Abstract
The cell wall of Staphylococcus aureus is characterized by an extremely high degree of cross-linking within its peptidoglycan (PGN). Penicillin-binding protein 4 (PBP4) is required for the synthesis of this highly cross-linked peptidoglycan. We found that wall teichoic acids, glycopolymers attached to the peptidoglycan and important for virulence in Gram-positive bacteria, act as temporal and spatial regulators of PGN metabolism, controlling the level of cross-linking by regulating PBP4 localization. PBP4 normally localizes at the division septum, but in the absence of wall teichoic acids synthesis, it becomes dispersed throughout the entire cell membrane and is unable to function normally. As a consequence, the peptidoglycan of TagO null mutants, impaired in wall teichoic acid biosynthesis, has a decreased degree of cross-linking, which renders it more susceptible to the action of lysozyme, an enzyme produced by different host organisms as an initial defense against bacterial infection.
Keywords: cell division, cell wall, penicillin-binding proteins, transpeptidases, lysozyme
The cell wall of Gram-positive bacteria is a highly complex network composed mainly of peptidoglycan (PGN) and teichoic acids (TAs), both essential to the maintenance of the structural integrity and shape of the bacterial cell. Peptidoglycan is a heterogeneous polymer of glycan chains cross-linked by short peptides of variable length and amino acid composition (1). Teichoic acids are phosphate-rich glycopolymers that can be either covalently linked to PGN (wall teichoic acids, or WTAs) or anchored to the cytoplasmic membrane (lipoteichoic acids, or LTAs) (2, 3).
Despite decades of study, it is still not well understood why pathogenic and nonpathogenic bacteria have TAs at their cell surface. TAs contribute to a variety of processes, including resistance to environmental stresses, such as heat (4) or low osmolarity (5), to antimicrobial peptides (6), antimicrobial fatty acids (7), cationic antibiotics (8), and lytic enzymes produced by the host, including lysozymes (9, 10). TAs also act as receptors for phage particles (11), and they can bind cationic groups (particularly magnesium ions), providing a reservoir of ions close to the bacterial surface that may be important for the activity of different enzymes (12). More recently, TAs have been proposed to be involved in cell division and morphogenesis (13). Lack of LTA synthesis in rod-shaped Bacillus subtilis cells causes defects in the formation of the division septum and in cell separation (14), whereas lack of WTAs results in round cells (15). Moreover, enzymes involved in LTA synthesis localize predominantly at the division sites of bacteria (14), and enzymes involved in the synthesis of WTAs localize in helical patterns (16) similar to the previously described patterns of PGN synthesis observed during elongation of B. subtilis cells (17). The observation that mutants lacking LTAs have altered autolysis rates (5, 18) or that interference with the synthesis of WTA in B. subtilis triggers the transcription of several genes involved in PGN synthesis (19), constitutes additional, indirect evidence that suggests a link between TAs and cell morphogenesis through an effect on the biosynthesis of PGNs, the correct assembly of which is required for proper cell division and morphogenesis.
To investigate whether the synthesis of WTAs is directly required for building a PGN macromolecule with the correct structure, we used Staphylococcus aureus as a model organism. S. aureus is a Gram-positive bacteria and a prominent pathogen in the community and healthcare settings, well known for its virulence and antibiotic resistance (20, 21). The main advantage of using S. aureus for studying PGNs is that it has only four penicillin-binding proteins (PBPs 1–4) instead of 12 or 16 that are present in the traditional model organisms Escherichia coli and B. subtilis, respectively (22). PBPs are enzymes involved in the final stages of PGN biosynthesis, which synthesize glycan chains (via transglycosylation reactions) and cross-link different glycan chains through short peptides (via transpeptidation reactions). The level of PGN cross-linking varies between bacterial species, and S. aureus has an unusually high degree of cross-linking (23), which is due mainly to the action of PBP4 (24, 25), and seems to require the long and flexible pentaglycine crossbridge that S. aureus cells use to connect two stem peptides from different glycan strands (23, 26). PBP4 is not essential for S. aureus viability. However, it has an important role in antibiotic resistance, as it is essential for the expression of β-lactam resistance in community-acquired methicillin-resistant strains (25). Interestingly, inactivation of PBP4 has been reported in laboratory step mutants and clinical isolates with intermediate vancomycin resistance (27–29), which have decreased levels of PGN cross-linking. This suggests that modulation of the degree of cross-linking is important for resistance to different antibiotics.
In this study, we describe a previously uncharacterized link between WTA biosynthesis and PGN biosynthesis. We found that WTAs act as temporal and spatial regulators of PGN metabolism, controlling the level of cross-linking by directing PBP4 to the division septum. We also showed that the highly cross-linked PGN that results from the regulated action of PBP4 is more resistant to enzymatic degradation by lysozyme. This increased resistance may be advantageous to S. aureus bacteria during interactions with host organisms that produce lysozyme as an initial defense against bacterial infection.
Results
TagO Null Mutant Has Decreased Levels of Peptidoglycan Cross-Linking.
