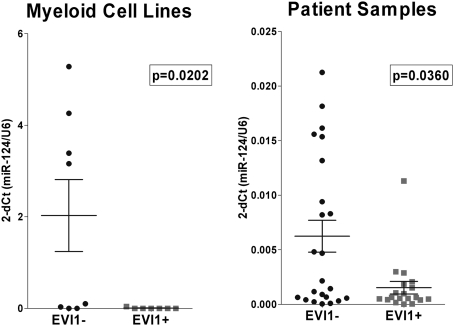
We read with great interest the work published by Dickstein et al. (1) showing that induced EVI1 expression in a murine model silences miR-124 expression by DNA methylation. The EVI1 gene codes for a transcription factor implicated in the development and progression of high-risk acute myeloid leukemia (AML) (2, 3). We quantify the expression of 250 microRNAs (miRNAs; TaqManHuman miRNA assay set) in 15 myeloid cell lines. Statistical analysis comparing cell lines with and without EVI1 protein identified miRNAs differentially expressed (B > 0). Among them, cell lines with EVI1 protein had no expression of hsa-miR-124, whereas most cell lines with no EVI1 had high hsa-miR-124 expression (Table 1). Interestingly, MEG-01 cells, with EVI1 overexpression and no protein, expressed low levels of hsa-miR-124 (Fig. 1 and Table 1). We first considered whether EVI1 was a target of hsa-miR-124. Transiently, transfection of pre–hsa-miR-124 in HEL and KU-812 cell lines, both with EVI1 protein and no hsa-miR-124 expression, showed a dramatic increase of hsa-miR-124; however, no changes in EVI1 expression either at the mRNA or protein level were detected. These results indicate that hsa-miR-124 does not regulate EVI1 expression. The results of Dickstein et al. (1) prompted us to examine whether DNA methylation could be responsible for the low expression of hsa-miR-124 in cell lines with EVI1 protein (1). We analyzed the methylation status of hsa-miR-124-1 by methylation-specific PCR as previously described (4). All of the cell lines analyzed that had low expression of hsa-miR-124 had the promoter methylated: four had EVI1, and three had no protein. Conversely, the two cell lines with high expression of hsa-miR-124 had no methylation and no EVI1 (Table 1). These results strongly support the hypothesis that EVI1 silences hsa-miR-124 expression by DNA methylation (1), although they would also indicate that, in some cases, the expression of hsa-miR-124 might be regulated by other mechanisms. To check the clinical importance of these results, we analyzed 42 AML patients, 19 of which had EVI1 overexpression (Table 1). Consistent with our results in cells lines, expression of EVI1 in patients was associated with decreased expression of hsa-miR-124 (P = 0.036), supporting that EVI1 could play a role in the transcriptional regulation of hsa-miR-124 (Fig. 1). Nevertheless, as in cell lines, some cases with low hsa-miR-124 expression had no EVI1 overexpression.
Table 1.
Quantification of the expression of hsa-miR-124 and methylation status of the hsa-miR-124-1 promoter region in 15 human myeloid cell lines
| Overexpression (qRT-PCR) |
||||||||||
| Cell line | Diagnosis | hsa-miR-124 (2-dct) | hsa-miR-124–1 (methylation status) | Evi1 protein | EVI1-1A | EVI1-1B | EVI1-1C | EVI1-1D | EVI1-3L | MDS1EVI1 |
| MV4-11 | AML-M5 | 4.2575 | UM | No | No | No | No | No | No | No |
| NOMO-1 | AML-M5 | 3.3870 | UM | No | No | No | No | No | No | No |
| MOLM-13 | AML-M5 | 3.1602 | nd | No | No | No | No | No | No | No |
| OCI-AML2 | AML-M4 | 5.2780 | nd | No | No | No | No | No | No | No |
| MEG-01 | CML-BP | 0.1022 | nd | No | No | Yes | No | No | No | No |
| KG-1 | AML-M6 | 0.0337 | M | No | No | No | No | No | No | No |
| Kasumi-1 | AML-M2 | 0.0000 | M | No | No | No | No | No | No | No |
| HL-60 | AML-M2 | 0.0000 | M | No | No | No | No | No | No | No |
| MUTZ-3 | AML-M4 | 0.0398 | M | Yes | Yes | Yes | Yes | Yes | No | No |
| KU-812 | CML-BP | 0.0009 | nd | Yes | No | Yes | No | No | No | No |
| TF-1 | AML-M6 | 0.0001 | M | Yes | Yes | Yes | Yes | Yes | No | No |
| HEL | AML-M6 | 0.0001 | M | Yes | Yes | Yes | Yes | Yes | No | No |
| K562 | CML-BP | 0.0001 | nd | Yes | Yes | Yes | No | No | No | Yes |
| F-36P | AML-M6 | 0.0001 | M | Yes | Yes | Yes | Yes | Yes | No | No |
| KYO-1 | CML-BP | 0.0001 | nd | Yes | No | Yes | Yes | No | No | No |
CML-BP, chronic myeloid leukemia blast phase; M, methylated; nd, no data; UM, unmethylated.
Fig. 1.
Diagram showing the expression of hsa-miR-124 in 15 myeloid cell lines and 42 samples of patients with AML. Data represents the mean ± SE.
Further studies are needed to fully clarify the role of hsa-miR-124 silencing in EVI1-positive AML. As shown in other malignancies, cyclin-dependent kinase 6 (CDK6) could be a target of hsa-miR-124 in AML. In the HEL cell line (hsa-miR-124-1–methylated and CDK6-expressed), we found that CDK4/6 inhibition by PD-0332991 induced dephosphorylation of retinoblastoma (Rb), with no changes in the protein levels of Rb or CDK6, and inhibited cell growth. These results may provide alternatives for future AML treatment. In conclusion, our results support the findings of Dickstein et al. (1), suggesting that a common mechanism for hsa-miR-124 down-regulation in hematological malignancies is the methylation of the promoter region of this gene and that EVI1 has a role in the transcriptional regulation of hsa-miR-124 expression.
Acknowledgments
This work was supported by Ministerio Ciencia Innovación (PI081687), Instituto de Salud Carlos III del Ministerio de Ciencia e Innovación–Red Temática de Investigación Cooperativa en Cáncer (RD06/0020/0078-RD06/0020/0111), and Union temporal de empresas (UTE) proyecto Centro de Investigación Médica Aplicada (CIMA).
Footnotes
The authors declare no conflict of interest.
References
- 1.Dickstein J, et al. Methylation and silencing of miRNA-124 by EVI1 and self-renewal exhaustion of hematopoietic stem cells in murine myelodysplastic syndrome. Proc Natl Acad Sci USA. 2010;107:9783–9788. doi: 10.1073/pnas.1004297107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nucifora G, Laricchia-Robbio L, Senyuk V. EVI1 and hematopoietic disorders: History and perspectives. Gene. 2006;368:1–11. doi: 10.1016/j.gene.2005.09.020. [DOI] [PubMed] [Google Scholar]
- 3.Wieser R. The oncogene and developmental regulator EVI1: Expression, biochemical properties, and biological functions. Gene. 2007;396:346–357. doi: 10.1016/j.gene.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 4.Agirre X, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–4453. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]