Abstract
We report the construction of two novel Escherichia coli strains (DH1lacdapD and DH1lacP2dapD) that facilitate the antibiotic-free selection and stable maintenance of recombinant plasmids in complex media. They contain the essential chromosomal gene, dapD, under the control of the lac operator/promoter. Unless supplemented with IPTG (which induces expression of dapD) or DAP, these cells lyse. However, when the strains are transformed with a multicopy plasmid containing the lac operator, the operator competitively titrates the LacI repressor and allows expression of dapD from the lac promoter. Thus transformants can be isolated and propagated simply by their ability to grow on any medium by repressor titration selection. No antibiotic resistance genes or other protein expressing sequences are required on the plasmid, and antibiotics are not necessary for plasmid selection, making these strains a valuable tool for therapeutic DNA and recombinant protein production. We describe the construction of these strains and demonstrate plasmid selection and maintenance by repressor titration, using the new pORT plasmid vectors designed to facilitate recombinant DNA exploitation.
INTRODUCTION
Antibiotics and antibiotic resistance genes have traditionally been used for the selection and maintenance of recombinant plasmids in hosts such as Escherichia coli, but their continued use is undesirable in many areas of biotechnology, such as gene therapy protocols that involve the direct introduction of plasmid DNA into patients. In the USA, federal regulations specifically discourage the use of selection markers that confer resistance to antibiotics in significant clinical use in order to avoid unnecessary risk of spread of antibiotic resistance traits to environmental microbes (1). Vectors should also be designed to minimise the amount of bacterial DNA they contain because short immunostimulatory sequences including CpG dinucleotides are present in plasmid backbones (2).
Widespread concern about antibiotic resistance genes in transgenic crops has recently helped to undermine public confidence in genetically modified food. In the UK, the Advisory Committee on Novel Foods declared that the β-lactamase gene (bla, conferring resistance to ampicillin) when present in transgenic corn posed an ‘unacceptable risk’ of transfer to the gut bacteria of livestock eating the corn (3). Where bla is present in transgenic crops, it serves no purpose and is a relic from plasmid selection during the cloning process. In their review on horizontal gene transfer, Dröge et al. (4) conclude: ‘effort should be put into the development of new strategies to remove antibiotic resistance determinants from transgenic constructs’.
In recombinant protein production, the antibiotic resistance gene present on the plasmid normally expresses the resistance enzyme constitutively at an unnecessarily high level. This invariably exerts a metabolic burden on the host cell, and can result in the resistance enzyme, as well as the antibiotic itself, being present as a contaminant in the required product. The removal of bla from a therapeutic plasmid has been shown to increase the efficiency of transgene expression 2-fold, and the removal of other vector sequences (including the lacZ α-complementation sequence) also resulted in enhanced transgene expression (5). Using a plasmid containing the kanamycin resistance gene kan and a recombinant gene of interest, Panayotatos (6) showed that a mutation causing a significant reduction of synthesis of kanamycin phosphotransferase resulted in a doubling of the recombinant protein production.
Existing antibiotic-free selection systems depend upon plasmid-borne gene expression of a protein or a tRNA that can be used in selection, and therefore they suffer from many of the same drawbacks as antibiotic resistance selections. The plasmid selection system that we have developed is based on a novel use of the phenomenon of repressor titration. Repressor titration has been used to identify and study natural operators (7), to isolate operator mutations (8) and to find ideal lac operator sequences from eukaryotic genomes (9).
To manipulate repressor titration and effect a plasmid selection pressure, the derepression of a negatively-regulated chromosomal operator/promoter system controlling a conditionally essential gene is achieved using a multicopy plasmid containing the same operator sequence. Under normal conditions, a repressor protein binds to the chromosomal operator and prevents transcription. The repressor is released when it binds to its inducer, which is often the substrate of the gene under control. However, when a molar excess of the operator sequence is present on a multicopy plasmid, it titrates the repressor from the chromosomal operator, allowing transcription to take place.
We previously described an E.coli strain, DH1lackan, in which the lac operator/promoter (lacOP) was used to control expression of kan on the E.coli chromosome (10). This strain allowed the selection of plasmids that possessed the lac operator sequences O1 and O3: these titrated the lac repressor allowing kan expression. Although DH1lackan enabled the selection of plasmids that did not contain antibiotic resistance genes, it was still dependent on kanamycin for the selection of transformants. Here we describe the construction and characterisation of E.coli strains DH1lacdapD and DH1lacP2dapD, which allow antibiotic-free plasmid selection and maintenance in complex media. These strains contain an ectopic copy of a dapD gene whose expression is controlled by a lac promoter. We demonstrate antibiotic-free plasmid selection and maintenance in these strains using the pORT plasmid vectors that facilitate the exploitation of repressor titration.
