Abstract
Protein arginine methylation, one of the most abundant and important posttranslational modifications, is involved in a multitude of biological processes in eukaryotes, such as transcriptional regulation and RNA processing. Symmetric arginine dimethylation is required for snRNP biogenesis and is assumed to be essential for pre-mRNA splicing; however, except for in vitro evidence, whether it affects splicing in vivo remains elusive. Mutation in an Arabidopsis symmetric arginine dimethyltransferase, AtPRMT5, causes pleiotropic developmental defects, including late flowering, but the underlying molecular mechanism is largely unknown. Here we show that AtPRMT5 methylates a wide spectrum of substrates, including some RNA binding or processing factors and U snRNP AtSmD1, D3, and AtLSm4 proteins, which are involved in RNA metabolism. RNA-seq analyses reveal that AtPRMT5 deficiency causes splicing defects in hundreds of genes involved in multiple biological processes. The splicing defects are identified in transcripts of several RNA processing factors involved in regulating flowering time. In particular, splicing defects at the flowering regulator FLOWERING LOCUS KH DOMAIN (FLK) in atprmt5 mutants reduce its functional transcript and protein levels, resulting in the up-regulation of a flowering repressor FLOWERING LOCUS C (FLC) and consequently late flowering. Taken together, our findings uncover an essential role for arginine methylation in proper pre-mRNA splicing that impacts diverse developmental processes.
PRMT5 is a type II protein arginine methyltransferase that catalyzes the formation of monomethylarginine and symmetric ω-NG, N′G-dimethylarginine (SDMA) (1). PRMT5 is highly conserved among yeast, animals, and higher plants, and is implicated in diverse cellular and biological processes, including transcriptional regulation, RNA metabolism (1, 2), ribosome biogenesis (3), Golgi apparatus structure maintenance (4), and cell cycle regulation (5). In mouse, fly, and worm, PRMT5 orthologs are involved in germ-cell formation, specification, and maintenance (6–10).
In mammalian cells, PRMT5 localizes to both the cytoplasm and the nucleus, and methylates multiple histone and nonhistone proteins (1). In the nucleus, PRMT5 usually functions in repressing target genes by methylating histone H4 Arginine 3 (H4R3), H3R8, as well as transcription factors/regulators (5, 11, 12). In the cytoplasm, PRMT5 mainly exists in a 20S methylosome complex, consisting of spliceosomal U snRNP (uridine-rich small nuclear riboucleoprotein particles) Sm proteins (including Sm B/B′, D1, D2, D3, E, F, and G), PRMT5, pICln, and WD repeat protein/MEP50 (13–15). In this complex, U1, 2, 4, 5 snRNP Sm proteins Sm B/B′, D1, D3, and U6 snRNP-specific Sm-like protein LSm4 are symmetrically dimethylated by PRMT5 (13, 16). Such methylation can increase the binding affinity of these Sm proteins for the downstream recipient, Survival Motor Neuron, the spinal muscular atrophy disease gene product (17, 18). Subsequently, the PRMT5- and Survival Motor Neuron-complexes cooperate to load the Sm proteins onto U snRNAs, forming U snRNPs (19, 20). As such, it is assumed that symmetric arginine dimethylation is essential for pre-mRNA splicing; however, only in vitro biochemical evidence has emerged (21), and to what extent PRMT5 affects splicing in vivo remains elusive.
AtPRMT5, an Arabidopsis homolog of human PRMT5, was defined as a type II enzyme for its ability to symmetrically dimethylate histone H4, H2A, and myelin basic protein in vitro (22). AtPRMT5 deficiency causes pleiotropic phenotypes, including delayed flowering, growth retardation, dark green and curled leaves, and reduced sensitivity to vernalization (22–24), implying a critical role for AtPRMT5 in regulating essential developmental processes in Arabidopsis. However, how AtPRMT5 impacts these biological processes is largely unknown.
In this study, we show that in addition to histones, AtPRMT5 methylates various nonhistone substrates, including RNA processing factors, U snRNP AtSmD1, D3, and AtLSm4 proteins. We also show that mutations in AtPRMT5 leads to splicing defects in hundreds of genes that are involved in multiple cellular and biological processes. In addition, transcripts of several RNA processing factors involved in regulating flowering time are identified to have splicing defects in atprmt5 mutants. We further demonstrate that splicing defects at the first intron of FLK (FLOWERING LOCUS KH DOMAIN) reduce its functional transcript and protein levels leading to the up-regulation of FLC (FLOWERING LOCUS C). Our findings uncover an important link between arginine methylation and proper pre-mRNA splicing.
Results
AtPRMT5 Methylates a Large Spectrum of Proteins.
