The cardiac IKs channel is a major repolarization current in the heart that responds rapidly and robustly to sympathetic nervous system stimulation to ensure adequate diastolic filling time in the face of accompanying accelerated heart rate. In cardiac myocytes, the IKs channel is a macromolecular complex composed of a pore-forming α (KCNQ1) subunit and modulatory β (KCNE1) subunit, as well as intercellular proteins critical for controlling the phosphorylation state of the complex (1). Although KCNQ1 alone assembles to form a voltage-gated potassium channel, the presence of KCNE1 is required to reproduce the kinetic properties of the native IKs channel, and KCNE1 coassembly is necessary to mediate the characteristic IKs response to sympathetic stimulation (2). The importance of the KCNE1 subunit is evidenced by its role in IKs channelopathies: mutation in either KCNQ1 or KCNE1 can underlie congenital long QT syndrome, and the presence of the β subunit is reported to be required for the gain-of-function phenotype in two KCNQ1 mutations implicated in congenital atrial fibrillation (3, 4). Among the questions surrounding the nature of the KCNQ1–KCNE1 association is that of the stoichiometry of the α and β subunits. Because of its homology with the remainder of the Kv channel superfamily, KCNQ1 (Kv7.1) is thought to form channels consisting of four α subunits (5). The number of KCNE1 subunits that can associate with the KCNQ1 tetramer, however, is a matter of some debate. In a study presented in PNAS, Nakajo et al. (6) confirm that KCNQ1 indeed forms tetrameric channels that can coassemble with one to four KCNE1 β subunits and show evidence for distinct gating properties associated with different stoichiometries.
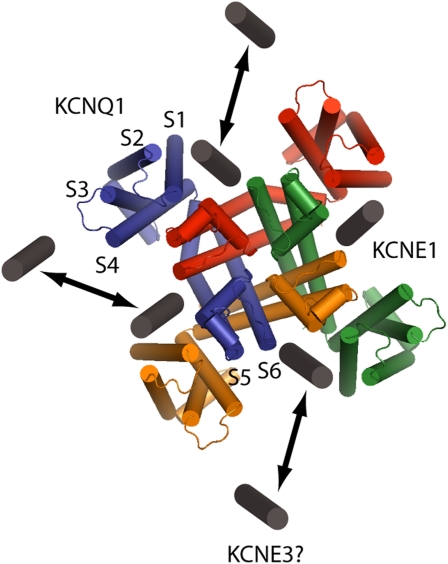
The study of KCNQ1–KCNE1 interactions began with the discovery that IKs is composed of KCNQ1 and KCNE1 subunits and that the functional characteristics of currents carried by KCNQ1 alone are very different from those mediated by KCNQ1–KCNE1 (7, 8). In fact, each member of the KCNE family (KCNE1 to –5) has now been shown to interact with KCNQ1, with strikingly different consequences for channel conductance and gating (9). Coassembly with KCNE1 uniquely reproduces the properties of the native IKs channel by increasing KCNQ1 channel conductance, slowing time course of activation, positively shifting voltage-dependent activation, and slowing the time course of deactivation. Although the presence of KCNE1 in the channel complex is vital to mediate kinetic properties important for physiology, it is also implicated in pathological states of the channel because point mutations in KCNE1 can confer defects in IKs that underlie heritable arrhythmias (10). Despite the dramatic changes KCNE1 imposes on KCNQ1, details of this interaction have been revealed slowly and remain a topic of interest. Efforts to locate the KCNE1 protein in the KCNQ1 channel complex have relied on biochemical methods because no crystal structure exists for KCNQ1 in the presence or absence of KCNE1. Early work showed that KCNE1 interacts directly with the S6 pore-forming helix of KCNQ1 (11). An emergent consensus based on homology modeling and disulfide crosslinking studies suggests that KCNE1 sits between the voltage-sensing domain and pore domain of adjacent KCNQ1 subunits (Fig. 1) (12–14) and that the intracellular end of the single transmembrane KCNE1 helix interacts with the KCNQ1 S4–S5 linker (15). Given this proposed binding site for KCNE1 within the KCNQ1 structure, it follows that four binding sites are possible within a fourfold symmetrical KCNQ1 tetramer.
