Abstract
We introduce a novel algorithm for actin filament segmentation in a 2D TIRFM image sequence. We treat the 2D time-lapse sequence as a 3D image volume and propose an over-grown active surface model to segment the body of a filament on all slices simultaneously. In order to locate the two ends of the filament on the over-grown surface, a novel 2D spatiotemporal domain is created based on the resulting surface. Two 2D active contour models deform in this domain to locate the two filament ends accurately. Evaluation on TIRFM image sequences with very low SNRs and comparison with a previous method demonstrate the accuracy and robustness of this approach.
1 Introduction
Actin proteins spontaneously assemble into long polymers to build networks and bundles of filaments that are used by cells to move and change shape. An experimental method to study the kinetics of single actin filament growth in vitro is total internal reflection microscopy (TIRFM) [1], [2]. The information on growth kinetics is used to develop mathematical models that describe the dependence of actin filament elongation rate and fluctuations on the concentration of actin and cofactors [1], and the mechanisms that cells use to regulate the dynamics of the actin cytoskeleton [2]. In order to study kinetics based on image information, an essential step is to segment actin filaments in TIRFM image sequences.
There has been previous research on segmentation of actin filaments or microtubules (MT). Li et al. [3] proposed a slice-by-slice Stretch Open Active Contour model to segment and to track actin filaments in a time-lapse sequence. Hadjidemetriou et al. [4] minimized an image-based energy function to segment MTs using consecutive level sets methods. Saban et al. [5] automatically detected MT tips in the first frame and tracked tips by searching for the closest match in subsequent frames. Sargin et al. [6] located tip locations using second order derivative of Gaussian filtering and iteratively calculated geodesic paths to segment MT bodies. However, the above methods process an image sequence slice by slice and do not take advantage of the time-lapse sequence's temporal information.
Other methods of tracking filament tips employ Particle Filters (PF) [7], [8]. However, the two methods ignore the supporting information from filament bodies and may fail to locate a tip away from the filament body. To solve this problem, Li et al. [9] used an over-grown active contour model to segment the filament body slice by slice and proposed a 1D PF to track filament tips across time. This method performs well in real time scenarios. Occasionally, this method generates some errors, such as segmenting two filaments as one, because it does not fully utilize all available temporal information: (i) it segments the filament body slice by slice and thus does not take into consideration of the location of filament bodies across time slices; and (ii) when tracking the tip in time t, only information prior to t (1, · · · , t − 2, t − 1) is considered. This is due to the PF's Bayesian tracking framework.
Since TIRFM image sequences are taken prior to data analysis, accuracy is more important than speed. In this paper, we treat a 2D time-lapse image sequence as a 3D spatiotemporal (2D spatial and 1D temporal) image volume. In such 3D domain, we use an over-grown active surface model to segment filament bodies. Given a reference point on resulting curves of all time slices, we can calculate the distances of all points along the resulting curves with respect to that reference point. An additional 2D spatiotemporal (1D spatial and 1D temporal) domain can be created by combining this distance-to-reference-point dimension with the time dimension. An active contour model in this 2D domain is then used to locate filament ends. In this proposed method, segmentation of the filament at time t depends on the information of the whole sequence (1, · · · , t − 1, t + 1, · · · , n). In evaluation on experimental images, this method shows more accurate segmentation performance over a previous method [3]. The overall framework of this method is illustrated in Fig. 1.
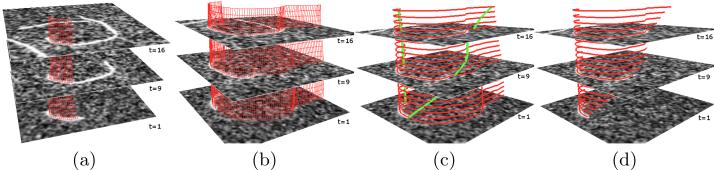
Fig. 1.
An overview of the proposed method. (a) Initialization of the active surface. (b) The resulting over-grown active surface of (a) (from a different viewpoint). (c) The resulting over-grown curves (red) on all slices and two active contours (green) across temporal domain for locating filament ends. (d) Final segmentation results on all slices.