To test whether the presence of WTAs is required to build a PGN macromolecule with the correct structure, we deleted the tagO gene from the chromosome of the S. aureus NCTC8325-4 strain, leaving no antibiotic resistance markers, to produce the strain NCTCΔtagO. TagO is the first enzyme in the teichoic acid biosynthetic pathway, catalyzing the transfer of GlcNAc-1–phosphate from cytoplasmic UDP-linked precursors to the C55-P lipid anchor bactoprenol (30). NCTCΔtagO did not produce detectable levels of TAs (Fig. S1) and showed several phenotypes previously described for staphylococcal mutants impaired in WTA biosynthesis (4), such as a larger cell diameter than wild-type cells, aggregation in clusters (Fig. S1), temperature sensitivity, and resistance to infection by phage 80α.
As a first approach to detecting changes in PGN structure due to the deletion of tagO, we purified PGN from NCTCΔtagO and its parental strain NCTC8325-4 and tested its susceptibility to the action of lysozyme, an enzyme that cuts PGN between the N-acetylmuramic acid and N-acetylglucosamine residues of the glycan chains. S. aureus is known for its intrinsic ability to resist lysozyme due to modifications of its PGN, such as O-acetylation of N-acetylmuramic acid residues and attachment of teichoic acids, which prevent access of the enzyme to its substrate (10, 31). We treated cell walls from both the wild-type and the TagO mutant with hydrofluoric acid, which removes teichoic acids and O-acetyl groups, and incubated the resulting “naked” PGN with lysozyme. The PGN of NCTCΔtagO was more susceptible to lysozyme than that of NCTC8325-4 (Fig. 1D), suggesting differences in PGN structure between the two strains.
Fig. 1.
Highly cross-linked muropeptides, resulting from PBP4 activity, are less abundant in the peptidoglycan of a S. aureus ΔtagO mutant. (A) HPLC analysis of mutanolysin-digested PGN of the parental strain NCTC8325-4 and mutants NCTCΔtagO and NCTCΔpbpD. Arrow points to highly cross-linked muropeptide species, which are less abundant in the mutants lacking TagO and PBP4. (I–V) Muropeptide species from monomers to pentamers. (B) Western blot analysis, using a specific anti-PBP4 antibody, of membrane proteins from the same three strains showing that PBP4 is expressed at wild-type levels in the NCTCΔtagO background. (C) Analysis of membrane proteins labeled with Bocillin-FL and separated by SDS/PAGE, showing that PBP4 is able to bind Bocillin-FL in the NCTCΔtagO background. (D) Lysozyme hydrolysis of purified PGN from NCTC8325-4, NCTCΔtagO, and NCTCΔpbpD, followed by monitoring the decrease in absorbance at OD600nm, and showing that PGN with a lower degree of cross-linking had an increased susceptibility to lysozyme.
The PGN muropeptide composition of NCTCΔtagO and NCTC8325-4 was then analyzed by HPLC, which revealed a reduced level of PGN cross-linking in NCTCΔtagO when compared with the parental strain NCTC8325-4, with the concomitant accumulation of monomeric and dimeric muropeptides (Fig. 1A and Fig. S2). This result suggests that TAs may be involved in the later stages of PGN maturation, which include the introduction of extra cross-links between the glycan strands.
Decreased Levels of Peptidoglycan Cross-Linking in a TagO Null Mutant Result from Delocalization of PBP4.
The secondary (high-level) cross-linking of S. aureus results mainly from the activity of PBP4, as inactivation of pbpD gene, encoding PBP4, leads to the disappearance of the highly cross-linked muropeptide species (24, 25) that typically elute as a broad peak at the end of the HPLC chromatogram (Fig. 1A, arrow). Interestingly, the composition of PGN purified from NCTCΔtagO was very similar to that of the PGN from NCTCΔpbpD (Fig. 1A). This observation led us to test if PBP4 was altered in the tagO null mutant. Sequencing of the pbpD gene (including its promoter region) in NCTCΔtagO did not identify any mutations, and Western blot analysis with specific anti-PBP4 antibodies showed that levels of PBP4 were identical in NCTCΔtagO and in its parental strain NCTC8325-4 (Fig. 1B). Furthermore, binding of Bocillin-FL (a fluorescent derivative of penicillin V and a substrate analog for PBPs) to the staphylococcal PBPs showed that the binding of PBP4 to this substrate analog was not altered in NCTCΔtagO cells when compared with the parental strain NCTC8325-4 (Fig. 1C).
We previously showed that inhibition of cell-wall synthesis in S. aureus by β-lactam antibiotics results in delocalization of PBP2 (32). Therefore, we decided to test whether the lack of high-level peptidoglycan cross-linking in the ΔtagO background was the result of incorrect localization of PBP4. For that purpose, we constructed S. aureus RN4220 strains expressing a C-terminal YFP fusion to PBP4 from its native chromosomal locus and under the control of its native promoter. RN4220 background was used because it is the only S. aureus strain that can be efficiently transformed with foreign DNA. The transformation efficiency of NCTC8325-4 is extremely low, and the ΔtagO mutation renders it resistant to phages such as 80α, preventing transduction and impairing the introduction of additional constructs into this background. When the PBP4–YFP fusion was expressed in the RN4220 parental background (RNPBP4YFP), it localized to the division septum (Fig. 2), where cell-wall synthesis has been reported to take place in S. aureus (33). However, when the same fusion was expressed in the ΔtagO background (RNΔtagOPBP4YFP), PBP4 was observed all around the cellular membrane, with no specific accumulation at the division septum (Fig. 2). This effect was not general to all staphylococcal PBPs, as PBP1 did not lose its septal localization in a ΔtagO background (Fig. S3).