Our previous study showed that AtPRMT5 catalyzes symmetric dimethylation of H4R3 (H4R3me2s) in vitro (22). To test whether loss of function of AtPRMT5 affects global H4R3me2s levels in vivo, we used an anti-H4R3me2s antibody to probe acidic extracts from atprmt5 mutants and wild-type Columbia (Col); however, no significant change was detected at H4 (Fig. S1). Nevertheless, some additional bands were only observed in Col but not in atprmt5 mutants, suggesting the cross-reaction of the anti-H4R3me2s antibody with many SDMA-containing nonhistone proteins, which are putative in vivo substrates of AtPRMT5.
To identify these substrates of AtPRMT5 in vivo, a proteomic approach of 2D electrophoresis (2-DE) combined with immunoblotting analysis was employed using anti-H4R3me2s and two symmetric arginine dimethyl-specific antibodies, SYM10 (21) and SYM11 (25). Through this method, 29 protein spots, for which the immuoblotting signals were completely lost in atprmt5 mutants, were initially found (Fig. S2) and subsequently identified by MALDI-TOF MS as representing 26 proteins (Table S1). These substrates were involved in multiple biological networks, including response to stress, response to abiotic and biotic stimulus, and response to metal ion, as well as DNA, RNA, and protein metabolism (Table S1), which is consistent with the pleiotropic phenotypes of atprmt5 mutants.
Among the methylated proteins identified by MS, 15% are RNA binding or processing factors. Arabidopsis glycine-rich RNA-binding protein 7 (AtGRP7) and AtGRP8 were detected by anti-H4R3me2s or SYM10 antibodies in Col, but not in atprmt5 mutants (Fig. 1A and Table S1), and further analysis proved that Arginine 141 (R141) in both AtGRP7 and AtGRP8 were methylated (Fig. S3). In vitro methyltransferase activity assay also confirmed that they were substrates of AtPRMT5 (Fig. 1B), indicating that they are bona fide AtPRMT5 substrates. AtGRP7 and AtGRP8 are homologs of human heterogeneous nuclear ribonucleoprotein A1 (hnRNP A1) and hnRNP A2/B1 that negatively regulate pre-mRNA splicing by competing with splicing factors (26). AtGRP7 and AtGRP8 have been also reported to autoregulate and reciprocally crossregulate the splicing of their pre-mRNAs (27).
Fig. 1.
AtPRMT5 methylates certain RNA processing-related proteins. (A) Identification of in vivo substrates of AtPRMT5. The 2-DE stained by Coomassie blue is shown on the top. Methylation status of proteins was detected with indicated antibodies (lower three panels). Spots marked by circles indicate the disappearance of methylation in atprmt5 mutants. Arrows indicate the corresponding proteins identified by MS. (B) Methyltransferase activity of GST and GST-AtPRMT5 with GST-AtGRP7, GST-AtGPR8 fusion proteins. (C) Methyltransferase activity of GST-AtPRMT5 with AtSmD1a, AtSmD1b, AtSmD3a, AtSmD3b and AtLSm4 fusion proteins. Autoradiography indicates AtPRMT5 methylates these AtSm and AtLSm proteins. “+” represents GST-AtPRMT5; “–” represents GST control. (D) In vivo methylation status of AtSmD1b and AtLSm4 in Col and atprmt5. AtSmD1b-GFP and AtLSm4-GFP immunoprecipitated by anti-GFP monoclonal antibody were immunoblotted with SYM10 antibody for arginine methylation and anti-GFP polyclonal antibody for loading controls.
In addition to hnRNPs, it has been reported that U snRNP Sm core proteins are affected by arginine methylation inhibitors (21, 28). In Arabidopsis, SmD1, D3, and LSm4 are conserved (Fig. S4). AtSmD1 proteins (AtSmD1-a and AtSmD1-b) and AtSmD3 proteins (AtSmD3-a and AtSmD3-b) are encoded by duplicated genes (26). Similar to human Sm protein, the C-terminal regions of AtSmD1, AtSmD3, and AtLSm4 proteins contain several RG di-peptides (Fig. S4). Methyltransferase activity assay showed that AtSmD1, D3, and AtLSm4 proteins could be methylated by AtPRMT5 in vitro (Fig. 1C). Consistently, methylation levels of AtSmD1b and AtLSm4 in atprmt5 mutants were largely decreased compared with those in Col (Fig. 1D). This conserved modification from mammals to plants highlights the importance of SDMA in Sm and LSm proteins.
Mutation of AtPRMT5 Leads to Widespread Splicing Defects.