MATERIALS AND METHODS
Reagents and media
Standard reagents and kits were obtained from reputable suppliers and suppliers’ instructions were followed. Standard molecular biology techniques and microbiological media were as described in Ausubel et al. (11). Additions to bacterial growth media were: kanamycin (50 µg ml–1), DAP (80 µg ml–1), ampicillin (100 µg ml–1) and IPTG (0.1 mM).
Genetic manipulations
Escherichia coli strain DH5α (Gibco BRL) was used as a routine cloning host and JC7623 (12) and DH1 (13) were used for transformation by linearised plasmid and P1 transduction, respectively. In the latter case a RecA+ plasmid was present in recA– DH1 to allow homologous recombination, after which plasmid-free recA– segregants of the appropriate genotype were isolated. In principle, the genetic manipulations of the E.coli chromosome involved manipulating the appropriate chromosome region inserted into a plasmid (for example, pUC18; 14), modifying it as desired, and then reintroducing the modified DNA by homologous recombination into the chromosome using linearised DNA and strain JC7623, which is recBC sbcBC and supports homologous recombination of linear DNA (15). P1 transduction was then used to transfer the required locus to the final strain. Details of all of the constructions are available on request.
DH1ΔdapD was deleted for the complete chromosomal dapD gene but had all bounding sequences intact. The dapD gene was replaced by the kan gene of pUC4K (16). The deletion replacement was first made in a recombinant plasmid, which was linearised and used to transform JC7623. P1 transduction was used to transfer the ΔdapD::kan locus to DH1 containing the RecA+ plasmid pPE13 (17). Loss of the plasmid led to RecA– DH1ΔdapD.
The ectopic lac-dapD locus was inserted into the dif region (18,19) of the chromosome of DH1ΔdapD using homologous recombination with linear DNA to produce DH1lacdapD, containing a wild-type lac promoter, and DH1lacP2dapD, containing a mutant lac promoter that exhibits reduced gene expression. Both these strains were selected by their ability to grow on IPTG plates. However, upon inoculation into LB broth, DH1lacdapD was also able to grow without IPTG present. This was due to a very low level of leaky expression from the lac promoter providing sufficient dapD expression for DAP-independent growth. The genotype of DH1lacdapD and DH1lacP2dapD is: recA endA1 gyrA96 thi1 hsdr17 supE44 relA1 Δ(dapD)::kan hipA::lac-dapD. These two strains differ only in that the former contains the wild-type lac promoter and the latter, the 2 nt change in lacP to give lacP2 (Fig. 1).
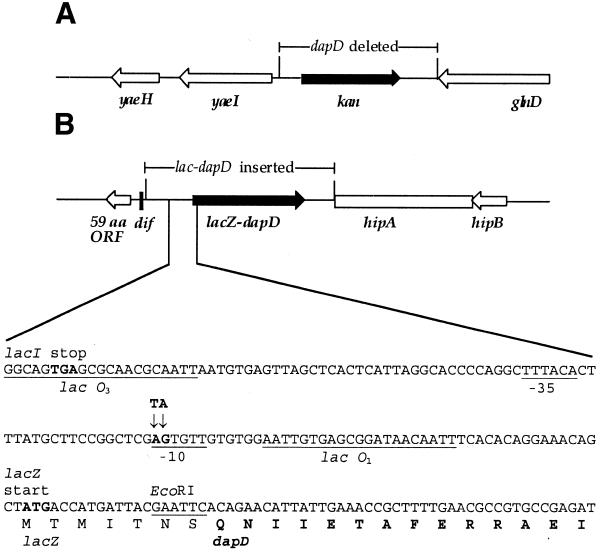
Figure 1.
Modifications to the chromosome of DH1lacP2dapD. (A) In the dapD locus, the dapD gene is completely replaced by kan. (B) The lacOP-dapD fusion is inserted into the dif locus, and the nucleotide sequence of the promoter region is shown, including the mutations –11 A→G and –12 T→A that converted DH1lacdapD into DH1lacP2dapD. The first four amino acids of dapD are replaced by the first seven from lacZ in the fusion protein.
