Abstract
Recognition and repair of cellular damage is crucial if organisms are to survive harmful environmental conditions. In mammals, the Keap1 protein orchestrates this response, but how it perceives adverse circumstances is not fully understood. Herein, we implicate NO, Zn2+, and alkenals, endogenously occurring chemicals whose concentrations increase during stress, in this process. By combining molecular modeling with phylogenetic, chemical, and functional analyses, we show that Keap1 directly recognizes NO, Zn2+, and alkenals through three distinct sensors. The C288 alkenal sensor is of ancient origin, having evolved in a common ancestor of bilaterans. The Zn2+ sensor minimally comprises H225, C226, and C613. The most recent sensor, the NO sensor, emerged coincident with an expansion of the NOS gene family in vertebrates. It comprises a cluster of basic amino acids (H129, K131, R135, K150, and H154) that facilitate S-nitrosation of C151. Taken together, our data suggest that Keap1 is a specialized sensor that quantifies stress by monitoring the intracellular concentrations of NO, Zn2+, and alkenals, which collectively serve as second messengers that may signify danger and/or damage.
Organisms in all kingdoms of life respond to environmental insults by up-regulating expression of cytoprotective genes encoding drug-metabolizing enzymes and antioxidant and repair proteins (1). This classic adaptation has been designated the environmental stress response (ESR) (2). The physiological significance of the ESR is profound; by minimizing cellular damage, it influences lifespan (3), plays a role in tumor suppression (4), and decreases susceptibility to degenerative diseases (5). Yet, the mechanisms by which mammalian cells recognize stress in order to initiate the ESR are still not fully understood.
The mammalian ESR is orchestrated primarily through two components, Kelch-like ECH-associated protein 1 (Keap1), a broad complex, tramtrack, bric-a-brac (BTB)-Kelch adaptor protein, and nuclear factor-erythroid 2-related factor 2 (Nrf2), a cap’n’collar (CNC) basic-region leucine zipper (bZIP) transcription factor that controls the expression of genes containing an electrophile response element (EpRE),* in their regulatory regions. Together, these two proteins form a negative-feedback loop, in which Keap1 acts as a stress sensor, whereas Nrf2 serves as the effector of the ESR. Thus, in the absence of stress, Keap1 represses Nrf2 by promoting its ubiquitylation upon the Cul3-Rbx1 ubiquitin ligase holoenzyme (6) but when Keap1 perceives stress, its ability to ubiquitylate and turnover Nrf2 is reduced, allowing the CNC-bZIP factor to accumulate and initiate the ESR (7).
The outstanding question regarding the function of Keap1 remains: How does it recognize environmental stress? The difficulty in answering this question lies in the number of stimuli that provoke the ESR. It has long been known that this response is stimulated by exposure to many electrophiles and heavy metals (8). Much less appreciated is the fact that the ESR is also initiated in response to physical insults, including heat, UV, and shear stress (9–11), and biotic stimuli, such as bacterial and viral infections (12, 13). The problem this diversity of stimuli presents for Keap1 has not been widely acknowledged because the protein is frequently viewed solely as a sensor of xenobiotic electrophiles. It has therefore been thought that the ability of such chemicals to covalently modify thiol groups provides a satisfactory explanation for the action of Keap1 (14). However, although it is true that most thiols in Keap1 become adducted when it is exposed to electrophiles in vitro (reviewed in ref. 15), it is unclear whether these modifications occur to a significant extent in vivo. Moreover, the hypothesis that Keap1 reacts directly with xenobiotic electrophiles fails to explain its responsiveness to physical and biotic stimuli or to heavy metals.
We sought a principle other than broad reactivity with electrophiles to explain how Keap1 perceives environmental stress. We postulated that Keap1 might have evolved to recognize macromolecular breakdown products or other endogenous chemicals that accumulate in stressed cells. Herein, we substantiate this idea by revealing that Keap1 possesses three sensors, one for each of the endogenous, stress-associated chemicals, nitric oxide (NO), Zn2+, and alkenals (a class of α, β-unsaturated carbonyls produced by lipid peroxidation).
Results
Evolution of Keap1 and Neofunctionalization.
It was previously reported that two paralogous families of Keap1 proteins exist, Keap1a and Keap1b (16). We extended this earlier work by examining a larger number of genomes (listed in Table S1), which revealed that Keap1 evolved in a common ancestor of bilateran organisms, and that the gene duplication event that produced the Keap1b paralogue occurred on the stem leading to vertebrates (Fig. S1 A and C). Crucially, our analysis has also revealed that the Keap1b family of proteins has gained a unique stress sensor (see later).
Selection of Chemicals to Investigate Stress Recognition by Keap1.