Fig. 2.
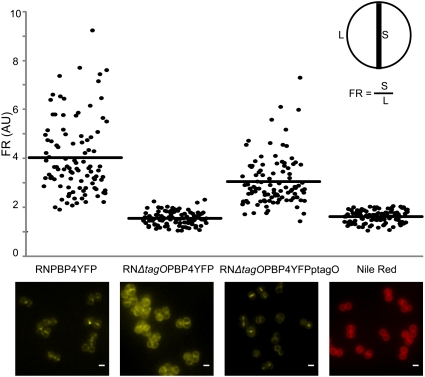
Septal localization of PBP4 is lost in a tagO null mutant. Microscopy images and quantification of septum versus lateral membrane fluorescence (fluorescence ratio, FR) of PBP4–YFP in a wild-type background (RNPBP4YFP), a ΔtagO background (RNΔtagOPBP4YFP), and a ΔtagO mutant complemented with plasmid-encoded tagO (RNΔtagOPBP4YFPptagO). Also shown are RNPBP4YFP cells labeled with membrane dye Nile Red, which is homogeneously distributed in the cell membrane. Quantification was performed in 100 cells displaying closed septa for each strain. Horizontal lines correspond to average FR values. FR values over 2 indicate preferential septal localization whereas FR values equal to or under 2 indicate that a protein is dispersed over the cell surface. p values < 10−7. Scale bar: 1 μm.
To quantify the extent of delocalization of PBP4 in the absence of the TagO protein, we calculated the ratio of fluorescence measured at the septum versus the fluorescence measured at the “lateral” wall (Fig. 2). If a fluorescent protein or dye (such as Nile Red membrane stain) is homogeneously distributed over the entire cell membrane, the intensity of the fluorescent signal at the septum (which contains two membranes) should be approximately twice the fluorescence at the lateral membrane. It follows that, if a fluorescent protein is specifically accumulated at the division septum, then the ratio of fluorescence at the septum versus the lateral wall should be higher than 2. When this ratio was calculated for PBP4–YFP in the parental strain RNPBP4YFP, we obtained an average value of 4.0 ± 1.52 whereas a value of 1.6 ± 0.27 was obtained for the ΔtagO mutant RNΔtagOPBP4YFP, indicating that the specific accumulation of PBP4 at the septum in wild-type cells was completely lost if the TagO protein was absent. Furthermore, complementation of RNΔtagOPBP4YFP, with plasmid encoded TagO (but not with the empty plasmid vector), restored the correct localization of PBP4 at the septum (Fig. 2).
PBP4 Is Recruited to the Division Septum Later than TagO.
The simplest explanation for the dependence of PBP4 on TagO for septal localization would be that TagO localizes to the division septum and recruits PBP4 by protein–protein interaction. In accordance, we found that a TagO–GFP fusion localized at the division septum (Fig. 3A and Fig. 4D). To study the dynamics of PBP4 and TagO recruitment to the septum, namely to determine if they arrived at the septum at the same time, we constructed strain RNTagOPBP4, which expresses both TagO–GFP and PBP4–mCherry fusion proteins. We analyzed over 3,000 cells and determined the localization of TagO and PBP4 in ≈900 cells that were in the initial stages of septum synthesis. In this subpopulation, TagO arrived at the septum before PBP4 (TagO was seen as two septal spots corresponding to a septal ring whereas PBP4 was not yet present at the septum) or was found “ahead” of PBP4 (TagO was found across the entire septum whereas PBP4 was still seen as two septal spots) in 44.3% of the cells (Fig. 3B). PBP4 arrived at the septum before TagO or was found ahead of TagO in only 4.4% of the cells. In 51.2% of the cells, both proteins had the same localization. The fact that TagO and PBP4 recruitment to the septum occurs at different times suggests that PBP4 is not recruited to the septum by direct protein–protein interaction with TagO. Further indication came from studies with a bacterial two-hybrid system (34), which failed to detect any interaction between PBP4 and TagO (Fig. S4).
Fig. 3.
TagO protein is recruited to the division septum before PBP4. (A) The strain RNTagOPBP4, expressing simultaneously TagO–GFP and PBP4–mCherry protein fusions, was analyzed by fluorescence microscopy. (Top) Results showing that TagO reaches the division septum before PBP4 in several cells. (Bottom) Cells in which PBP4 and TagO colocalize at the septum. Scale bar: 1 μm. (B) Localization of TagO–GFP and PBP4–mCherry was analyzed in 902 cells in the early stages of septum formation. Localization of each protein was assigned to three sequential stages: scattering around the entire membrane; localization in a ring around the division plane, usually seen as two spots; and localization over the entire closed septum, usually seen as a line across the cell. In 44% of the cells, TagO was found at the septum “ahead” of PBP4, meaning that either TagO is already at the septum, seen as two spots, whereas PBP4 is still scattered around the cell membrane or that TagO is already across the entire septum, whereas PBP4 is still in a ring around the division septum.