Construction of the pORT vectors
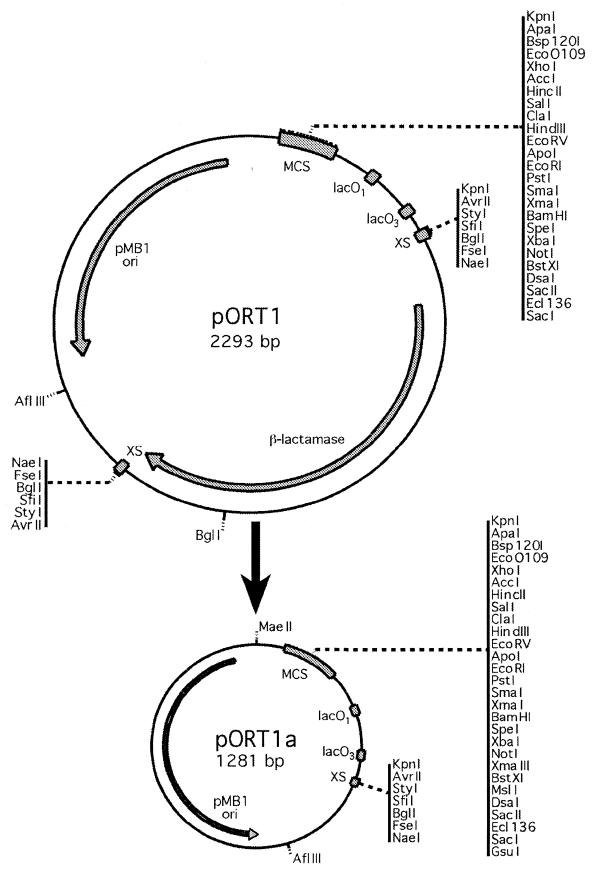
These were constructed from pBluescript SK(+) (Stratagene) by cleaving at all eight MaeII sites and self-ligating the largest fragment (1258 bp). This contained the pMB1 origin of replication, lacOP and truncated lacZ sequences, but no bla gene. This plasmid, pMAEII,was transformed into DH1lacdapD and selected by repressor titration on LB agar. Two primers were designed with the 3′ ends homologous to regions flanking bla, and the 5′ ends containing recognition sites for six restriction endonucleases: AvrII, StyI, SfiI, BglI, FseI and NaeI (underlined). Primer AMPSTOP (5′-GGCCGGCCCGGCCTAGGGTCTGACAGTTACCAATGCTTAATC-3′) included the bla stop codon (bold), and primer AMPKPNI (5′-GGGGTACCTAGGCCGGGCCGGCCGCACTTTTCGGGGAAATGTGCGC-3′) contained a KpnI site (bold) to determine insert orientation. These were used to amplify bla by PCR from pBluescript SK(+). bla was then cloned as a blunt fragment into PvuII-cleaved pMaeII. Plasmid vectors were produced (2293 bp each) with bla in both orientations (pORT1 and pORT2). The six flanking restriction sites were termed ‘excision sites’ (XSs), and cutting with any one of these enzymes and religating removed bla and generated a single XS. This was done with FseI and the resulting DNA was transformed into DH1lacdapD to generate 1281 bp pORT1a and pORT2a (Fig. 2 shows this for pORT1).
Figure 2.
Cloning vectors pORT1 and pORT1a. They contain a large multi-cloning site (MCS) flanked by M13 forward and reverse sequencing primer sites, and a minimal backbone with a pMB1 ori and lacOP. The bla gene in pORT1 is flanked by two XSs, containing directly repeated recognition sites for six restriction endonucleases. Cleaving with any one of these, followed by religation produces the bla-free minimal vector pORT1a, which can be selected by repressor titration. The single XS generated can be used as an alternative polylinker in pORT1a. Indicated restriction sites shown are unique, with the exception of KpnI and those in the XS of pORT1.
RESULTS
Construction and characterisation of strains suitable for plasmid selection by antibiotic-free repressor titration
In the construction of the repressor titration strains, the first stage was the complete deletion of dapD from the chromosome of DH1 to generate DH1ΔdapD. The conditionally essential gene dapD is normally located at 0.185 Mb on the E.coli chromosome (20). dapD encodes tetrahydrodipicolinate N-succinyltransferase, which catalyses the conversion of tetrahydrodipicolinate and succinyl-CoA to N-succinyl-α-amino-ɛ-keto-pimilate and CoA in the lysine/diaminopimelate pathway (21,22). DAP cross-links peptidoglycan in the bacterial cell wall (23) and is a precursor for lysine biosynthesis (24). Mutants with disrupted dapD are therefore DAP and lysine auxotrophs. As lysine is present in the rich media often used to grow E.coli (such as LB broth) and DAP is not, dapD mutants will undergo cell lysis unless DAP is added to the medium (25).