2 Spatiotemporal Active-Surface for Filament Body Segmentation
We treat the time-lapse 2D image sequence as a 3D spatiotemporal (2D spatial and 1D temporal) image volume, where each point has coordinates (x, y, t), where x and y are the spatial coordinates, and t is the discrete time coordinate. One important observation of filament behavior is that filament bodies tend to remain static across time. Thus, if we connect points on the same filament across time, they form a smooth surface in the (x, y, t) domain. Based on this observation, we create an open active surface model [10] not in a pure 3D spatial domain but a 3D spatiotemporal domain to segment the filament in all slices simultaneously (Fig. 1.(b)).
2.1 Surface Initialization and Registration
Human interaction is necessary in this application since there is a limited number of filaments of interest in a sequence. Users specify a short line (2-3 clicks) close to the filament of interest on the first slice and use a 2D open active contour model [3] to fit only to a part of the filament. Then this initial short curve is used to fit a part of the same filament in all slices. The resulting curves on all slices are registered using the ICP method [11] to compensate for image drift [2] between contiguous frames. Since the drift between two contiguous frames is usually small, the ICP method generates satisfactory registration performance. We then create a grid mesh as the initial surface by connecting points on curves in all slices (Fig. 1.(a)).
2.2 The Over-grown Spatiotemporal Active-Surface Model
After being initialized, an active surface can be defined by a mapping from parameters p and q to the 3D spatiotemporal domain, ,
| (1) |
We seek to minimize its overall energy E3D:
| (2) |
where the internal energy is represented by the first 5 derivative terms where the elasticity is weighed by (w10, w01), rigidity by (w20, w02), and the resistance to twist by w11. These terms help to keep the active surface smooth. is the external energy, which is derived from the image and other external constraints. In our case, the external energy generates forces to attract the active surface to fit the central line of the filament.
A local minimum of E3D can be obtained by iteratively solving its associated Euler-Lagrange equations [10],
| (3) |
where r̄k is the vector consisting of values of r(p, q) for all p and q at iteration k, I is the identity matrix, τ is the step size parameter, and A3D is a sparse matrix encoding the structure of the surface mesh and weights of the internal energy (details on A3D can be found in [10]). The initial vector r̄0 is given by the initial surface (Section 2.1). With this model and properly defined external energy, the resulting surface would locate the central line of a filament in all slices.
However, the computation time for the 3D surface increases significantly compared with 2D active contour models. The intersections between the active surface and slices are also difficult to calculate. A solution is to use a simplified 3D surface model similar to that proposed in [10]. The degrees of freedom of the deformation are limited to two components instead of three by constraining the time component t, which corresponds to the discrete time coordinate, to depend linearly only on one parameter. The external force along the time dimension then becomes zero. Therefore, the surface we seek is now represented by a sequence of parametrized curves. We have r(p, q) = (x(p, q), y(p, q), q) so that for a given slice index q, there corresponds an open curve parametrized by p lying on the qth slice of the 3D spatiotemporal volume.
Compared to slice-by-slice approaches in [9], this new active surface model better utilizes temporal information to constrain the segmentation results. For instance, if a filament is missing or hardly observed in a slice, the surface bridges the neighboring curves, creating a smooth surface. In contrast, the previous methods may fail or generate large errors in this scenario.
2.3 The External Energy
The external energy consists of an image term and a stretching term, i.e., . As we mentioned above, the external force of the constrained surface is always zero along time dimension. Therefore, we can calculate it slice by slice. The image term represents the image constraints. Since we want to locate the bright ridges in images, we directly use the Gaussian-filtered or Anisotropic Diffusion filtered image of the qth slice as the image term of the qth slice, i.e., Eimg(r(p, q)) = Gσ * I(r(p, q)), p ∈ [0, 1], for any q. The gradient vectors of such an image term, ∇Eimg, point toward the center of filaments and attract the active surface to fit the filament.
On the one hand, with the image term only, the active surface would only fit a part of the filament because there is no force attracting the active surface to the ends of filaments. On the other hand, filament ends usually have very low SNR and might be difficult to distinguish during the segmentation process. One important observation is that an over-grown surface almost always covers tips of the filament [9]. Based on this observation, we let the active surface over-grow in both directions and distinguish the ends on the resulting over-grown surface in a subsequent step (Section 3). To make the surface over-grow, we add stretching forces to the boundaries of the active surface:
| (4) |
where rp is the derivative of r with respect to p, rp(p, q)/|rp(p, q)| is the direction pointing outwards along the surface, and kstr is the stretching weight and only needs to be large enough to let the surface over-grow. Fig. 2.(a) illustrates the stretching forces. After every 3-4 iterations, the stretching force is removed for one iteration to let the active surface better fit to the filament. We deform the surface until intensity values corresponding to all boundary points, r(0, q) and r(1, q), ∀ q ∈ [0, 1], are lower than the foreground intensity value, which is estimated in the surface initialization step (Section 2.1). An over-grown active surface is shown in Fig. 1.(b).