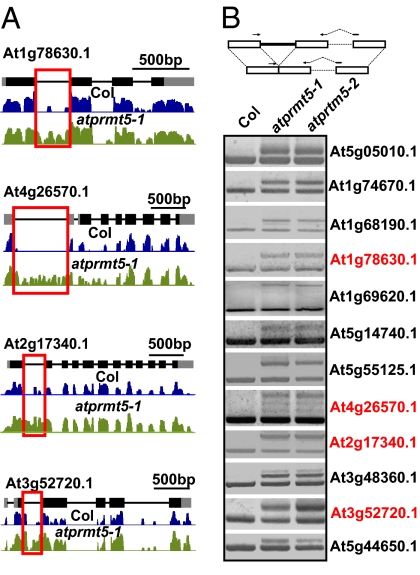
To test the functional consequences of AtPRMT5 on RNA metabolism, we performed ultrahigh-throughput RNA sequencing (RNA-seq) on the SOLiD (sequencing by oligonucleotide ligation and detection) platform to examine the global pre-mRNA splicing changes between Col and atprmt5 mutants. From Col and atprmt5 libraries, respectively, 44.8 and 41.3 million of single-end reads (35 nucleotides) were produced, in which 19.8 million and 17.0 million contiguous reads were aligned to the Arabidopsis genome; 1.5 million and 1.4 million reads were aligned to the annotated splice junctions (Table S2). Approximately 91% of total matched reads were mapped to unique loci, representing 13,407 and 13,703 intron-containing genes in Col and atprmt5 mutants, respectively [(reads per kilobase of exons per million mapped reads) RPKM > 3] (Table S2). Compared with Col, 648 intron retention events out of 600 genes were represented by aberrant transcripts in atprmt5 mutants (P < 0.01, intron reads coverage > 80%), demonstrating that AtPRMT5 regulates genome-wide pre-mRNA splicing. The intron retention events (P < 0.01, intron reads coverage ≥ 95%) in atprmt5 mutants were listed in Dataset S1. Fourteen of 16 selected genes were further validated by RT-PCR using intron-flanking primers (Table S3), in which the corresponding retained introns were detected in atprmt5 mutants, but not in Col (Fig. 2).
Fig. 2.
AtPRMT5 is required for pre-mRNA splicing. (A) Four examples of mRNAs with splicing defects detected by RNA-seq. Annotated gene structures are shown (Top), with thick lines representing exons and thin lines representing introns. Wiggle plots representing the normalized reads coverage in a logarithmic scale (log2) are shown in blue for Col (Middle) and in green for atprmt5 mutants (Bottom). The red frames indicate the retained introns. (B) Validation of the intron retention events in 12 genes by RT-PCR. The upper and lower bands represent the unspliced and spliced forms, respectively. The wiggle plots of genes marked in red are shown in A. The diagrammatic presentation of primer position is shown in the top panel. The dashed lines in the primers represent primers crossing introns that are normally spliced to avoid genomic DNA contamination.
In a previous study, Laubinger et al. showed that SERRATE (SE) and cap-binding complex proteins (CBP80/ABH1 and CBP20) are involved in pre-mRNA splicing, and the first intron is prone to being retained in cbp20, cbp80 (abh1-753), and se mutants, which is consistent with their functions of 5′ cap binding (29). Here the distributions of retained introns did not show such preference in atprmt5 mutants (Fig. S5). In addition, only 13 of 600 aberrantly spliced genes overlapped with those affected in se, cbp80, and cbp20 mutants, indicating that AtPRMT5 regulates splicing differently from SE and CBP80. We also found that pre-mRNAs with more introns tended to undergo aberrant splicing in atprmt5 mutants. The median number of introns in intron-containing genes is four in the Arabidopsis genome (30), whereas that of the affected transcripts in atprmt5 mutants is eight. A similar result was observed in survival motor neuron-deficiency mammals as well (31), indicating a widely conserved mechanism among eukaryotes that pre-mRNAs with more introns (exons) are susceptible to splicing changes.
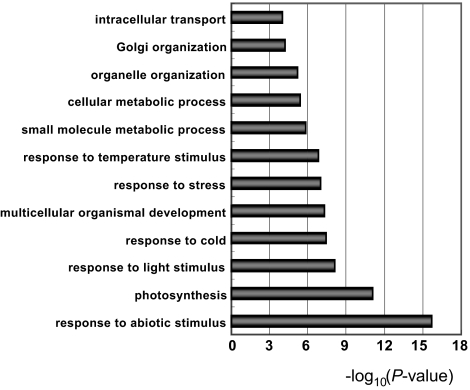
Gene Ontology (GO) analysis of the genes undergoing aberrant splicing in atprmt5 mutants revealed that they were involved in many biological processes, including response to abiotic stimulus (P = 2.09 × 10−16), photosynthesis (P = 8.75 × 10−12), response to light (P = 8.24 × 10−9) and temperature (P = 1.49 × 10−7), response to stress (P = 1.03 × 10−7), and so on (Fig. 3), suggesting that AtPRMT5 is involved in various biological processes through regulating pre-mRNA splicing, consistent with the pleiotropic phenotypes of atprmt5 mutants.