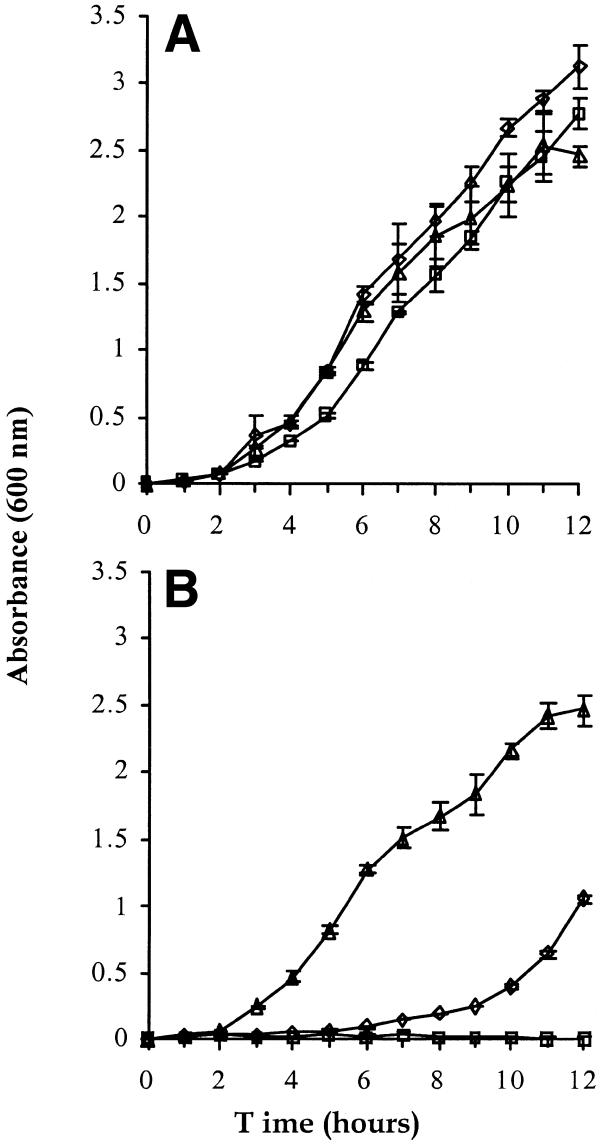
The first strain constructed (DH1lacdapD) had the endogenous dapD gene completely deleted and the ectopic dapD under the control of the wild-type lac promoter. The dif locus was chosen as a suitable ectopic location (1.59 Mb) (20) for the insertion of the lacOP–dapD fusion, as in the orientation shown there are no genes within 35 kb upstream which could cause transcriptional readthrough and thus dapD expression in the absence of induction or repressor titration (Fig. 1). DH1lacdapD grows in liquid culture lacking DAP when uninduced by either IPTG or repressor titration (Fig. 3A), suggesting that basal expression of tetrahydrodipicolinate N-succinyltransferase from the lac promoter was sufficient to synthesise enough DAP for growth. Nevertheless it was possible to use this strain for selection of plasmids after transformation by repressor titration (see below).
Figure 3.
Growth of (A) DH1lacdapD and (B) DH1lacP2dapD. Inoculum cultures were grown overnight in LB broth (with DAP added to the cultures not transformed with pORT1a) and 200 µl pelleted, washed three times to remove residual DAP and resuspended in 1.0 ml media. This was used to inoculate experimental cultures to an initial optical density of A600 = 0.004, and cultures were incubated at 37°C with constant agitation. Optical density was measured hourly by absorbance at 600 nm. Squares, no additives; diamonds, +IPTG; triangles, contain pORT1a. Data points are the means and error bars represent standard deviations of triplicate experiments.
In order to make a strain whose growth in the absence of DAP was dependent on derepression of lacP, two mutations were introduced to reduce lac promoter activity and basal expression of tetrahydrodipicolinate N-succinyltransferase, yet allowing sufficient expression for good growth after induction by IPTG or repressor titration. The first mutation, –11 A→G, virtually abolishes expression from lacP, but the second mutation, –12 T→A, shifts the transcription start site to –1 and restores β-galactosidase activity to ∼23% that of wild-type in the presence of activated CAP and 2–3% under conditions of catabolite repression during growth in glucose (26). The strain carrying this promoter-down mutation, DH1lacP2dapD, was dependent on IPTG or repressor titration for growth in liquid media lacking DAP (Fig. 3B). Although DH1lacP2dapD grew in liquid media in the presence of IPTG or DAP, its growth on solid media containing DAP and/or IPTG was restricted to areas of the plate containing a high inoculum: few isolated single colonies were present. In contrast, transformation with a lacO-containing plasmid led to single colonies that grew well on solid media, thereby allowing the use of this strain for plasmid selection by repressor titration.