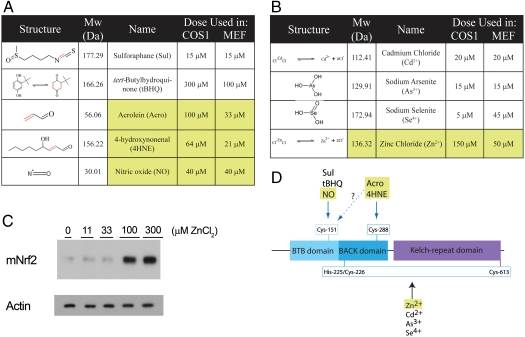
Our conclusions about how Keap1 perceives environmental stress in general were reached after studying how it recognizes in particular a select panel of five electrophiles (Fig. 1A) and four inorganic compounds (Fig. 1B). Five chemicals [Sulforaphane (Sul), tert-butylhydroquinone (tBHQ), Cd2+, As3+, and Se4+] are exogenous in origin. The remaining four chemicals [two alkenals—acrolein (Acro) and 4-hydroxy-2-nonenal (4HNE)—NO, and Zn2+] occur endogenously and accumulate in stressed cells (17–19). With the exception of Se4+, all chemicals chosen for study were previously shown to stimulate Nrf2 activity (8, 20–23), and we found that they all caused endogenous Nrf2 protein to accumulate in mouse fibroblasts [Fig. 1C (Zn2+) and Fig. S2, remaining chemicals, except NO (see later)].
Fig. 1.
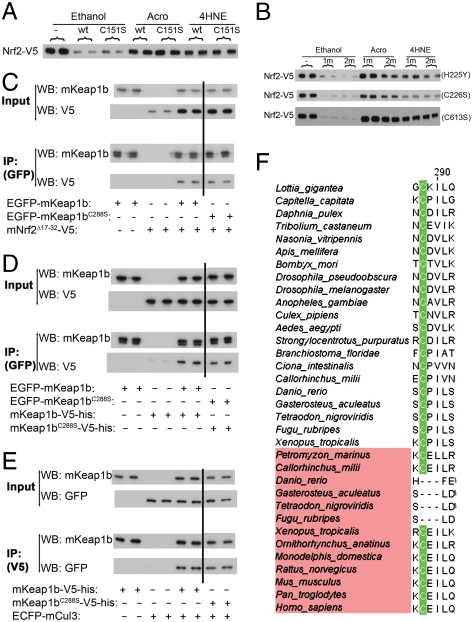
Chemical analysis of mKeap1b. Structures and concentrations of electrophiles (A) and inorganic compounds (B) used to treat cells. The chemical tBHQ is shown in equilibrium with its quinone derivative. Endogenously occurring chemicals are highlighted in yellow. (C) Samples prepared from mouse embryonic fibroblasts treated with the indicated doses of ZnCl2 for 2 h were blotted for Nrf2 and actin. D, A cartoon of mKeap1b indicating its BTB, BACK and Kelch domains, the three sensors identified in this paper, and the chemicals that trigger them. Although it is likely that Acro and 4HNE trigger the C151-based sensor, this conjecture remains to be formally proven.
In Vivo Modification of Mouse Keap1b at Two Sites.
We devised a mass-spectrometry assay, described in SI Text, section 1, to examine whether Keap1 cysteine thiols are modified in vivo by the selected compounds. The data for ectopic mouse (m)Keap1b purified from COS1 cells exposed to four electrophiles (Sul, tBHQ, Acro, 4HNE) and one nonelectrophile (Cd2+) are summarized in Fig. S3. Precursor ions derived from peptides covering 60% of the protein were identified by mass fingerprinting and sequenced.
Stress-elicited cysteine modifications were confined to two residues of mKeap1b (peptide sequencing data supporting this conclusion are provided in Fig. S4). First, C151 in the BTB domain of mKeap1b was altered in cells exposed to the electrophiles tBHQ, Acro, and 4HNE but not the heavy metal Cd2+. The isothiocyanate Sul also modified C151, though the product was too unstable to be detected by mass spectrometry. Instead, a biotin-switch technique (BST) was used to convert unstable cysteine adducts to stable biotin-cysteine adducts, and by precipitating biotinylated mKeap1b with streptavidin beads (Fig. S5A), we confirmed that mKeap1b was adducted in cells exposed to Sul; substitution of C151 with Ser suggested this single residue to be a major site of modification (Fig. S5B). Second, the alkenals, but not Sul or tBHQ, were also capable of modifying C288, a residue present in the BACK domain of mKeap1b.
Based on these chemical data, we hypothesized that C151 may sense several types of electrophile, whereas C288 regonizes only alkenals. As inorganic Cd2+ modified neither C151 nor C288, we surmised that it is sensed through some other mechanism. These inferences are summarized in the model of Keap1 sensing, shown in Fig. 1D, which is explored below.