Fig. 3.
Gene ontology terms enriched in genes with splicing defects in atprmt5 mutants.
AtPRMT5 Regulates Pre-mRNA Splicing of Several RNA Processing-Related Flowering Time Regulators.
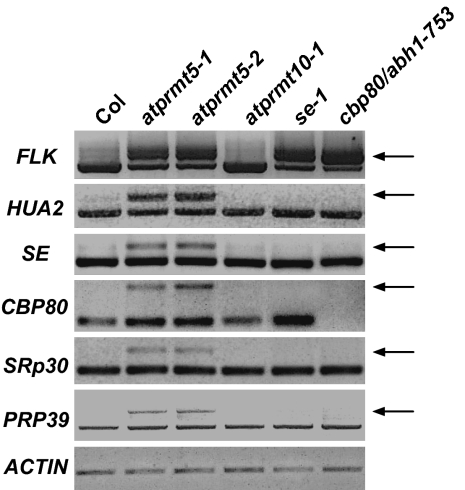
Among all of the defects of atprmt5 mutants, FLC-dependent late flowering is one of the most important phenotypes, because flowering time determination is essential for successful sexual reproduction. In the complicated flowering regulation networks, many RNA processing factors were reported to affect flowering time through either posttranscriptional or unknown mechanisms (32). Here, some transcripts encoding RNA processing factors were identified to have splicing defects in atprmt5 mutants, including HUA2 (a floral repressor that functions in pre-mRNA processing of AGAMOUS) (33, 34), FLK (a KH motif-containing putative RNA binding protein) (35, 36), SRp30 (a serine-arginine rich RNA binding protein involved in pre-mRNA splicing through 5′ splice site selection) (37), PRP39 (a homolog of the yeast mRNA processing protein Prp39p that affects 5′ splice site selection) (38, 39), SE (40), CBP80, and CBP20 (29). Compared with Col and atprmt10 mutants (mutation of a plant-specific asymmetric arginine methyltransferase causing a late flowering phenotype independent of AtPRMT5) (41), the corresponding introns of these mRNAs are inefficiently spliced in atprmt5 mutants (Fig. 4). It is worth noting that mutations in SE, CBP80, and CBP20 also cause aberrant splicing of FLK (29). However, except for FLK, other identified mRNAs were fully spliced in se-1 and cbp80 mutants (Fig. 4), which further confirms that the involvement of AtPRMT5 in splicing is independent of SE and CBP80. These RNA processing factors function differently in both RNA processing and flowering pathways. Therefore, AtPRMT5 regulates the splicing of these RNA processing factors, which might subsequently affect mRNA metabolism and flowering time, and the late flowering phenotype of atprmt5 mutants is an integrative effect of splicing defects of diverse genes in distinct cellular processes rather than an effect on a single target.
Fig. 4.
Analysis of splicing defects in RNA processing-related flowering regulators in atprmt5, atprmt10-1, se-1, and cbp80/abh1-753 mutants. The bands marked by arrows represent the forms with retained introns.
FLK Splicing Defects Reduce Functional Transcript and Protein Levels of FLK.
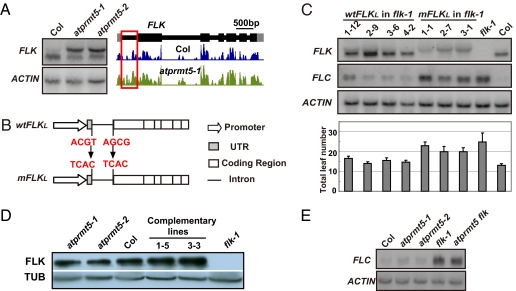
Our previous study showed the up-regulation of total transcripts of FLK and HUA2 in atprmt5 mutants (22). Here, the results indicated that the increase of these total transcripts was caused by accumulation of the unspliced transcripts. In particular, the functional transcript of FLK, an autonomous pathway gene that promotes flowering by repressing FLC expression, decreased 30% in atprmt5 mutants, but the unspliced transcript with intron 1 retention (named as FLK long transcript, FLKL) increased dramatically (Figs. 4 and 5A). Therefore, we hypothesize that the up-regulation of FLC in atprmt5 mutants is caused by misregulation of FLK splicing.
Fig. 5.
AtPRMT5 regulates FLK splicing and flowering time. (A) RNA blot and RNA-seq analysis of the expression and splicing patterns of FLK, with ACTIN as a control. The annotated gene structure and wiggle plots of FLK from Col and atprmt5 are shown (Right). (B) Schematic representation of wild-type FLKL (wtFLKL) and mutated FLKL (mFLKL). In mFLKL, the splice sites were mutated, as shown by the arrows. (C) Expression levels of FLK and FLC (Upper) and average total leaf number at flowering of flk-1 transgenic plants with wtFLKL and mFLKL (Lower) under long-day photo-period conditions. ACTIN was used as a loading control and error bars show SDs. (D) Reduction of FLK protein levels in atprmt5 mutants. Immunoblotting was performed using anti-FLK antibody, with Tubulin as a loading control. Lines “1–5” and “3–3” represent individual lines for AtPRMT5 rescued with the AtPRMT5 transgene. (E) RNA blot analysis of FLC mRNA levels in Col, atprmt5-1, atprmt5-2, flk-1, and atprmt5-2 flk-1 mutants.