Novel vectors for use in repressor titration
The pORT vectors (Fig. 2) were designed for use in repressor titration strains, but any other lacO-containing, high copy number plasmid can be selected by repressor titration and stably maintained. The use of plasmids pORT1 and pORT2 allows initial transformations and recombinant DNA manipulations to be carried out in any host strain using ApR as a selection (most of the lacZ α-peptide was removed in the construction of these plasmids to reduce non-essential DNA to a minimum, so blue/white selection is not possible). Then bla is removed as a final step prior to transformation into DH1lacP2dapD or DH1lacdapD. This is achieved by exploiting the six restriction endonuclease recognition sites that flank bla in direct repeat. Three of these (AvrII, SfiI and FseI) have rare cleavage sites, so the probability of all being present in an insert is very low.
Growth of DH1lacdapD and DH1lacP2dapD
The overall growth in liquid medium lacking DAP of uninduced DH1lacdapD was slightly less than when IPTG was present, apparently as a consequence of a lag in reaching full log growth, underlining the failure to repress expression from wild-type lacOP completely in this strain (Fig. 3A). DH1lacdapD containing pORT1a grew initially as fast as the IPTG-induced control although in late log phase its growth rate reduced and after 24 h A600 was ∼3.2 as compared to ∼4.0 for the plasmid-free control.
DH1lacP2dapD containing pORT1a showed an identical growth behaviour to DH1lacdapD containing pORT1a (Fig. 3B): perhaps this reduced growth during late log and stationary phase growth is a consequence of the burden of maintaining the high copy number pORT1a plasmid, despite it not expressing any protein products. DH1lacP2dapD did not grow at all without IPTG (Fig. 3B). Although the initial rate of growth of IPTG-induced DH1lacP2dapD was lower than that of DH1lacP2dapD containing pORT1a and IPTG-induced DH1lacdapD, at 24 h the densities of the plasmid-free cultures were identical (A600 ∼4.0). It seems likely that the apparent lag in the growth of the IPTG-induced DH1lacP2dapD is a consequence of some lysis induced by DAP limitation during the short period between DAP removal by washing and the effective in vivo action of IPTG. As both the leaky and non-leaky IPTG-induced cultures reached similar final cell densities, we conclude that the concentration of IPTG used allowed sufficient dapD expression in DH1lacP2dapD for this not to be a growth-limiting factor in liquid medium.
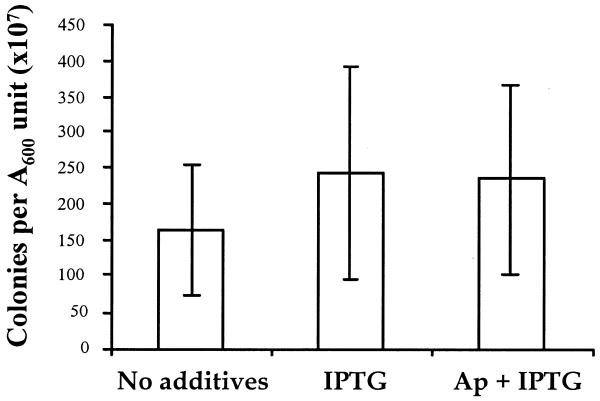
Figure 4 shows the number of plasmid-containing colonies obtained by plating out equal numbers of cells of DH1lacP2dapD (pORT1) (as determined by A600 measurements) from cultures grown by daily subculture over 5 days (∼47 generations). These cultures were grown under three conditions: repressor titration selection (no additives), no selection (IPTG) and antibiotic selection (Ap + IPTG). There is no significant difference in the number of colonies from these three cultures, indicating that the plasmid was not lost, even when grown with IPTG when repressor titration is no longer effective. Therefore, pORT1 is a stable plasmid suitable for use as a recombinant DNA vector.
Figure 4.
Propagation of pORT1 in DH1lacP2dapD. DH1lacP2dapD(pORT1) was grown in LB broth by repeated subculture (at 37°C with constant agitation) with no additives, with IPTG and with IPTG + Ap. Each culture was subcultured at intervals of 24 h (inoculum density A600 = 0.01) over 5 days. A dilution series was prepared from a volume of the final cultures equivalent to 0.1 A600 units and plated on LB agar + Ap. The results represent the mean number of colonies counted from triplicate experiments when plated at a 10–6 dilution (error bars represent standard deviations).
Plasmid selection by repressor titration after plasmid transformation
The ability of DH1lacdapD to act as a host for plasmid transformation during selection by repressor titration was compared with the efficiency of transformation by antibiotic selection (ApR) using pUC18, and pUC18ΔlacO, deleted for a PvuII fragment that removes 322 bp containing lacO1 and lacO3. pORT1a was also used to demonstrate selection of an antibiotic gene-free plasmid. The LB agar plates used contained Ap, Ap + IPTG and no additives, and the results of these experiments are shown in Table 1.