Fig. 4.
Synthesis of teichoic acids, and not TagO protein itself, is required for PBP4 recruitment to the division septum. (A) Quantification of septum versus lateral membrane fluorescence (fluorescence ratios, FR) and fluorescence microscopy images for PBP4–YFP protein fusion in ΔtagO mutants complemented with wild-type TagO protein (RNΔtagOPBP4YFPptagOwt) or with different TagO mutants (RNΔtagOPBP4YFPptagOD87A, D88A, D87A/D88A, G152A, and N198A). Quantification was performed in 100 cells that displayed closed septa for each strain. Horizontal lines correspond to average FR values. FR values over 2 indicate septal localization, and FR values equal to or under 2 indicate that a protein is dispersed over the cell surface. p values < 10−7. Scale bar: 1 μm. (B) WTAs were isolated from RNΔtagOptagO and RNΔtagOptagOD87A, D88A, D87A/D88A, G152A, and N198A and analyzed by native PAGE stained with alcian blue/silver stain. Mutations of the aspartic acids and glycine residues led to a decrease or absence of the WTAs. (C) Comparison between the levels of WTA and the degree of PBP4 localization to the division septa (calculated as described in Materials and Methods) indicates a strong correlation between the amount of WTA present in the cell and the ability of PBP4 to localize at the septum. (D) Fluorescence microscopy images of RNTagOwtGFP and RNTagOG152AGFP showing that the TagOG152A–GFP fusion localizes to the division septum, similarly to the GFP fusion to the wild-type TagO protein. Scale bar: 1 μm.
Synthesis of Teichoic Acids, and Not TagO Protein Itself, Is Required for PBP4 Recruitment to the Division Septum.
To elucidate whether it was the presence of the TagO protein at the septum or the activity of the TagO protein (also implying the presence of teichoic acids) at the septum that was required for PBP4 recruitment, we constructed a series of strains expressing TagO proteins with single amino acid changes, with the aim of selecting mutants with loss of TagO activity (i.e., lack of TA production) but with the ability to correctly localize at the septum when fused to GFP protein (suggesting that the protein may be correctly folded). Four conserved residues of TagO (D87, D88, G152, N198) were individually substituted with alanine residues, and a double mutant in which D87 and D88 were simultaneously substituted with alanines was also constructed. The different TagO proteins (wild type and mutants) were expressed from the replicative pMAD plasmid (35) under the control of the native tagO promoter in the RNΔtagO background and tested for their ability to catalyze TA synthesis (Fig. 4B). Expression of TagOD87A and TagOD87A/D88A did not result in the production of detectable amounts of TAs; expression of TagOD88A and TagOG152A led to the production of significantly reduced amounts of TAs (less than 25% of wild-type levels); and expression of TagON198A resulted in a small decrease in the amount of TAs produced when compared with expression of wild-type TagO protein (74% of wild-type levels). The localization of PBP4–YFP was then determined in cells expressing either TagOwt or the different TagO mutants, and we found that PBP4 was unable to localize correctly at the division septum in the four mutants with significantly reduced levels of WTAs (Fig. 4A). Moreover, there is a direct correlation between the amount of WTA production and the fraction of PBP4 recruited to the division septum (Fig. 4C).
It was possible that PBP4 delocalization in strains expressing TagO proteins with lower or no activity was not due to the lack of TAs at the septum, but to the fact that mutated TagO proteins were degraded or had lost their septal localization. We therefore selected TagOG152A for further localization studies because it was still able to produce small amounts of WTAs, implying that the protein was probably correctly folded. However, TagOG152A showed some of the phenotypes characteristic of tagO mutants, such as phage resistance, decreased cross-linking, or cell clustering (Fig. S5), implying that the amount of WTA produced was not sufficient to fully complement the phenotype of RNΔtagO. When TagOG152A was fused to GFP and expressed in RN4220, the protein correctly localized to the septum, similarly to TagOwt (Fig. 4D). Furthermore, introduction of the G152A mutation in TagO–GFP did not result in reduced levels of expression of the fluorescent protein (Fig. S5). The fact that TagOG152A had lower activity but maintained correct folding and localization, and was unable to recruit PBP4 to the septum, strongly suggests that the presence of WTA, and not the presence of the TagO protein itself, was required for PBP4 localization.
Discussion
Teichoic acids have been reported to be involved in cell growth, cell division, and morphogenesis (4, 5, 36), but their exact role in these processes remains unknown. In this study, we show that WTAs have a fundamental role in PGN metabolism as they modulate the degree of cross-linking by temporally and spatially regulating the recruitment of PBP4 to the site of cell-wall synthesis, the division septum.