To test this hypothesis, we generated two FLK cDNA constructs containing the first intron, namely wtFLKL (wild-type FLKL) and mFLKL (mutated FLKL), driven by the endogenous FLK promoter. Both constructs had identical sequences except for the 5′ and 3′ splice sites of the first intron (Fig. 5B), in which mFLKL was mutated and expected to produce a long transcript with an unspliced first intron. Both wtFLKL and mFLKL were transformed into flk-1 mutants. As expected, RNA blot analysis showed that unlike wtFLKL, which can be fully spliced, mFLKL accumulated an unspliced FLKL transcript, which mimics the large transcript in atprmt5 mutants (Fig. 5C) (RNA blot for FLK). The transgenic plants with wtFLKL, but not mFLKL, rescued the late-flowering phenotype of flk-1 mutants with low FLC expression, indicating that wtFLKL was fully functional; but the unproductive splicing form FLKL could not repress FLC expression and hence failed to promote flowering (Fig. 5C). In addition, FLK protein levels were low in atprmt5 compared with Col and atprmt5 mutants rescued with AtPRMT5 (Fig. 5D). Thus, in atprmt5 mutants, FLKL could not compensate for the decrease in FLK mRNA, resulting in the reduction of both functional mRNA and protein levels of FLK. To determine whether the derepression of FLC in atprmt5 may be caused by deficiency of FLK splicing, we analyzed the atprmt5 flk-1 double mutants. FLC mRNA levels in the atprmt5 flk-1 double mutants were similar to those in flk-1 single mutants, indicating that the derepression of FLC in atprmt5 was partially caused by deficiency of FLK splicing and FLK is a major target for AtPRMT5 to regulate FLC expression (Fig. 5E).
Discussion
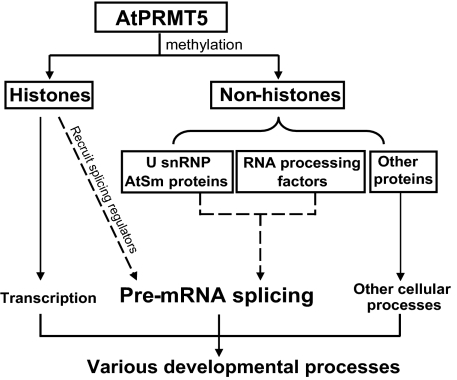
Our analysis provides biochemical and genetic evidence showing that AtPRMT5 methylates diverse substrates and globally affects pre-mRNA splicing under normal growth conditions. This finding is consistent with the pleiotropic phenotypes of atprmt5 mutants. Our findings uncover a general role of AtPRMT5 in pre-mRNA splicing and RNA metabolism, as well as the participation of AtPRMT5 in response to diverse environmental stimuli and in multiple developmental processes. Therefore, we propose that AtPRMT5 methylates histones and many nonhistone proteins. Through histone modifications, AtPRMT5 could directly regulate transcription at the target loci. Histone H4R3 modifications may also provide a mark for coupling the regulation of transcription and alternative splicing similar to the H3K36me3 mark reported recently (42). Through many other nonhistone modifications, AtPRMT5 could directly or indirectly affect pre-mRNA splicing. Therefore, the involvement of AtPRMT5 in plant developmental processes is the integrative effects attributed to the accurate splicing of diverse genes that are not restricted to any single target (Fig. 6).
Fig. 6.
A diagram illustrating the essential role of AtPRMT5 in pre-mRNA splicing.
RNA splicing is a key posttranscriptional regulatory mechanism, conferring extensive proteomic and regulatory diversity to provide more complex genome information. Accurate pre-mRNA splicing, modulated by cis-acting elements and trans-acting factors, is crucial for proper gene expression and normal cellular functions. In humans, disrupted splicing can cause various diseases directly. For example, spinal muscular atrophy (SMA), a common genetic motor neuron-degenerative disease, is caused by mutations in the Survival Motor Neurons (SMN) gene involved in snRNP assembly (43). SMN deficiency results in widespread splicing defects and, therefore, SMA is a general splicing disease (31). SMN may not be a direct substrate of PRMT5; however, Sm B/B′, D1, and D3 are symmetrically dimethylated by PRMT5, increasing the binding affinity between Sm proteins and SMN-complex to promote U snRNP assembly (44). Although direct evidence between PRMT5 and pre-mRNA splicing is lacking in humans, given the fact that PRMT5 is an evolutionarily conserved protein and symmetric arginine dimethylation of Sm proteins by PRMT5 is required for snRNP assembly (20), it will not be surprising if mutations in human PRMT5 cause widespread splicing defects, which may be because of the multiple functions for PRMT5.