Table 1. Plasmid selection by repressor titration.
| |
Transformation efficiency: colonies/fmol
DNA |
||
| No additives | Ap | Ap + IPTG | |
| pUC18 | 618.02 | 587.36 | 575.73 |
| pUC18ΔlacO | 9.90 | 0 | 632.39 |
| pORT1a | 589.69 | 0 | 0 |
Calcium-competent DH1lacdapD (100 µl aliquots) were transformed with pUC18, pUC18ΔlacO and pORT1a by adding 0.5 µg plasmid DNA, heat shocking at 42°C for 45 s, and incubating at 37°C in 900 µl LB broth containing IPTG for 1 h. Cells were spun down and washed to remove IPTG and resuspended in 1.0 ml LB broth, then plated on LB agar (100 µl of a 1/100 dilution of the transformation cultures) containing no additives, Ap or Ap + IPTG. Results represent the means of three separate experiments in which transformations were plated in triplicate. Transformation efficiencies are displayed in colonies per femtomole of plasmid DNA. A negative control (no DNA) had the same number of colonies as those for pUC18ΔlacO with no additives, but no colonies with Ap present.
When DH1lacdapD was transformed with pUC18, transformants were obtained on all three plates at equivalent numbers, as pUC18 confers both ApR and repressor titration. To confirm that colonies growing with no Ap present were indeed pUC18 transformants, 100 colonies were streaked onto Ap plates, where they all re-grew. Transformants of pUC18ΔlacO grew on Ap + IPTG plates at an equivalent number to those of pUC18, thus Ap + IPTG represents antibiotic selection alone. There was no growth on Ap plates, as without the lacO sequences, repressor titration could not function. The few colonies seen in the absence of Ap did not contain plasmids since they failed to re-grow on Ap + IPTG plates. A negative control transformation was also performed (no DNA added) and similar colony numbers were seen as for pUC18ΔlacO with no additives. These were due to leaky dapD expression in DH1lacdapD, caused by compensating mutations allowing cells to grow in the absence of IPTG, as negative control colonies did form single colonies when re-streaked onto plates with no additives.
pORT1a, being devoid of the bla gene, produced no transformants where Ap was present, but transformants were obtained in its absence at similar numbers to pUC18. It should be noted that transformation efficiencies are expressed in colonies per femtomole DNA, as smaller plasmids would be present at a higher concentration per unit mass. The values for pUC18 were in the range of 105 transformants per µg DNA.
DISCUSSION
We have created two strains, DH1lacdapD and DH1lacP2dapD, which allow the selection and propagation of plasmids free from the burden of plasmid gene expression that leads to antibiotic resistance, or indeed any other expression of protein encoding or RNA genes from the plasmid vector. This avoids the problems associated with the use of antibiotics and antibiotic resistance genes in therapeutic protein or DNA vaccine production, gene therapy or genetic engineering applications.
Deletion of chromosomal dapD was required rather than simple insertional inactivation, as there was concern that a partially deleted dapD may undergo homologous recombination with the lacOP-dapD cassette that had been integrated at an ectopic position, thereby restoring wild-type dapD. Note that during the genetic constructions, strains were proficient in homologous recombination until the last step when the RecA+ plasmid was removed, in order that the desired recombinational events could occur.
Transformation efficiencies in DH1lacdapD by repressor titration and antibiotic selection are equivalent, and calcium-competent cells were used as we have seen higher and more consistent transformation efficiencies with these cells than with electro-competent DH1lacdapD.
The lacP–dapD fusion contains a 21 bp natural lactose operator, O1, located between the lacP and the start codon of lacZ. An additional 21 bp ‘pseudo-operator’, O3, is also present 92 bp upstream of O1 and includes the stop codon of lacI. O1 is the major target for LacI binding, although deletion of O3 reduces repression 50-fold (27). Plasmids that we have selected by repressor titration so far (e.g. pUC, pORT) are high copy number and contain both O1 and O3. For gene therapy and the construction of transgenic organisms, the absence of non-essential expressed sequences and reduction in the size of the plasmid backbone of pORT1a is highly desirable for the reasons discussed earlier, as well as to facilitate the cloning and maintenance of large DNA fragments.