The synthesis of PGN in S. aureus occurs mainly through the action of PBPs 1–4. PBP1 is a monofunctional transpeptidase, essential for cell viability and required for septation and cell separation at the end of cell division (37). It localizes to the division septum through a mechanism that is independent of its ability to bind its substrate (38). PBP2 is an essential bifunctional transglycosylase and transpeptidase that plays a central role in the ability of bacteria to express their resistance to antibiotics (39) and localizes to the division septum in a way that is dependent on its ability to recognize the translocated substrate (32). PBP3 and PBP4 are nonessential, monofunctional transpeptidases whose localization has not yet been studied in detail (22).
We have shown here that in wild-type cells PBP4 can be found at the septum of S. aureus, similarly to PBP1 and PBP2. However, in a different way from these two proteins, recruitment of PBP4 to the septum is dependent upon the synthesis of WTAs. In S. aureus strains lacking TagO, the first enzyme in the teichoic acid biosynthesis pathway, PBP4 no longer accumulates specifically at the division septum, but instead is dispersed over the entire cell membrane. Concomitantly with PBP4 delocalization, the level of PGN cross-linking in ΔtagO mutants is severely decreased, a phenotype also observed by Schlag et al. (40) while this manuscript was in preparation.
Recruitment of PBP4 to the septum does not seem to occur via a direct protein–protein interaction with TagO because (i) PBP4 and TagO do not interact in a bacterial two-hybrid screening, (ii) the two proteins do not colocalize in 49% of the cells in early stages of septum synthesis, and (iii) the presence of (inactive) TagO protein properly localized at the septum is not sufficient to keep PBP4 at that location. Instead, recruitment of PBP4 to the septum seems dependent on the septal synthesis of WTAs. If this synthesis is abolished, either by complete removal of TagO or by generating TagO point mutants, which lose their activity while maintaining correct localization at the septum, then PBP4 loses its septal localization and becomes unable to perform its function in the synthesis of highly cross-linked PGN. The fact that an intact PBP4 is unable to perform its function when incorrectly localized may be due to the substrate being found only at the septum or to the lateral PGN exhibiting a different structure when compared with the septal PGN, which may not allow the addition of further cross-links between the glycan strands.
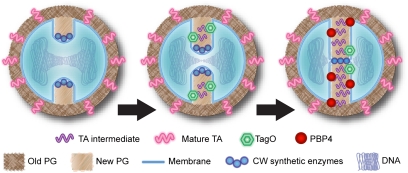
On the basis of the results described in this work, we propose that teichoic acid synthesis functions not only as a spatial cue, but also as a temporal cue, for PBP4 recruitment to the division septum. Fig. 5 illustrates a model in which the initial cell-wall synthetic machinery is recruited to the division septum in the early stages of its formation. TagO, and most likely the remaining enzymes involved in WTA biosynthesis, are recruited to the septum and initiate WTA biosynthesis, which functions as a temporal indication that early PGN biosynthesis is complete and that PGN can be further processed to become highly cross-linked. PBP4 (which, as we have shown, arrives at the septum later than TagO) is then recruited to the septum, where it takes over the last steps of PGN synthesis, performing the final weaving of the PGN mesh. Importantly, it is likely that recruitment of PBP4 is not mediated by fully synthesized/mature WTAs, which are present throughout entire surface of S. aureus (but may not yet be present at the septum), but rather by an immature form of WTA corresponding to an intermediate of WTA biosynthesis, which is encountered only at the septum. One hypothesis would be that the addition of d-alanyl groups to the WTA backbone, catalyzed by the enzymes encoded in the dltABCD operon, would be essential for binding of PBP4 to the WTA because the natural substrate of PBP4 is the d-alanyl-d-alanine terminus of the peptidoglycan muropeptide precursor. However, we have deleted the dltABCD operon from RN4220 and shown that it has no significant effect on localization of PBP4 (Fig. S6).
Fig. 5.
Model for the role of teichoic acids synthesis in PBP4 recruitment to the septum. The early cell-wall synthetic machinery assembles at the division site, leading to the synthesis of new PGN, with low levels of cross-linking (Left). TagO (together with other WTA synthetic enzymes) is recruited to the septum by an unknown mechanism, leading to the synthesis of intermediate molecules in TA biosynthesis (Center). These intermediates (or another cellular component dependent on TA biosynthesis) function as a temporal and spatial cue for PBP4 recruitment to the division septum, allowing the synthesis of highly cross-linked PGN to occur in a regulated manner (Right).
The fact that synthesis of WTA and PGN share the same lipid carrier—bactoprenol—for their precursors led us to think of an alternative model for PBP4 delocalization. Bactoprenol is usually found in limiting amounts in the cell. Therefore, inhibition of WTA synthesis could increase the availability of bactoprenol for PGN synthesis, leading to an increase in the metabolic flux toward PGN synthesis. This could then result in the increased synthesis, and possibly delocalization, of lipid II (bactoprenol linked to a dissacharide-pentapeptide with a pentaglycine crossbridge), the substrate of PBPs, which could be the driving force for PBP4 delocalization. We have tested this hypothesis by purifying the lipid-linked PGN precursors (Fig. S7). The fact that there was no detectable accumulation of lipid II in the ΔtagO mutant led us to rule out changes in lipid II concentrations as the cause for PBP4 delocalization. Therefore, although we cannot formally rule out the possibility that delocalization of PBP4 results not from the absence of WTA intermediates, but from other cellular changes that are themselves caused by the depletion of WTA intermediates, we currently favor the model depicted in Fig. 5.