Molecular Basis for the Reactivity of C151 with Electrophiles in Vivo.
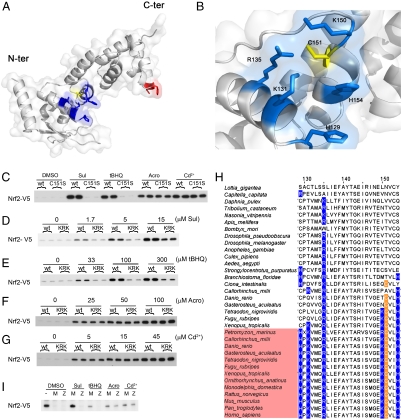
Although only confirmed for 4HNE, several observations suggest that all four tested electrophiles directly adduct to C151 (SI Text, section 2). We modeled amino acids 1–240 of mKeap1b and found that C151 was adjacent to H129, K131, R135, K150, and H154 (Fig. 2 A and B). This model suggests the reactivity of C151 is due to these proximate basic amino acids reducing the pKa of its thiol group through coulombic interactions, enabling it to exist as an anion at physiological pH.
Fig. 2.
Characterization of the C151-based sensor. (A) Homology model of amino acids 59–240 of mKeap1b. (B) A scaled-up view of C151 (yellow) highlighting its juxtaposition with H129, K131, R135, K150, and H154 (blue). (C–G) A plasmid expressing wild-type mKeap1b (WT), mKeap1bC151S (C151S), or mKeap1b bearing triple K131, R135, and K150 to Met substitutions (KRK) was cotransfected into COS1 cells with plasmids expressing mNrf2-V5 and β-galactosidase (β-gal) (a control for transfection efficiency). Blotting for mNrf2-V5 was performed 2 h after chemical treatment. The blots shown are typical of n≥3 independent experiments. (H) Sequence alignment of Keap1 homologues highlighting the amino acid signature of the C151-based sensor. Keap1b paralogues are named in pink. The remaining sequences are of Keap1a proteins. (I) An empty plasmid (−) or one expressing either mKeap1b (M) or zKeap1a (Z) was cotransfected into Keap1b-/- mouse embryonic fibroblast cells with plasmids expressing mNrf2-V5 and β-gal. Blotting for mNrf2-V5 was performed 2 h after chemical treatment (n = 2 independent experiments).
C151 and its five adjacent basic amino acids jointly comprise a sensor that is required for Keap1 to detect some—but not all—electrophiles. For example, as anticipated from previous work (24), mKeap1bC151S did not recognize Sul or tBHQ, and the mutant protein continued to turnover mNrf2-V5 in the presence of these electrophiles (Fig. 2C). Moreover, the ability of mKeap1bKRK (mKeap1b with K131, R135, and K150 replaced by Met residues) to sense both electrophiles was blunted in comparison with the wild-type protein (Fig. 2 D and E), even though it retained C151. In contrast, however, mKeap1bC151S remained fully responsive to Acro (Figs. 2C), as was mKeap1bKRK (Fig. 2F). Finally, inorganic Cd2+ inactivated Keap1 regardless of the presence or absence of C151 and its adjacent basic residues (Fig. 2 C and G).
The modeled structure of the mKeap1b BTB domain suggests the protein fold surrounding C151 might be relatively unstable because of charge repulsion between the proximate basic residues. The thiolate anion presumably stabilizes this protein fold through its energetically favorable interactions with these cations. It follows that removal of the negative charge, via adduction of C151 with electrophiles, is likely to lead to local structural changes in mKeap1b that might prevent ubiquitylation of Nrf2. Consistent with this model, we found that replacing C151 with an Ala (which, unlike Ser, cannot engage the surrounding basic residues in coulombic interactions) generates a nonfunctional mKeap1b molecule that fails to destabilize mNrf2-V5 in Keap1b-/- fibroblasts (Fig. S6A). This failure occurred despite mKeap1bC151A being able to homodimerize, interact with Cul3, and recruit the CNC-bZIP factor (Fig. S6 B–D). We conclude that a thiolate anion at position 151 in mKeap1b is essential for Nrf2 turnover, though a partial negative charge also appears to suffice. Elimination of this negative charge by chemical adduction inactivates mKeap1b.
Nitric Oxide is an Endogenously Produced Trigger for the C151-Based Sensor.