A low level of tetrahydrodipicolinate N-succinyltransferase expression from the wild-type lac promoter of DH1lacdapD occurs even when uninduced, which allows growth in liquid culture (but not single colony formation on solid media) without DAP. This was overcome by introducing mutations into the lac promoter that reduced expression, thereby generating DH1lacP2dapD, a strain that requires DAP, IPTG or repressor titration for its growth. DH1lacP2dapD differs from DH1lacdapD in only two nucleotides, which represent mutations to the lac promoter controlling dapD. On solid media, DH1lacP2dapD will only form single colonies if transformed with a lacO-containing plasmid. The inability of DH1lacP2dapD to grow well on solid media in the absence of repressor titration is a consequence of its recA– status, since derivatives of this strain carrying either of two different RecA+, lacO– plasmids grew well on plates, as did a plasmid-free derivative of DH1lacP2dapD that contained a chromosomal RecA+ allele. DH1ΔdapD also did not grow well on plates supplemented with DAP unless the strain was RecA+. Previous studies of ΔdapD mutants have been restricted to RecA+ strains (28–30). We believe it likely that this inability to form single colonies on solid media is a consequence of increased DNA damage under these conditions, which is particularly detrimental to recA– strains. Consistent with this view is the observation that E.coli mutants that lack superoxide dismutase (SOD) contained a compensatory mutation in dapD, thereby relieving their growth defects resulting from SOD deficiency (30). This report proposed that dipicolinate, which is the immediate precursor of tetrahydrodipicolinate and can also be produced by tetrahydrodipicolinate oxidation, chelates iron lost from superoxide-damaged iron–enzyme clusters and thus allows iron to be recycled. It may be that the accumulation of precursors in the dapD mutants leads to an effective increase in cytoplasmic iron concentrations in SOD+ strains, thereby increasing DNA damage as a consequence of the Fenton reaction. This could lead to the inviability of the recA– strain.
DH1lacP2dapD has a significantly lower and more variable transformation efficiency compared to DH1lacdapD, perhaps because of its greater sensitivity to DAP depletion. Nevertheless, it is possible to select plasmid-containing cells by repressor titration using DH1lacP2dapD as Figure 3 demonstrates. We recommend that DH1lacdapD be used for the construction of antibiotic gene-free plasmids at the stage of antibiotic resistance gene removal, while DH1lacP2dapD is used as a final production strain. As DH1lacP2dapD is the first non-leaky, inducible DAP auxotroph, it may also be useful as a model strain for assessing dapD as a potential antibiotic target.
Since these repressor titration strains retain the chromosomal kan gene used to replace the deleted dapD gene, the next step is to create a derivative deleted for kan as well as dapD, and which therefore contains no antibiotic resistance gene. We conclude that repressor titration is a highly effective means of selecting plasmid-containing cells after genetic transformation and we are confident that repressor titration will be an extremely effective mechanism for maintaining plasmids that may not otherwise be stably maintained.
References
- 1.Murphy D.B. and Epstein,S.L. (1998) Guidance for Industry. Guidance for Human Somatic Cell Therapy and Gene Therapy. Food and Drug Administration, Rockville, MD.
- 2.Sato Y., Roman,M., Tighe,H., Lee,D., Corr,M., Nguyen,M.D., Silverman,G.J., Lotz,M., Carson,D.A. and Raz,E. (1996) Immunostimulatory DNA sequences necessary for effective intradermal gene immunization. Science, 273, 352–354. [DOI] [PubMed] [Google Scholar]
- 3.Wadman M. (1996) Genetic resistance spreads to consumers. Nature, 383, 564. [DOI] [PubMed] [Google Scholar]
- 4.Dröge M., Pühler,A. and Selbitschka,W. (1998) Horizontal gene transfer as a biosafety issue: A natural phenomenon of public concern. J. Biotechnol., 64, 75–90. [DOI] [PubMed] [Google Scholar]
- 5.Hartikka J., Sawdey,M., Cornefert-Jensen,F., Margalith,M., Barnhart,K., Nolasco,M., Vahlsing,H.L., Meek,J., Marquet,M., Hobart,P., Norman,J. and Manthorpe,M. (1996) An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum. Gene Ther., 7, 1205–1217. [DOI] [PubMed] [Google Scholar]
- 6.Panayotatos N. (1988) Recombinant protein production with minimal-antibiotic-resistance vectors. Gene, 74, 357–363. [DOI] [PubMed] [Google Scholar]
- 7.Mata-Gilsinger M., Ritzenthaler,P. and Blanco,C. (1983) Characterization of the operator sites of the Exu regulon in Escherichia coli K-12 by operator-constitutive mutations and repressor titration. Genetics, 105, 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irani M., Orosz,L., Busby,S., Taniguchi,T. and Adhya,S. (1983) Cyclic AMP-dependent constitutive expression of gal operon: Use of repressor titration to isolate operator mutations. Proc. Natl Acad. Sci. USA, 80, 4775–4779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simons A., Tils,D., von Wilken-Bergmann,B. and Müller-Hill,B. (1984) Possible ideal lac operator: Escherichia coli lac operator-like sequences from eukaryotic genomes lack the central G·C pair. Proc. Natl Acad. Sci. USA, 81, 1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams S.G., Cranenburgh,R.M., Weiss,A.M., Wrighton,C.J., Sherratt,D.J. and Hanak,J.A.J. (1998) Repressor titration: a novel system for selection and maintenance of recombinant plasmids. Nucleic Acids Res., 26, 2120–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ausubel F.M., Brent,R., Kingston,R.E., Moore,D.D., Seidman,J.G., Smith,J.A. and Struhl,K. (eds) (1994) Current Protocols in Molecular Biology. John Wiley and Sons, Inc., New York, NY.