Interestingly, while this manuscript was in preparation, Schlag and colleagues (40) reported that WTAs are involved in targeting the bifunctional autolysin Atl to the septum. The authors propose an exclusion strategy in which mature WTAs, which are present throughout the mature cell wall but absent (or in lower concentration) at the septum, would prevent binding of Atl to the old cell wall but not to the septal region. Therefore, WTAs may play a key role not only in the regulation of the secondary cross-linking of PGN, but also in regulating the cleavage of the PGN macromolecule, coordinating (or temporally and spatially restricting) both its synthesis and degradation/autolysis.
Why does S. aureus require such fine-tuning of the level of the cross-linking of its PGN? One possibility may be that careful regulation of the timing of PGN cross-linking may be required to ensure the covalent attachment of different molecules to the PGN. Delaying the production of highly cross-linked PGN would permit the introduction of bulky glycopolymers, such as WTAs or large proteins, through the assembled PGN. Afterward, staphylococcal cells would promote cross-linking of PGN to high levels, which, as we have shown, renders it more resistant to lysozyme, an enzyme produced by hosts as a defense against bacterial pathogens.
Materials and Methods
Bacterial Strains and Growth Conditions.
The bacterial strains used in this study are listed in Table S1, and the details of their construction are described in SI Materials and Methods. Primers used in this study are listed in Table S2. Strains and plasmids used in the bacterial two hybrid assays are listed in Table S3. S. aureus strains were grown at 30 °C in tryptic soy broth medium (Difco) supplemented with appropriate antibiotics when required (erythromycin 10 μg/mL or kanamycin 50 μg/mL and neomycin 50 μg/mL; Sigma-Aldrich) and transformed by electroporation as previously described (41). E. coli strains were grown at 37 °C in Luria–Bertani medium (Difco) supplemented with 100 μg/mL of ampicillin (Sigma-Aldrich).
Wall Teichoic Acid Analysis.
WTAs were extracted by alkaline hydrolysis from overnight cultures, analyzed by native polyacrylamide gel electrophoresis, and visualized by combined alcian blue/silver staining, as previously described (42). ImageJ software was used to quantify the percentage of WTA produced by each strain (43). The signal intensity of each lane was quantified and normalized against the corresponding value for the wild type (considered as 100%).
Peptidoglycan Purification and Analysis.
PGN from NCTC8325-4, NCTCΔtagO, and NCTCΔpbpD was prepared from exponentially growing cells as previously described (44) and as detailed in SI Materials and Methods.
Detection of PBPs.
Membrane protein extracts were prepared from exponentially growing cells as previously described (28) and as detailed in SI Materials and Methods.
Fluorescence Microscopy.
S. aureus strains were grown to midexponential phase and observed by fluorescence microscopy on a thin layer of 1% agarose in PBS. When necessary, cells were stained with membrane dye Nile Red (3 μg/mL; Molecular Probes). Images were obtained using a Zeiss Axio Observer.Z1 microscope equipped with a Photometrics CoolSNAP HQ2 camera (Roper Scientific using Metamorph software (Meta Imaging series 7.5) and analyzed using Image J software (43).
Fluorescence ratio (FR) was determined by quantifying the fluorescence at the center of the division septa (only cells with closed septa were considered for this analysis) divided by the fluorescence at the lateral wall. Average background fluorescence was subtracted from both values. Quantification was performed for at least 100 cells with complete septa for each strain. The percentage of PBP4 localized at the division septa (Fig. 4C) was calculated from the ratio of the FR value obtained for each RNΔtagOPBP4YFPptagOmut strain divided by the FR value obtained for RNΔtagOPBP4YFPptagOwt.
Supplementary Material
Acknowledgments
We thank Dr. D.-J. Scheffers for critical reading of the manuscript, Dr. H. Komatsuzawa and Dr. M. Sugai (Hiroshima University, Japan) for the generous gift of PBP1 and PBP4 antibodies, and Dr. G. Karimova (Institut Pasteur, France) for the generous gift of the BTH plasmids. This work was funded by Fundação para a Ciência e Tecnologia through research Grants POCI/SAU-IMI/56501/2004 and PTDC/SAU-MII/75696/2006 (to S.R.F.), POCI/BIA-MIC/67845/2006 and PTDC/BIA-MIC/099151/2008 (to M.G.P.) and fellowships SFRH/BD/28440/2006 (to M.L.A.), SFRH/BD/41119/2007 (to P.M.P.), SFRH/BPD/23838/2005 (to J.Y.), SFRH/BPD/23812/2005 (P.R.), and SFRH/BD/38732/2007 (H.V.), and a European Molecular Biology Organization long-term fellowship (ALTF 1042-2007) (to J.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1004304107/-/DCSupplemental.