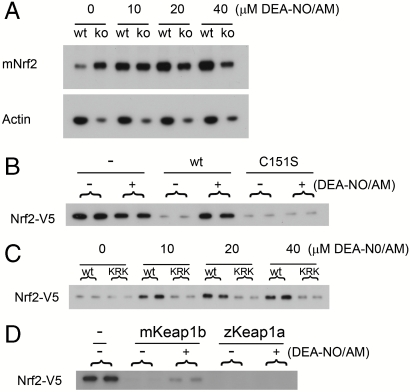
We found that acetoxymethylated diethylamine-NONOate (DEA-NO/AM), a cell-permeable NO donor molecule, produced a dose-dependent increase in the level of Nrf2 protein in wild-type fibroblasts but not in Keap1b-/- fibroblasts (Fig. 3A). Although mass spectrometry failed to reveal any modified mKeap1b thiol groups after treatment of COS1 cells with DEA-NO/AM, use of the BST revealed an unstable modification of C151 (Fig. S5B). Although the ability of the BST to discriminate between S-nitrosothiols and other thiol modifications has been questioned (25), these data are most simply explained by the suggestion that mKeap1b perceives NO through S-nitrosation of C151. Consistent with this hypothesis, we found that mNrf2-V5 failed to accumulate in response to DEA-NO/AM in cells expressing mKeap1bC151S (Fig. 3B) or mKeap1bKRK (Fig. 3C). S-nitrosation of C151 presumably proceeds by transnitrosation, possibly via dinitrosyliron complexes (26).
Fig. 3.
NO is an endogenous trigger of the C151-based sensor. (A) WT or Keap1b-/- (ko) mouse embryonic fibroblast (MEF) cells were exposed to DEA-NO/AM for 2 h before blotting for Nrf2 or actin. (B and C) A plasmid expressing mKeap1b (WT), mKeap1C151S (C151S), or mKeap1b bearing triple K131, R135 and K150 to Met substitutions (KRK) was cotransfected into COS1 cells with plasmids expressing mNrf2-V5 and β-galactosidase (β-gal). Cells were treated with DEA-NO/AM for 2 h before blotting for mNrf2-V5. (D) An empty plasmid (−) or one expressing mKeap1b or zKeap1a was cotransfected into Keap1b-/- MEF cells with plasmids expressing mNrf2-V5 and β-gal. Blotting for mNrf2-V5 was performed 2 h after DEA-NO/AM treatment. All blots are typical of n≥3 independent experiments, except for blot D where n = 2.
C151-Based Sensor Evolved in Vertebrates to Sense NO.
Although C151 in mKeap1b reacts with Sul, tBHQ, Acro, 4HNE, and NO, two separate lines of evidence suggest that it evolved to sense NO rather than electrophiles in general.
Firstly, NO is a more specific trigger of the C151-based sensor than is Sul, as it forms adducts with C151 without modifying other cysteines in mKeap1b or in other proteins (Fig. S5 B and C). In contrast, adduction of C151 by Sul was accompanied by modification of many other cysteines, not only in mKeap1b but in numerous other proteins, also (Fig. S5 B and C). Moreover, the environment surrounding C151 in mKeap1b shows evidence of selection to facilitate specifically S-nitrosation of the thiol; the ability of NO—but not Sul or tBHQ—to trigger the C151-centered sensor was absolutely dependent upon the presence of K131, R135, and K150 (compare Fig. 2 D and E with Fig. 3C).
Secondly, the set of amino acids that collectively form the C151-based sensor is only found in Keap1b proteins (Fig. 2H), suggesting that it is absent from the more ancient Keap1a family. This hypothesis was confirmed for zKeap1a, which did not perceive Sul or tBHQ (Fig. 2I) or NO (Fig. 3D). The C151-based sensor is therefore a recent evolutionary adaptation that is unlikely to have evolved to sense electrophiles in general, because such chemicals have existed in the environment since long before the sensor first appeared. Instead, the origin of this sensor coincides with a vertebrate-specific expansion of the neuronal nitric oxide synthase (nNOS) gene (Fig. S1 B and C). As expansion of the NOS family was accompanied by a more pervasive role for NO as a signaling molecule (18), we suggest that the C151-based sensor evolved specifically to recognize NO. Therefore, we refer to it as the NO sensor.
Keap1 is a Zn2+ Sensor.