Fig. 1.
The KCNQ1–KCNE1 channel complex. Top-down view of the homology structure of KCNQ1 from Kang et al. (14), with putative docking sites for one to four KCNE1 β subunits. Transmembrane spanning domains (S1…S6) labeled for one of four α subunits show the location of KCNE1 between voltage-sensing (S1–S4) and pore (S5, S6) domains of adjacent KCNQ1 subunits. KCNE3 may dock in the same cleft in KCNE1/KCNE3 heteromultimeric channels.
Previous investigations into the stoichiometry of KCNQ1–KCNE1 have either proposed a fixed KCNQ1:KCNE1 stoichiometry of 4:2 (16, 17) or that multiple stoichiometries are possible (18). Nakajo et al. present a new study (6) of the association between KCNQ1 and KCNE1 using a single-molecule fluorescence bleaching technique to count subunits. They are able to observe different numbers of bleaching steps in different single-channel complexes, suggesting a flexible stoichiometry in KCNQ1–KCNE1 channels, and they observe that the bleaching profile of these channels depends on the relative expression of the two genes. Importantly, data from this study suggest that IKs channel complexes are functionally different depending on their KCNQ1:KCNE1 stoichiometry, representing a unique putative mechanism of IKs regulation based on KCNE1 expression. In addition, these investigators find that addition of KCNE3 to channels with a set KCNQ1:KCNE1 stoichiometry of 4:2 reduces current, possibly by binding to one or both of the remaining unoccupied binding sites, thus extending the number of potential mechanisms for functional modulation of IKs within the heart to include addition of other KCNE proteins to the IKs channel complex.
These provocative results lead to a more complicated view of β subunit modulation of KCNQ1, raising new questions about how KCNE1 and other KCNE proteins assemble to make different flavors of IKs channels. One series of questions might focus on where and how KCNE2 to –5 assemble to modulate KCNQ1 channels. Are their differential effects a result of distinct binding sites on the KCNQ1 structure, or do all KCNE proteins bind in the same cleft and make subtly different interactions with the KCNQ1 channel to account for their different modulatory properties? Additionally, although the study by Nakajo et al. (6) illustrates the possible stoichiometries of this channel, it remains to be determined how IKs channel complexes form in the heart. In particular, is there heterogeneity in expression of KCNE1 among different cardiac cell types, and if so, would this lead to channels with tissue-specific functional properties? Might other KCNE proteins participate in these tissue-specific complexes? Might the makeup and thus the functional properties of IKs change in response to a particular disease state or in response to physiological stimuli? In fact, two studies looking at expression of these genes suggest that the ratio of mRNA for KCNQ1 and KCNE1 (as well as other KCNEs) may vary in a tissue-dependent manner within the heart (9, 19), perhaps offering a way for IKs to participate uniquely in repolarization for cell types with profoundly different action potential morphologies. The study by Nakajo et al. (6) demonstrates a unique mechanism for modulation of IKs current and as such adds to the understanding of the molecular interactions underlying the critical function of the KCNQ1–KCNE1 channel complex.
Footnotes
The authors declare no conflict of interest.
See companion article on page 18862.