References
- 1.Schleifer KH, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Neuhaus FC, Baddiley J. A continuum of anionic charge: Structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol Mol Biol Rev. 2003;67:686–723. doi: 10.1128/MMBR.67.4.686-723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weidenmaier C, Peschel A. Teichoic acids and related cell-wall glycopolymers in Gram-positive physiology and host interactions. Nat Rev Microbiol. 2008;6:276–287. doi: 10.1038/nrmicro1861. [DOI] [PubMed] [Google Scholar]
- 4.Vergara-Irigaray M, et al. Wall teichoic acids are dispensable for anchoring the PNAG exopolysaccharide to the Staphylococcus aureus cell surface. Microbiology. 2008;154:865–877. doi: 10.1099/mic.0.2007/013292-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oku Y, et al. Pleiotropic roles of polyglycerolphosphate synthase of lipoteichoic acid in growth of Staphylococcus aureus cells. J Bacteriol. 2009;191:141–151. doi: 10.1128/JB.01221-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peschel A, et al. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem. 1999;274:8405–8410. doi: 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 7.Kohler T, Weidenmaier C, Peschel A. Wall teichoic acid protects Staphylococcus aureus against antimicrobial fatty acids from human skin. J Bacteriol. 2009;191:4482–4484. doi: 10.1128/JB.00221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peschel A, Vuong C, Otto M, Götz F. The D-alanine residues of Staphylococcus aureus teichoic acids alter the susceptibility to vancomycin and the activity of autolytic enzymes. Antimicrob Agents Chemother. 2000;44:2845–2847. doi: 10.1128/aac.44.10.2845-2847.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins LV, et al. Staphylococcus aureus strains lacking D-alanine modifications of teichoic acids are highly susceptible to human neutrophil killing and are virulence attenuated in mice. J Infect Dis. 2002;186:214–219. doi: 10.1086/341454. [DOI] [PubMed] [Google Scholar]
- 10.Bera A, et al. Influence of wall teichoic acid on lysozyme resistance in Staphylococcus aureus. J Bacteriol. 2007;189:280–283. doi: 10.1128/JB.01221-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chatterjee AN. Use of bacteriophage-resistant mutants to study the nature of the bacteriophage receptor site of Staphylococcus aureus. J Bacteriol. 1969;98:519–527. doi: 10.1128/jb.98.2.519-527.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heptinstall S, Archibald AR, Baddiley J. Teichoic acids and membrane function in bacteria. Nature. 1970;225:519–521. doi: 10.1038/225519a0. [DOI] [PubMed] [Google Scholar]
- 13.Xia G, Kohler T, Peschel A. The wall teichoic acid and lipoteichoic acid polymers of Staphylococcus aureus. Int J Med Microbiol. 2010;300:148–154. doi: 10.1016/j.ijmm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 14.Schirner K, Marles-Wright J, Lewis RJ, Errington J. Distinct and essential morphogenic functions for wall- and lipo-teichoic acids in Bacillus subtilis. EMBO J. 2009;28:830–842. doi: 10.1038/emboj.2009.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.D'Elia MA, Millar KE, Beveridge TJ, Brown ED. Wall teichoic acid polymers are dispensable for cell viability in Bacillus subtilis. J Bacteriol. 2006;188:8313–8316. doi: 10.1128/JB.01336-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Formstone A, Carballido-López R, Noirot P, Errington J, Scheffers D-J. Localization and interactions of teichoic acid synthetic enzymes in Bacillus subtilis. J Bacteriol. 2008;190:1812–1821. doi: 10.1128/JB.01394-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel RA, Errington J. Control of cell morphogenesis in bacteria: Two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- 18.Fedtke I, et al. A Staphylococcus aureus ypfP mutant with strongly reduced lipoteichoic acid (LTA) content: LTA governs bacterial surface properties and autolysin activity. Mol Microbiol. 2007;65:1078–1091. doi: 10.1111/j.1365-2958.2007.05854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.D'Elia MA, et al. Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem Biol. 2009;16:548–556. doi: 10.1016/j.chembiol.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- 21.de Lencastre H, Oliveira D, Tomasz A. Antibiotic resistant Staphylococcus aureus: A paradigm of adaptive power. Curr Opin Microbiol. 2007;10:428–435. doi: 10.1016/j.mib.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheffers D-J, Pinho MG. Bacterial cell wall synthesis: New insights from localization studies. Microbiol Mol Biol Rev. 2005;69:585–607. doi: 10.1128/MMBR.69.4.585-607.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gally D, Archibald AR. Cell wall assembly in Staphylococcus aureus: Proposed absence of secondary crosslinking reactions. J Gen Microbiol. 1993;139:1907–1913. doi: 10.1099/00221287-139-8-1907. [DOI] [PubMed] [Google Scholar]