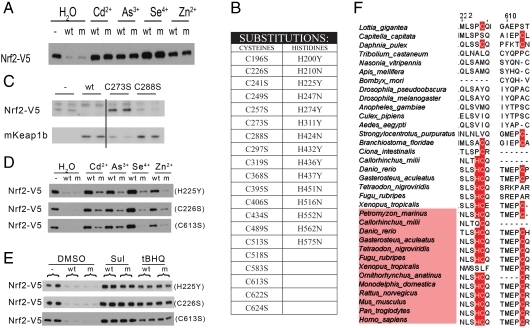
The existence of a second sensor in mKeap1b dedicated to recognizing certain metal(loid)s is clear from our finding that C151 is not required for the protein to sense Cd2+, As3+, Se4+, and Zn2+ (Fig. 4A). It was previously reported that the protein coordinates Zn2+ in a 1∶1 complex (27), suggesting to us that Keap1 contains a single Zn2+ sensor. Three-dimensional reconstruction from single particle EM images has revealed that Keap1’s BACK and Kelch domains form a single globular structural unit (28). We identified a center for coordination chemistry within this unit by systematically substituting noncoordinating amino acids (Fig. 4B) for each of the Cys and His residues in both domains. As expected, this procedure yielded a number of inactive mutants, mKeap1bC249S, mKeap1bC273S (Fig. 4C), mKeap1bC288S (Fig. 4C), and mKeap1bH311Y, which were uninformative. However, characterization of the remaining substituted proteins (Fig. 4D and Fig. S7) revealed three residues, H225, C226 and C613, that were individually required for mKeap1b to sense Cd2+, As3+, Se4+, and Zn2+ (Fig. 4D), but not Sul nor tBHQ (Fig. 4E). The H225/C226 dyad is represented in red in our partial model of mKeap1b (Fig. 2A), where the two residues are only 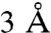 apart. Although this dyad and C613 cannot be located precisely within the Keap1 EM structure, they must exist in close proximity as an intramolecular disulfide bond forms between C226 and C613 when cells are treated with hydrogen peroxide (29).
apart. Although this dyad and C613 cannot be located precisely within the Keap1 EM structure, they must exist in close proximity as an intramolecular disulfide bond forms between C226 and C613 when cells are treated with hydrogen peroxide (29).
Fig. 4.
The Zn2+ sensor. (A) An empty plasmid (−) or one expressing mKeap1b (WT), or mKeap1bC151S (m) was cotransfected into COS1 cells with plasmids expressing mNrf2-V5 and β-galactosidase (β-gal). Blotting for mNrf2-V5 was performed 2 h after chemical treatment. (B) Amino acid substitutions introduced into the BACK and Kelch domains of mKeap1b. (C) An empty plasmid (−) or one expressing mKeap1b (WT), mKeap1bC273S (C273S), or mKeap1bC288S (C288S) was cotransfected into Keap1b-/- mouse embryonic fibroblast cells with plasmids expressing mNrf2-V5 and β-gal. Blotting was performed 24 h later. Two lanes have been cropped from the middle of these blots, indicated by the vertical line, as they are not relevant to this paper. (D and E) An empty plasmid (−) or one expressing either mKeap1b (WT), or a mutated form of mKeap1b (m) was cotransfected into COS1 cells with plasmids expressing mNrf2-V5 and β-gal. Cells were treated with chemicals for 2 h before blotting for the stated proteins. Three separate substitutions were evaluated (H225Y, C226S, and C613S). (F) The amino acid signature of the Zn2+ sensor is highlighted in red. All immunoblots shown are representative of n≥3 independent experiments.
Our data define a previously unrecognized metal(loid) sensor in mKeap1b that minimally comprises the H225/C226 dyad and C613. It enables mKeap1b to recognize Cd2+, As3+, Se4+, and Zn2+, presumably through coordination chemistry. Because Zn2+ is endogenous to cells, it is the most likely trigger of this sensor, for which reason we designate it the Zn2+ sensor. Phylogenetic data (Fig. 4F) suggest the Zn2+ sensor evolved prior to the appearance of the Keap1b family; it thus predates the NO sensor and this hypothesis was confirmed by demonstrating that zKeap1a recognizes Cd2+ (Fig. 2I).
Alkenal Sensor.
Mutational ablation of the NO sensor, either alone (Fig. 5A) or in conjunction with the Zn2+ sensor (Fig. 5B), had no impact upon the ability of mKeap1b to recognize Acro and 4HNE, suggesting the BTB-Kelch protein possesses a third sensor. C288 was previously implicated in stress-sensing by Keap1 (30) and we now suggest that it is an alkenal sensor because it was found to be modified specifically by this class of chemical (Figs. S3 and S4).
Fig. 5.
The alkenal sensor. (A) An empty plasmid (−) or one expressing mKeap1b (WT), or mKeap1bC151S (C151S) was cotransfected into COS1 cells with plasmids expressing mNrf2-V5 and β-galactosidase (β-gal). Blotting for mNrf2-V5 was performed 2 h after treatment of cells with chemicals. (B) An empty plasmid (−) or one expressing mKeap1bC151S (m) or mKeap1bC151S bearing an additional second substitution (2 m) was cotransfected into COS1 cells with plasmids expressing mNrf2-V5 and β-gal. Blotting for mNrf2-V5 was performed after cells had been treated with the stated chemicals for 2 h; the second substitution utilized in each experiment is identified to the right of each blot. (C–E) The indicated combinations of proteins were expressed in COS1 cells and EGFP-tagged or V5-tagged material was immunoprecipitated. Immunoprecipitates (IP) and input samples were blotted with the indicated antibodies. (F) C288 is highlighted in this sequence alignment. All immunoblots shown are representative of n≥3 independent experiments.