A genome-wide study reports that at least ∼42% of intron-containing genes in Arabidopsis are subject to alternative splicing under various stress conditions (45). It is well known that in addition to endogenous signals, proper growth and development of plants are also regulated by environmental cues, such as light, temperature, nutrition, and stress. As a trans-acting factor in pre-mRNA splicing by methylating certain RNA processing factors and U snRNP AtSmD1, D3, and AtLSm4 proteins, AtPRMT5 provides an extra layer of control for proper gene expression and transcriptome plasticity at different developmental stages. Based on the basic function of AtPRMT5 and the pleiotropic phenotypes of atprmt5 mutants, it can be predicted that AtPRMT5 is involved in many other physiological processes besides those we observed here.
By comparing the methylation status between Col and atprmt5 mutants, we successfully identified several dozen proteins as candidate substrates for AtPRMT5 in vivo. Because 2-DE immunoblotting methods show limited recovery of proteins with pI outside the range of 3 to 10, these identified proteins may only represent a small portion of the total AtPRMT5 substrates in vivo. Both in vivo and in vitro data suggest that AtPRMT5 acts upon a large spectrum of substrates and this important posttranslational modification results in diverse consequences, such as regulating the binding affinity between proteins or between proteins and nucleic acids. Therefore, it would be interesting to identify more substrates of AtPRMT5 to better understand how AtPRMT5 is involved in various cellular and developmental processes.
Materials and Methods
Plant Materials and Growth Conditions.
The atprmt5-1, atprmt5-2, and atprmt10-1 mutants and growth conditions were described previously (22, 41). The mutant cbp80 (SALK_016753) was from the SALK T-DNA mutant collection.
Proteomic Analysis.
The 2-DE and in-gel digestion analysis of total proteins from Col and atprmt5 plants were performed as described previously (46). The digested peptides were analyzed by MALDI-TOF MS (Bruker-Franzen) and liquid chromatography tandem-MS (Micromass). Immunoblotting analysis was performed using antibodies against H4R3me2s (ab5823; Abcam), symmetric dimethyl-Arginine SYM10 (07–412; Millipore), and SYM11 (07–413; Millipore).
Coimmunoprecipitation Assay.
Total proteins extracted from 12-d-old seedlings of AtSmD1b-GFP and AtLSm4-GFP transgenic plants were immunoprecipitated with anti-GFP monoclonal antibody (11814460001; Roche), and the immunoprecipitates were detected with anti-GFP polyclonal antibody (632460; Clontech) and antibody against symmetric dimethyl-arginine (SYM10; 07–412; Millipore).
RNA Sequencing.
Total RNAs were extracted from 12-d-old seedlings of wild type Col and atprmt5-1 plants with TRIzol Reagent (15596–026; Invitrogen). Polyadenylated RNAs were isolated using the Oligotex mRNA Midi Kit (70042; Qiagen). The RNA-seq libraries were constructed using SOLiD Whole Transcriptome Analysis Kit following the standard protocol (AB, Applied Biosystems) and sequenced on the Applied Biosystems SOLiD platform to generate high-quality single-end reads of 35 nt in length.
RNA-seq Data Analysis.
Arabidopsis genome sequences and annotated gene models were downloaded from TAIR9 (http://www.arabidopsis.org/). The raw reads were aligned to genome sequences using Bowtie 0.12.5 (47), trimming off a nucleotide each from the 5′ and 3′ ends and allowing up to two mismatches. All unmapped reads were aligned to splice junction sequences based on the TAIR9 gene model annotations with at least three bases on each side of the junction. Reads mapped to multiple locations were discarded and only uniquely mapped reads were used for subsequent analysis. The gene expression levels were measured in RPKM. The presence of more than 3 RPKM in total exons of a gene was the criterion for gene detection (48).
Fisher's Exact Tests in R (http://www.r-project.org/) was performed to identify differential representation of introns in wild-type Col and atprmt5 mutants using read counts from an intron and the corresponding two flanking exons. The cut-offs in minimal-read frequencies of intron and flanking exons are 5 and 4 RPKM, respectively. In total, introns with more than 80% reads coverage and two-sided P value less than 0.01 were regarded as the intron retention events.
GO Analysis.
GOEAST software was used for GO enrichment analysis (49).
Data Deposition.
The RNA sequencing dataset was deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo/) under accession no. GSE21323.