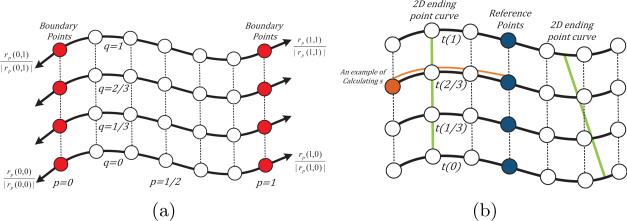
Fig. 2.
Illustration of the 3D surface mesh. Solid and dashed lines represent spatial and temporal connections between sampled points, respectively. (a) Stretching forces (black arrows) are added on the boundary points (red). (b) Illustration of the reference points (blue) and ending-point active contours (green). The brown curve gives an example of calculating the brown point's length s with respect to the reference point.
3 Spatiotemporal Active-Contour Model for Locating Filament Ends
3.1 The 2D Spatiotemporal Domain
After we obtained the over-grown active surface, we can constrain the search of filament ends along the resulting curve on every slice (red curves in Fig. 1.(c)). One simple way of locating ends might be to perform 1D Sobel filtering along each curve and to locate ends as locations with highest filtering response. But this operation ignores the relations between filament ends on different time slices: filament ends usually elongate at a consistent speed. Based on this observation, we create a 2D spatiotemporal active contour model to locate filament ends at all slices simultaneously (green curves in Fig. 1.(c)).
Given a reference point on the first slice's curve, we can obtain its nearest corresponding reference points on all other slices, such that every point on every curve is of length s away from the reference point along the curve (Fig. 2.(b)). The length s dimension, together with the time t dimension, create a 2D spatiotemporal domain. Every point (s, t) in this 2D domain implicitly specifies a possible 2D ending point (x(s), y(s)) of the filament on the tth slice. Therefore, if we connect the true ending points of a filament on different slices and map them to the (s, t) domain, these mappings form a smooth curve in the (s, t) domain.
3.2 The 2D Active Contour Model for Locating Filament Ends
The active contour is then defined by a mapping of parameter u to the 2D (s, t) domain, ,
Similarly to the above constrained 3D model, we constrain the time t dimension. The point on the tth slice can only deform along the resulting over-grown curve on the tth slice. Therefore, the internal force along t dimension should not be included in the model, and the external force along the t dimension is zero. Based on the constrained model and the above consistent growing speed assumption on filament ends, we define the 2D active contour's energy E2D as
| (5) |
where w1 and w2 weigh the first-order and second-order derivative terms of the internal energy. For the external force , because it is zero along the t dimension, we use the 1D Sobel filtering response along each over-grown curve as the external energy on that slice.
A local minimum of E2D can be solved iteratively:
| (6) |
where notations are similarly defined. Iteratively solving Eqn. (6) would deform the active contour to fit the desired filament end locations on all slices simultaneously (Fig. 1.(c)). We then cut the over-grown curves using those estimated filament end locations to generate the final segmentation results (Fig. 1.(d)).
3.3 Ending Point Initialization
Eqn. (6) only converges to a local minimum of E2D. An ending point curve needs to be initialized in the (s, t) domain. One straightforward strategy is to initialize ending points on the tth slice at locations on the over-grown curves with highest Sobel filtering response.
Prior knowledge, if available, can also be taken into consideration when initializing the ending point curve. Given an expected end growing speed m, we can initialize a straight line, (s1, 1), (s2, 2), · · · , (sn, n) in the (s, t) domain, where si+1 − si = m, ∀i ∈ 2, 3, · · · , n. We shift the straight line along the s dimension find the best initialization location with the highest Sobel filtering response.
4 Application to Experimental Images
We used two TIRFM image sequences from [1]. In these experiments, polymerization of muscle Mg-ADP-actin was monitored in the presence of varying concentrations of inorganic phosphate (Pi) and actin monomers. The pixel size was 0.17 μm. There were 20 images in sequence I and 34 images in sequence II. The time interval between frames was 30 sec in sequence I and 10 sec in II.