References
- 1.Nerbonne JM, Kass RS. Molecular physiology of cardiac repolarization. Physiol Rev. 2005;85:1205–1253. doi: 10.1152/physrev.00002.2005. [DOI] [PubMed] [Google Scholar]
- 2.Marx SO, et al. Requirement of a macromolecular signaling complex for beta adrenergic receptor modulation of the KCNQ1-KCNE1 potassium channel. Science. 2002;295:496–499. doi: 10.1126/science.1066843. [DOI] [PubMed] [Google Scholar]
- 3.Chen YH, et al. KCNQ1 gain-of-function mutation in familial atrial fibrillation. Science. 2003;299:251–254. doi: 10.1126/science.1077771. [DOI] [PubMed] [Google Scholar]
- 4.Hong K, et al. De novo KCNQ1 mutation responsible for atrial fibrillation and short QT syndrome in utero. Cardiovasc Res. 2005;68:433–440. doi: 10.1016/j.cardiores.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 5.Long SB, Campbell EB, Mackinnon R. Crystal structure of a mammalian voltage-dependent Shaker family K+ channel. Science. 2005;309:897–903. doi: 10.1126/science.1116269. [DOI] [PubMed] [Google Scholar]
- 6.Nakajo K, Ulbrich MH, Kubo Y, Isacoff EY. Stoichiometry of the KCNQ1 − KCNE1 ion channel complex. Proc Natl Acad Sci USA. 2010;107:18862–18867. doi: 10.1073/pnas.1010354107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanguinetti MC, et al. Coassembly of K(V)LQT1 and minK (IsK) proteins to form cardiac I(Ks) potassium channel. Nature. 1996;384:80–83. doi: 10.1038/384080a0. [DOI] [PubMed] [Google Scholar]
- 8.Barhanin J, et al. K(V)LQT1 and lsK (minK) proteins associate to form the I(Ks) cardiac potassium current. Nature. 1996;384:78–80. doi: 10.1038/384078a0. [DOI] [PubMed] [Google Scholar]
- 9.Lundquist AL, et al. Expression of multiple KCNE genes in human heart may enable variable modulation of I(Ks) J Mol Cell Cardiol. 2005;38:277–287. doi: 10.1016/j.yjmcc.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 10.Moss AJ, Kass RS. Long QT syndrome: From channels to cardiac arrhythmias. J Clin Invest. 2005;115:2018–2024. doi: 10.1172/JCI25537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melman YF, Um SY, Krumerman A, Kagan A, McDonald TV. KCNE1 binds to the KCNQ1 pore to regulate potassium channel activity. Neuron. 2004;42:927–937. doi: 10.1016/j.neuron.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Chung DY, et al. Location of KCNE1 relative to KCNQ1 in the I(KS) potassium channel by disulfide cross-linking of substituted cysteines. Proc Natl Acad Sci USA. 2009;106:743–748. doi: 10.1073/pnas.0811897106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu X, Jiang M, Hsu KL, Zhang M, Tseng GN. KCNQ1 and KCNE1 in the IKs channel complex make state-dependent contacts in their extracellular domains. J Gen Physiol. 2008;131:589–603. doi: 10.1085/jgp.200809976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang C, et al. Structure of KCNE1 and implications for how it modulates the KCNQ1 potassium channel. Biochemistry. 2008;47:7999–8006. doi: 10.1021/bi800875q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lvov A, Gage SD, Berrios VM, Kobertz WR. Identification of a protein-protein interaction between KCNE1 and the activation gate machinery of KCNQ1. J Gen Physiol. 2010;135:607–618. doi: 10.1085/jgp.200910386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Kim LA, Rajan S, Xu S, Goldstein SA. Charybdotoxin binding in the I(Ks) pore demonstrates two MinK subunits in each channel complex. Neuron. 2003;40:15–23. doi: 10.1016/s0896-6273(03)00570-1. [DOI] [PubMed] [Google Scholar]
- 17.Morin TJ, Kobertz WR. Counting membrane-embedded KCNE beta-subunits in functioning K+ channel complexes. Proc Natl Acad Sci USA. 2008;105:1478–1482. doi: 10.1073/pnas.0710366105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang W, Xia J, Kass RS. MinK-KvLQT1 fusion proteins, evidence for multiple stoichiometries of the assembled IsK channel. J Biol Chem. 1998;273:34069–34074. doi: 10.1074/jbc.273.51.34069. [DOI] [PubMed] [Google Scholar]
- 19.Gaborit N, et al. Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J Physiol. 2007;582:675–693. doi: 10.1113/jphysiol.2006.126714. [DOI] [PMC free article] [PubMed] [Google Scholar]