- 24.Łeski TA, Tomasz A. Role of penicillin-binding protein 2 (PBP2) in the antibiotic susceptibility and cell wall cross-linking of Staphylococcus aureus: Evidence for the cooperative functioning of PBP2, PBP4, and PBP2A. J Bacteriol. 2005;187:1815–1824. doi: 10.1128/JB.187.5.1815-1824.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Memmi G, Filipe SR, Pinho MG, Fu Z, Cheung A. Staphylococcus aureus PBP4 is essential for beta-lactam resistance in community-acquired methicillin-resistant strains. Antimicrob Agents Chemother. 2008;52:3955–3966. doi: 10.1128/AAC.00049-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strandén AM, Ehlert K, Labischinski H, Berger-Bächi B. Cell wall monoglycine cross-bridges and methicillin hypersusceptibility in a femAB null mutant of methicillin-resistant Staphylococcus aureus. J Bacteriol. 1997;179:9–16. doi: 10.1128/jb.179.1.9-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sieradzki K, Tomasz A. Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. J Bacteriol. 1999;181:7566–7570. doi: 10.1128/jb.181.24.7566-7570.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sieradzki K, Pinho MG, Tomasz A. Inactivated pbp4 in highly glycopeptide-resistant laboratory mutants of Staphylococcus aureus. J Biol Chem. 1999;274:18942–18946. doi: 10.1074/jbc.274.27.18942. [DOI] [PubMed] [Google Scholar]
- 29.Sieradzki K, Tomasz A. Alterations of cell wall structure and metabolism accompany reduced susceptibility to vancomycin in an isogenic series of clinical isolates of Staphylococcus aureus. J Bacteriol. 2003;185:7103–7110. doi: 10.1128/JB.185.24.7103-7110.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soldo B, Lazarevic V, Karamata D. tagO is involved in the synthesis of all anionic cell-wall polymers in Bacillus subtilis 168. Microbiology. 2002;148:2079–2087. doi: 10.1099/00221287-148-7-2079. [DOI] [PubMed] [Google Scholar]
- 31.Bera A, Herbert S, Jakob A, Vollmer W, Götz F. Why are pathogenic staphylococci so lysozyme resistant? The peptidoglycan O-acetyltransferase OatA is the major determinant for lysozyme resistance of Staphylococcus aureus. Mol Microbiol. 2005;55:778–787. doi: 10.1111/j.1365-2958.2004.04446.x. [DOI] [PubMed] [Google Scholar]
- 32.Pinho MG, Errington J. Recruitment of penicillin-binding protein PBP2 to the division site of Staphylococcus aureus is dependent on its transpeptidation substrates. Mol Microbiol. 2005;55:799–807. doi: 10.1111/j.1365-2958.2004.04420.x. [DOI] [PubMed] [Google Scholar]
- 33.Pinho MG, Errington J. Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol. 2003;50:871–881. doi: 10.1046/j.1365-2958.2003.03719.x. [DOI] [PubMed] [Google Scholar]
- 34.Karimova G, Pidoux J, Ullmann A, Ladant D. A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc Natl Acad Sci USA. 1998;95:5752–5756. doi: 10.1073/pnas.95.10.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnaud M, Chastanet A, Débarbouillé M. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl Environ Microbiol. 2004;70:6887–6891. doi: 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gründling A, Schneewind O. Synthesis of glycerol phosphate lipoteichoic acid in Staphylococcus aureus. Proc Natl Acad Sci USA. 2007;104:8478–8483. doi: 10.1073/pnas.0701821104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pereira SF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. Role of PBP1 in cell division of Staphylococcus aureus. J Bacteriol. 2007;189:3525–3531. doi: 10.1128/JB.00044-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pereira SF, Henriques AO, Pinho MG, de Lencastre H, Tomasz A. Evidence for a dual role of PBP1 in the cell division and cell separation of Staphylococcus aureus. Mol Microbiol. 2009;72:895–904. doi: 10.1111/j.1365-2958.2009.06687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pinho MG, de Lencastre H, Tomasz A. An acquired and a native penicillin-binding protein cooperate in building the cell wall of drug-resistant staphylococci. Proc Natl Acad Sci USA. 2001;98:10886–10891. doi: 10.1073/pnas.191260798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schlag M, et al. Role of staphylococcal wall teichoic acid in targeting the major autolysin Atl. Mol Microbiol. 2010;75:864–873. doi: 10.1111/j.1365-2958.2009.07007.x. [DOI] [PubMed] [Google Scholar]
- 41.Veiga H, Pinho MG. Inactivation of the SauI type I restriction-modification system is not sufficient to generate Staphylococcus aureus strains capable of efficiently accepting foreign DNA. Appl Environ Microbiol. 2009;75:3034–3038. doi: 10.1128/AEM.01862-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meredith TC, Swoboda JG, Walker S. Late-stage polyribitol phosphate wall teichoic acid biosynthesis in Staphylococcus aureus. J Bacteriol. 2008;190:3046–3056. doi: 10.1128/JB.01880-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abramoff MD, Magelhaes PJ, Ram SJ. Image processing with ImageJ. Biophoton Itl. 2004;11:36–42. [Google Scholar]
- 44.Filipe SR, Tomasz A, Ligoxygakis P. Requirements of peptidoglycan structure that allow detection by the Drosophila Toll pathway. EMBO Rep. 2005;6:327–333. doi: 10.1038/sj.embor.7400371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.