It is not possible to assess directly what impact chemical modification of C288 in mKeap1b has on Nrf2 turnover, because replacing this amino acid with a Ser inactivates the protein (Fig. 4C). Nevertheless, the fact that genetic modification of this residue prevents Nrf2 turnover by Keap1 suggests that chemical modification of C288 should do so also. This assertion is justified by the finding that mKeap1C288S folds correctly; it is indistinguishable from the wild-type protein in terms of its ability to dimerize and interact with both Nrf2 and Cul3 in cell lysates (Fig. 5 C–E).
We conclude that chemical modification of C288 suffices to prevent Nrf2 turnover. For this reason, we refer to it herein as the C288 alkenal sensor. Its importance is underscored by its early appearance in an ancestor of all bilaterans, and its retention in the Keap1 proteins of nearly all descendents examined (Fig. 5F).
Discussion
In this paper, we propose a general solution to the problem of how Keap1 perceives environmental stress, which involves alkenals, Zn2+ and NO serving as second messengers for cell damage or danger. Collectively, these three chemicals are well placed to serve such a role for the simple reason that their accumulation is a quasi-universal occurrence in cells exposed to environmental insults. For example, alkenals are produced upon lipid decomposition, which is an unavoidable consequence of cell stress (17). Similarly, free Zn2+ levels become elevated in cells following exposure to numerous chemical stressors (19), and when cells are exposed to physical and biotic stimuli (31). The source of this free Zn2+ appears to be oxidation and degradation of Zn2+-containing proteins (19). Lastly, NO is purposely produced by cells in response to a select set of stressful stimuli, such as bacterial infection or shear stress (18). We propose, therefore, that Keap1 quantifies environmental stress by monitoring and integrating the intracellular concentrations of NO, Zn2+, and alkenals.
Importantly, we have shown that Keap1 is equipped with a minimum of three sensors, one for each of the three chemically distinct signaling molecules. The C288 alkenal sensor appears to have arisen in an ancestor of all bilateran species and would have conferred upon Keap1 a broad capacity to sense environmental stress. The subsequent evolution of the Zn2+ sensor (H225/C226 and C613) would have enhanced the sensitivity of Keap1 proteins to such stress. Finally, the NO sensor (i.e., C151 along with H129, K131, R135, K150, and H154) emerged in the vertebrate-specific Keap1b family coincident with an expansion of the NOS protein family. Acquisition of this third sensor is likely to have benefited organisms because NO could then be used to initiate preemptive adaptation to certain sources of stress prior to the onset of cellular damage.
In order to gauge to what extent Keap1 is likely to rely upon NO, Zn2+, and alkenals to perceive environmental stress, it is helpful to consider two extreme scenarios. (i) Did the three sensors in Keap1 evolve specifically to recognize NO, Zn2+, and alkenals? (ii) Are these sensors promiscuous and their ability to recognize the three signaling molecules simply coincidental? We believe the weight of evidence favors the former scenario over the latter. For example, we have already remarked upon the apparent coevolution of the NO sensor with the expansion of the NOS gene family. Moreover, the alkenal sensor does not display promiscuity, as it failed to recognize either of the two xenobiotic electrophiles, Sul and tBHQ. There is also a sound theoretical reason for believing that the evolution of Keap1 was driven primarily by endogenous chemicals rather than xenobiotics. Furthermore, this reasoning can be extended to account for why Keap1 recognizes directly certain commonly encountered xenobiotics (see SI Discussion).
Although the present paper has introduced alkenals, Zn2+, and NO in the context of adaptation to stress, NO and Zn2+ act as second messengers in healthy cells also. For example, NO is produced in response to diverse physiological stimuli (32). Zn2+ too is an important signaling molecule in normal cells and its concentration can also be purposely altered by up-regulating the expression of Zn2+ import proteins (33). These considerations imply that Keap1-Nrf2 signaling might play a role in healthy as well as stressed cells. One plausible candidate role for the Keap1-Nrf2 pathway in healthy cells might be to act as a master orchestrator of redox cell signaling (34), via its ability to control the cell’s redox tone. Although speculative, such a role resonates with a growing literature indicating that genetic perturbation of Nrf2 activity in healthy cells affects cell differentiation and proliferation (see, for example, refs. 35 and 36), processes that are controlled in part by redox signaling networks (34).
In summary, we have described the structure, function, and evolution of a NO sensor within Keap1b proteins. We have also revealed the existence of two additional sensors, for Zn2+ and alkenals, within Keap1. The discovery that Keap1 acts as a hub for multiple second messengers provides a theoretical framework within which the ability of Keap1 proteins to perceive harmful conditions can be rationalized. It also hints at a crucial role for Keap1 in healthy cells.
