Naip5 in inflammasome activation by a conserved carboxy-terminal domain of flagellin. Nat Immunol. 2008;9:1171–1178. doi: 10.1038/ni.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Molofsky AB, et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med. 2006;203:1093–1104. doi: 10.1084/jem.20051659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zamboni DS, et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol. 2006;7:318–325. doi: 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- 11.Ren T, Zamboni DS, Roy CR, Dietrich WF, Vance RE. Flagellin-deficient Legionella mutants evade caspase-1- and Naip5-mediated macrophage immunity. PLoS Pathog. 2006;2:e18. doi: 10.1371/journal.ppat.0020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisburg WG, et al. Phylogenetic diversity of the Rickettsiae. J Bacteriol. 1989;171:4202–4206. doi: 10.1128/jb.171.8.4202-4206.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seshadri R, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci USA. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chien M, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 15.Cazalet C, et al. Evidence in the Legionella pneumophila genome for exploitation of host cell functions and high genome plasticity. Nat Genet. 2004;36:1165–1173. doi: 10.1038/ng1447. [DOI] [PubMed] [Google Scholar]
- 16.Beare PA, et al. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun. 2009;77:642–656. doi: 10.1128/IAI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lührmann A, Roy CR. Coxiella burnetii inhibits activation of host cell apoptosis through a mechanism that involves preventing cytochrome c release from mitochondria. Infect Immun. 2007;75:5282–5289. doi: 10.1128/IAI.00863-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voth DE, Howe D, Heinzen RA. Coxiella burnetii inhibits apoptosis in human THP-1 cells and monkey primary alveolar macrophages. Infect Immun. 2007;75:4263–4271. doi: 10.1128/IAI.00594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voth DE, et al. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. 2009;191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sunayama J, et al. Physical and functional interaction between BH3-only protein Hrk and mitochondrial pore-forming protein p32. Cell Death Differ. 2004;11:771–781. doi: 10.1038/sj.cdd.4401418. [DOI] [PubMed] [Google Scholar]
- 22.Kamal A, Datta K. Upregulation of hyaluronan binding protein 1 (HABP1/p32/gC1qR) is associated with Cisplatin induced apoptosis. Apoptosis. 2006;11:861–874. doi: 10.1007/s10495-006-5396-4. [DOI] [PubMed] [Google Scholar]
- 23.Lindsten T, et al. The combined functions of proapoptotic Bcl-2 family members bak and bax are essential for normal development of multiple tissues. Mol Cell. 2000;6:1389–1399. doi: 10.1016/s1097-2765(00)00136-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jiang J, Zhang Y, Krainer AR, Xu RM. Crystal structure of human p32, a doughnut-shaped acidic mitochondrial matrix protein. Proc Natl Acad Sci USA. 1999;96:3572–3577. doi: 10.1073/pnas.96.7.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Itahana K, Zhang Y. Mitochondrial p32 is a critical mediator of ARF-induced apoptosis. Cancer Cell. 2008;13:542–553. doi: 10.1016/j.ccr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jantsch J, et al. Small interfering RNA (siRNA) delivery into murine bone marrow-derived dendritic cells by electroporation. J Immunol Methods. 2008;337:71–77. doi: 10.1016/j.jim.2008.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.