Additional methods are available in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Chentao Lin (University of California Los Angeles) and Dr. Miltos Tsiantis (University of Oxford) for providing flk-1 and se-1 seeds, respectively, and the Arabidopsis Biological Resource Center for providing SALK T-DNA insertion lines. This work was supported by National Basic Research Program of China Grants 2009CB941500 and 2005CB522400 (to X.C.), National Natural Science Foundation of China Grants 90919033 and 30771209 (to C.L.) and 30930048 and 30921061 (to X.C.), and Chinese Academy of Sciences Grant KSCX2-YW-N-047.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The RNA sequencing dataset was deposited in the Gene Expression Omnibus database at NCBI (accession no. GSE21323).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009669107/-/DCSupplemental.
References
- 1.Bedford MT, Clarke SG. Protein arginine methylation in mammals: Who, what, and why. Mol Cell. 2009;33:1–13. doi: 10.1016/j.molcel.2008.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu C, Lu F, Cui X, Cao X. Histone methylation in higher plants. Annu Rev Plant Biol. 2010;61:395–420. doi: 10.1146/annurev.arplant.043008.091939. [DOI] [PubMed] [Google Scholar]
- 3.Ren J, et al. Methylation of ribosomal protein S10 by protein-arginine methyltransferase 5 regulates ribosome biogenesis. J Biol Chem. 2010;285:12695–12705. doi: 10.1074/jbc.M110.103911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Z, et al. PRMT5 regulates Golgi apparatus structure through methylation of the golgin GM130. Cell Res. 2010;20:1023–1033. doi: 10.1038/cr.2010.56. [DOI] [PubMed] [Google Scholar]
- 5.Jansson M, et al. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- 6.Gonsalvez GB, Rajendra TK, Tian L, Matera AG. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol. 2006;16:1077–1089. doi: 10.1016/j.cub.2006.04.037. [DOI] [PubMed] [Google Scholar]
- 7.Yang M, et al. Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis. PLoS Genet. 2009;5:e1000514. doi: 10.1371/journal.pgen.1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ancelin K, et al. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- 9.Vagin VV, et al. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anne J, Ollo R, Ephrussi A, Mechler BM. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development. 2007;134:137–146. doi: 10.1242/dev.02687. [DOI] [PubMed] [Google Scholar]
- 11.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Amente S, et al. Identification of proteins interacting with the RNAPII FCP1 phosphatase: FCP1 forms a complex with arginine methyltransferase PRMT5 and it is a substrate for PRMT5-mediated methylation. FEBS Lett. 2005;579:683–689. doi: 10.1016/j.febslet.2004.12.045. [DOI] [PubMed] [Google Scholar]
- 13.Friesen WJ, et al. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol Cell Biol. 2001;21:8289–8300. doi: 10.1128/MCB.21.24.8289-8300.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meister G, et al. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol. 2001;11:1990–1994. doi: 10.1016/s0960-9822(01)00592-9. [DOI] [PubMed] [Google Scholar]
- 15.Friesen WJ, et al. A novel WD repeat protein component of the methylosome binds Sm proteins. J Biol Chem. 2002;277:8243–8247. doi: 10.1074/jbc.M109984200. [DOI] [PubMed] [Google Scholar]
- 16.Brahms H, Meheus L, de Brabandere V, Fischer U, Lührmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B' and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA. 2001;7:1531–1542. doi: 10.1017/s135583820101442x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friesen WJ, Massenet S, Paushkin S, Wyce A, Dreyfuss G. SMN, the product of the spinal muscular atrophy gene, binds preferentially to dimethylarginine-containing protein targets. Mol Cell. 2001;7:1111–1117. doi: 10.1016/s1097-2765(01)00244-1. [DOI] [PubMed] [Google Scholar]
- 18.Côté J, Richard S. Tudor domains bind symmetrical dimethylated arginines. J Biol Chem. 2005;280:28476–28483. doi: 10.1074/jbc.M414328200. [DOI] [PubMed] [Google Scholar]
- 19.Meister G, Fischer U. Assisted RNP assembly: SMN and PRMT5 complexes cooperate in the formation of spliceosomal UsnRNPs. EMBO J. 2002;21:5853–5863. doi: 10.1093/emboj/cdf585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neuenkirchen N, Chari A, Fischer U. Deciphering the assembly pathway of Sm-class U snRNPs. FEBS Lett. 2008;582:1997–2003. doi: 10.1016/j.febslet.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 21.Boisvert FM, et al. Symmetrical dimethylarginine methylation is required for the localization of SMN in Cajal bodies and pre-mRNA splicing. J Cell Biol. 2002;159:957–969. doi: 10.1083/jcb.200207028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pei Y, et al. Mutations in the type II protein arginine methyltransferase AtPRMT5 result in pleiotropic developmental defects in Arabidopsis. Plant Physiol. 