Materials and Methods
Cell Culture, Transfections, Chemical Challenge, and β-Galactosidase Assay.
COS1 and mouse embryonic fibroblast cells were cultured and transfected as described previously (37). Cells were challenged with chemicals not less than 24 h after plating. Sul was obtained from LKT laboratories. Acro, tBHQ, Cd2+, Zn2+, As3+, and Se4+ were obtained from Sigma chemicals. 4HNE was purchased from Cayman Chemicals. Diethylamine-NONOate and diethylamine-NONOate/AM were obtained from Calbiochem. The β-galactosidase assay was carried out as described previously (7).
Addition Techniques.
Analysis of mKeap1b by liquid chromatography-MS/MS, use of the BST, plasmids, immunoblots and immunoprecipitations, bioinformatics analyses, and molecular modeling can be found in SI Material and Methods (see also Fig. S8).
Supplementary Material
Acknowledgments.
We thank Prof. Masayuki Yamamoto for Keap1-/- mouse embryonic fibroblast cells. This work was supported by Cancer Research UK Grants C4909/A5942 and C4909/A9990 (to J.D.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. M.P.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1007387107/-/DCSupplemental.
*The EpRE is also referred to as the antioxidant response element.
References
- 1.Kultz D. Molecular and evolutionary basis of the cellular stress response. Annu Rev Physiol. 2005;67:225–257. doi: 10.1146/annurev.physiol.67.040403.103635. [DOI] [PubMed] [Google Scholar]
- 2.Gasch AP. Comparative genomics of the environmental stress response in ascomycete fungi. Yeast. 2007;24:961–976. doi: 10.1002/yea.1512. [DOI] [PubMed] [Google Scholar]
- 3.Sykiotis GP, Bohmann D. Keapl/Nrf2 signaling regulates oxidative stress tolerance and lifespan in Drosophila. Dev Cell. 2008;14:76–85. doi: 10.1016/j.devcel.2007.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh A, et al. Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 2006;3:1865–1876. doi: 10.1371/journal.pmed.0030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sykiotis GP, Bohmann D. Stress-activated cap‘n’collar transcription factors in aging and human disease. Sci Signal. 2010;3:1–22. doi: 10.1126/scisignal.3112re3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi A, et al. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McMahon M, Itoh K, Yamamoto M, Hayes JD. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. J Biol Chem. 2003;278:21592–21600. doi: 10.1074/jbc.M300931200. [DOI] [PubMed] [Google Scholar]
- 8.Prestera T, Holtzclaw WD, Zhang Y, Talalay P. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc Natl Acad Sci USA. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang SW, et al. Heat stress activates interleukin-8 and the antioxidant system via Nrf2 pathways in human dental pulp cells. J Endodont. 2009;35:1222–1228. doi: 10.1016/j.joen.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Ali F, et al. Induction of the cytoprotective enzyme heme oxygenase-1 by statins is enhanced in vascular endothelium exposed to laminar shear stress and impaired by disturbed flow. J Biol Chem. 2009;284:18882–18892. doi: 10.1074/jbc.M109.009886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruber F, et al. NF-E2-related factor 2 regulates the stress response to UVA-1-oxidized phospholipids in skin cells. Faseb J. 2010;24:39–48. doi: 10.1096/fj.09-133520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rushworth SA, Chen XL, Mackman N, Ogborne RM, O’Connell MA. Lipopolysaccharide-induced heme oxygenase-1 expression in human monocytic cells is mediated via Nrf2 and protein kinase C. J Immunol. 2005;175:4408–4415. doi: 10.4049/jimmunol.175.7.4408. [DOI] [PubMed] [Google Scholar]
- 13.Burdette D, Olivarez M, Waris G. Activation of transcription factor Nrf2 by hepatitis C virus induces the cell-survival pathway. J Gen Virol. 2010;91:681–690. doi: 10.1099/vir.0.014340-0. [DOI] [PubMed] [Google Scholar]
- 14.Kensler TW, Wakabayashi N, Biswal S. Cell survival responses to environmental stresses via the Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol. 2007;47:89–116. doi: 10.1146/annurev.pharmtox.46.120604.141046. [DOI] [PubMed] [Google Scholar]
- 15.Holland R, et al. Prospective type 1 and Type 2 disulfides of Keap1 protein. Chem Res Toxicol. 2008;21:2051–2060. doi: 10.1021/tx800226m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li L, et al. Molecular evolution of Keap1. Two Keap1 molecules with distinctive intervening region structures are conserved among fish. J Biol Chem. 2008;283:3248–3255. doi: 10.1074/jbc.M708702200. [DOI] [PubMed] [Google Scholar]