- 12.Lloyd R.G. and Buckman,C. (1985) Identification and genetic analysis of sbcC mutations in commonly used recBC sbcB strains of Escherichia coli K-12. J. Bacteriol ., 164, 836–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol., 166, 557–580. [DOI] [PubMed] [Google Scholar]
- 14.Yanisch-Perron C., Vieira,J. and Messing,J. (1985) Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene, 33, 103–119. [DOI] [PubMed] [Google Scholar]
- 15.Vieira J. and Messing,J. (1982) The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene, 19, 259–268. [DOI] [PubMed] [Google Scholar]
- 16.Jasin M. and Schimmel,P. (1984) Deletion of an essential gene in Escherichia coli by site-specific recombination with linear DNA fragments. J. Bacteriol., 159, 783–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hickson I.D., Gordon,R.L., Tomkinson,A.E. and Emmerson,P.T. (1981) A temperature sensitive RecA protein of Escherichia coli. Mol. Gen. Genet., 184, 68–72. [DOI] [PubMed] [Google Scholar]
- 18.Leslie N.R. and Sherratt,D.J. (1995) Site-specific recombination in the replication terminus region of Escherichia coli: functional replacement of dif. EMBO J., 14, 1561–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Black D.S., Kelly,A.J., Mardis,M.J. and Moyed,H.S. (1991) Structure and organisation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. J. Bacteriol ., 173, 5732–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blattner F., Plunkett,G.I., Bloch,C., Perna,N., Burland,V., Riley,M., Collado-Vides,J., Glasner,J., Rode,C., Mayhew,G., Gregor,J., Davis,N., Kirkpatrick,H., Goeden,M., Rose,D., Mau,B. and Shao,Y. (1997) The complete genome sequence of Escherichia coli K-12. Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 21.Richaud C., Richaud,F., Martin,C., Haziza,C. and Patte,J.C. (1984) Regulation of expression and nucleotide sequence of the Escherichia coli dapD gene. J. Biol. Chem., 259, 14824–14828. [PubMed] [Google Scholar]
- 22.Gilvarg C. (1961) N-succinyl-α-amino-ɛ-ketopimelic acid. J. Biol. Chem., 236, 1429–1431. [PubMed] [Google Scholar]
- 23.Work E. (1950) A new naturally occurring amino-acid. Nature, 165, 74–75. [DOI] [PubMed] [Google Scholar]
- 24.Davis B.D. (1952) Biosynthetic interrelations of lysine, diaminopimelic acid, and threonine in mutants of Escherichia coli. Nature, 169, 534–536. [DOI] [PubMed] [Google Scholar]
- 25.Degryse E. (1991) Development of stable, genetically well-defined conditionally viable Escherichia coli strains. Mol. Gen. Genet., 227, 49–51. [DOI] [PubMed] [Google Scholar]
- 26.Karls R., Schulz,V., Jovanovich,S.B., Flynn,S., Pak,A. and Reznikoff,W.S. (1989) Pseudorevertants of a lac promoter mutation reveal overlapping nascent promoters. Nucleic Acids Res., 17, 3927–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oehler S., Eismann,E.R., Krämer,H. and Müller-Hill,B. (1990) The three operators of the lac operon cooperate in repression. EMBO J., 9, 973–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bukhari A.I. and Taylor,A.L. (1971) Genetic analysis of diaminopimelic acid- and lysine-requiring mutants of Escherichia coli. J. Bacteriol ., 105, 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Degryse E. (1991) Stability of a host-vector system based on complementation of an essential gene in Escherichia coli. J. Biotechnol., 18, 29–40. [DOI] [PubMed] [Google Scholar]
- 30.Maringanti S. and Imlay,J.A. (1999) An intracellular iron chelator pleiotropically suppresses enzymatic and growth defects of superoxide dismutase-deficient Escherichia coli. J. Bacteriol ., 181, 3792–3802. [DOI] [PMC free article] [PubMed] [Google Scholar]