2007;144:1913–1923. doi: 10.1104/pp.107.099531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang X, et al. SKB1-mediated symmetric dimethylation of histone H4R3 controls flowering time in Arabidopsis. EMBO J. 2007;26:1934–1941. doi: 10.1038/sj.emboj.7601647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmitz RJ, Sung S, Amasino RM. Histone arginine methylation is required for vernalization-induced epigenetic silencing of FLC in winter-annual Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:411–416. doi: 10.1073/pnas.0710423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boisvert FM, Côté J, Boulanger MC, Richard S. A proteomic analysis of arginine-methylated protein complexes. Mol Cell Proteomics. 2003;2:1319–1330. doi: 10.1074/mcp.M300088-MCP200. [DOI] [PubMed] [Google Scholar]
- 26.Wang BB, Brendel V. The ASRG database: Identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 2004;5:R102. doi: 10.1186/gb-2004-5-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schöning JC, Streitner C, Meyer IM, Gao Y, Staiger D. Reciprocal regulation of glycine-rich RNA-binding proteins via an interlocked feedback loop coupling alternative splicing to nonsense-mediated decay in Arabidopsis. Nucleic Acids Res. 2008;36:6977–6987. doi: 10.1093/nar/gkn847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, Dreyfuss G. In vivo and in vitro arginine methylation of RNA-binding proteins. Mol Cell Biol. 1995;15:2800–2808. doi: 10.1128/mcb.15.5.2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laubinger S, et al. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reddy AS. Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu Rev Plant Biol. 2007;58:267–294. doi: 10.1146/annurev.arplant.58.032806.103754. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, et al. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Terzi LC, Simpson GG. Regulation of flowering time by RNA processing. Curr Top Microbiol Immunol. 2008;326:201–218. doi: 10.1007/978-3-540-76776-3_11. [DOI] [PubMed] [Google Scholar]
- 33.Doyle MR, et al. HUA2 is required for the expression of floral repressors in Arabidopsis thaliana. Plant J. 2005;41:376–385. doi: 10.1111/j.1365-313X.2004.02300.x. [DOI] [PubMed] [Google Scholar]
- 34.Cheng Y, Kato N, Wang W, Li J, Chen X. Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell. 2003;4:53–66. doi: 10.1016/s1534-5807(02)00399-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mockler TC, et al. Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci USA. 2004;101:12759–12764. doi: 10.1073/pnas.0404552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim MH, et al. A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell. 2004;16:731–740. doi: 10.1105/tpc.019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lopato S, et al. atSRp30, one of two SF2/ASF-like proteins from Arabidopsis thaliana, regulates splicing of specific plant genes. Genes Dev. 1999;13:987–1001. doi: 10.1101/gad.13.8.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lockhart SR, Rymond BC. Commitment of yeast pre-mRNA to the splicing pathway requires a novel U1 small nuclear ribonucleoprotein polypeptide, Prp39p. Mol Cell Biol. 1994;14:3623–3633. doi: 10.1128/mcb.14.6.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang C, et al. The Arabidopsis thaliana AT PRP39-1 gene, encoding a tetratricopeptide repeat protein with similarity to the yeast pre-mRNA processing protein PRP39, affects flowering time. Plant Cell Rep. 2007;26:1357–1366. doi: 10.1007/s00299-007-0336-5. [DOI] [PubMed] [Google Scholar]
- 40.Grigg SP, Canales C, Hay A, Tsiantis M. SERRATE coordinates shoot meristem function and leaf axial patterning in Arabidopsis. Nature. 2005;437:1022–1026. doi: 10.1038/nature04052. [DOI] [PubMed] [Google Scholar]
- 41.Niu L, Lu F, Pei Y, Liu C, Cao X. Regulation of flowering time by the protein arginine methyltransferase AtPRMT10. EMBO Rep. 2007;8:1190–1195. doi: 10.1038/sj.embor.7401111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Luco RF, et al. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang GS, Cooper TA. Splicing in disease: Disruption of the splicing code and the decoding machinery. Nat Rev Genet. 2007;8:749–761. doi: 10.1038/nrg2164. [DOI] [PubMed] [Google Scholar]
- 44.Chari A, et al. An assembly chaperone collaborates with the SMN complex to generate spliceosomal SnRNPs. Cell. 2008;135:497–509. doi: 10.1016/j.cell.2008.09.020. [DOI] [PubMed] [Google Scholar]
- 45.Filichkin SA, et al. Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 2010;20:45–58. doi: 10.1101/gr.093302.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang BC, et al. Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics. 2006;6:2555–2563. doi: 10.1002/pmic.200500657. [DOI] [PubMed] [Google Scholar]
- 47.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 49.Zheng Q, Wang XJ. GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008;36(Web Server issue):W358–W363. doi: 10.1093/nar/gkn276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.