- 17.Dargel R. Lipid-peroxidation—a common pathogenetic mechanism. Exp Toxicol Pathol. 1992;44:169–181. doi: 10.1016/S0940-2993(11)80202-2. [DOI] [PubMed] [Google Scholar]
- 18.Derakhshan B, Hao G, Gross SS. Balancing reactivity against selectivity: The evolution of protein S-nitrosylation as an effector of cell signaling by nitric oxide. Cardiovasc Res. 2007;75:210–219. doi: 10.1016/j.cardiores.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kroncke KD. Cellular stress and intracellular zinc dyshomeostasis. Arch Biochem Biophys. 2007;463:183–187. doi: 10.1016/j.abb.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 20.Alam J, et al. Nrf2, a cap‘n’collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 21.Kwak MK, Kensler TW, Casero RA., Jr Induction of phase 2 enzymes by serum oxidized polyamines through activation of Nrf2: Effect of the polyamine metabolite acrolein. Biochem Biophys Res Commun. 2003;305:662–670. doi: 10.1016/s0006-291x(03)00834-9. [DOI] [PubMed] [Google Scholar]
- 22.Moellering D, et al. The induction of GSH synthesis by nanomolar concentrations of NO in endothelial cells: A role for gamma-glutamylcysteine synthetase and gamma-glutamyl transpeptidase. FEBS Lett. 1999;448:292–296. doi: 10.1016/s0014-5793(99)00371-3. [DOI] [PubMed] [Google Scholar]
- 23.Levonen AL, et al. Cellular mechanisms of redox cell signalling: Role of cysteine modification in controlling antioxidant defences in response to electrophilic lipid oxidation products. Biochem J. 2004;378:373–382. doi: 10.1042/BJ20031049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang B, Chen C. An ascorbate-dependent artifact that interferes with the interpretation of the biotin switch assay. Free Radical Bio Med. 2006;41:562–567. doi: 10.1016/j.freeradbiomed.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 26.Bosworth CA, Toledo JC, Jr, Zmijewski JW, Li Q, Lancaster JR., Jr Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc Natl Acad Sci USA. 2009;106:4671–4676. doi: 10.1073/pnas.0710416106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dinkova-Kostova AT, Holtzclaw WD, Wakabayashi N. Keap1, the sensor for electrophiles and oxidants that regulates the phase 2 response, is a zinc metalloprotein. Biochemistry. 2005;44:6889–6899. doi: 10.1021/bi047434h. [DOI] [PubMed] [Google Scholar]
- 28.Ogura T, et al. Keap1 is a forked-stem dimer structure with two large spheres enclosing the intervening, double glycine repeat, and C-terminal domains. Proc Natl Acad Sci USA. 2010;107:2842–2847. doi: 10.1073/pnas.0914036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fourquet S, Guerois R, Biard D, Toledano MB. Activation of NRF2 by nitrosative agents and H2O2 involves KEAP1 disulfide formation. J Biol Chem. 2010;285:8463–8471. doi: 10.1074/jbc.M109.051714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dinkova-Kostova AT, et al. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci USA. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saydam N, Georgiev O, Nakano MY, Greber UF, Schaffner W. Nucleo-cytoplasmic trafficking of metal-regulatory transcription factor 1 is regulated by diverse stress signals. J Biol Chem. 2001;276:25487–25495. doi: 10.1074/jbc.M009154200. [DOI] [PubMed] [Google Scholar]
- 32.Dudzinski DM, Igarashi J, Greif D, Michel T. The regulation and pharmacology of endothelial nitric oxide synthase. Annu Rev Pharmacol. 2006;46:235–276. doi: 10.1146/annurev.pharmtox.44.101802.121844. [DOI] [PubMed] [Google Scholar]
- 33.Cousins RJ, Liuzzi JP, Lichten LA. Mammalian zinc transport, trafficking, and signals. J Biol Chem. 2006;281:24085–24089. doi: 10.1074/jbc.R600011200. [DOI] [PubMed] [Google Scholar]
- 34.Jones DP. Redefining oxidative stress. Antioxid Redox Signaling. 2006;8:1865–1879. doi: 10.1089/ars.2006.8.1865. [DOI] [PubMed] [Google Scholar]
- 35.Reddy NM, et al. Genetic disruption of the Nrf2 compromises cell-cycle progression by impairing GSH-induced redox signaling. Oncogene. 2008;27:5821–5832. doi: 10.1038/onc.2008.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao F, et al. Nrf2 promotes neuronal cell differentiation. Free Radical Bio Med. 2009;47:867–879. doi: 10.1016/j.freeradbiomed.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McMahon M, Thomas N, Itoh K, Yamamoto M, Hayes JD. Dimerization of substrate adaptors can facilitate cullin-mediated ubiquitylation of proteins by a “Tethering” mechanism—a two-site interaction model for the Nrf2-Keap1 complex. J Biol Chem. 2006;281:24756–24768. doi: 10.1074/jbc.M601119200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.