4.1 Evaluation and Comparison with the Previous Method [9]
For both sequences, we set w01 = w10 = 0.1, w11 = 0, w02 = w20 = 0.02, w1 = 0.01, w2 = 0.05, and kstr = 1. The average growing speed of filaments is used as the prior knowledge for ending point curve initialization (Section 3.3) and was set according to the average elongation rate estimated by a manual method [2].
The filament bodies were usually tracked accurately by our algorithm. Most of the errors are among two types: (i) intersected filaments may be mis-segmented as one filament (Fig. 3.(1)), and (ii) tip locations were wrongly estimated (Fig. 3.(3)). Therefore, we evaluated our algorithm by counting the number of mis-segmentation of filament bodies and by measuring errors on tip location estimation. We selected 15 and 5 actin filaments from image sequence I and II respectively to evaluate our algorithm. For all selected filaments, we manually labeled their two tips in each frame as ground truth and calculated L2 distances between the ground truth and our algorithm's results.
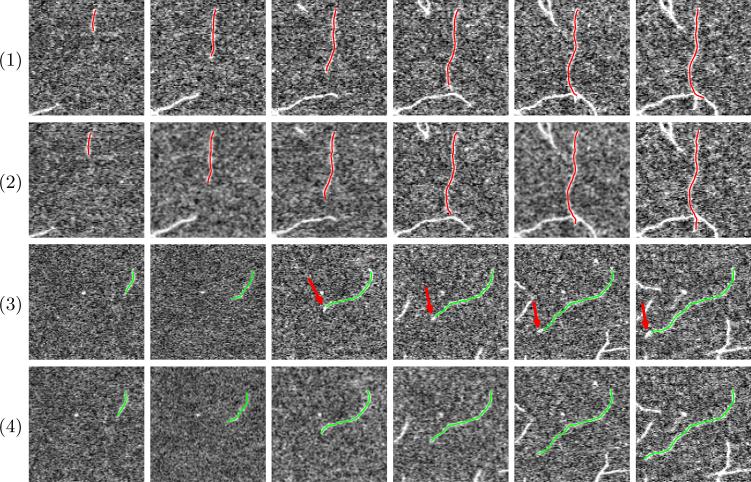
Fig. 3.
Two examples of comparison on filament segmentation in a time-lapse sequence. (Odd rows) segmentation results by the previous method [9]. (Even rows) segmentation results by our proposed method.
We also compared with a previous method [9], which only utilizes partial temporal information on filament tips. We show two examples of comparison with the previous method in Fig. 3. The first example shows the advantages of our spatiotemporal surface model. In Fig. 3.(1), the filament of interest intersects with another one. The previous method segments this filament slice by slice and mis-stretches to the other filament. Our proposed method utilizes information about the filament body at all slices and thus can better handle filament intersection (Fig. 3.(2)). The second example illustrates the advantages of the 2D spatiotemporal ending point curve. In Fig. 3.(3), the filament grows much faster than other filaments in the sequence. The prior knowledge on the growing speed is thus inaccurate. The previous method [9] generates large tip estimation errors because of the wrong prior knowledge (red arrows in Fig. 3.(3)). As shown in Fig. 3.(4), our proposed ending point curve better utilizes data from observation and is less sensitive to inaccurate prior knowledge, and thus it accurately segments the filament. Table 1 shows tracking error statistics of our algorithm in comparison to the previous method; our new algorithm clearly outperforms the previous method.
Table 1.
Tip tracking error statistics of selected filaments in both image sequences I and II. Smaller errors are shown in bold. (Unit: pixel)
| Sequnce | Mean | Maximum | Number of Mis-segmentation | |
|---|---|---|---|---|
| Proposed Method | I | 0.93 | 2.60 | 1 out of 225 |
| II | 1.47 | 6.01 | 3 out of 170 | |
| | ||||
| Previous Method [9] | I | 1.29 | 3.91 | 4 out of 225 |
| II | 1.73 | 5.83 | 4 out of 170 | |
5 Conclusions
In this paper, we present novel active surface and contour models for the segmentation of actin filaments in a time-lapse sequence. We solved the 2D tracking problem as a segmentation problem with smoothness constraints in a higher spatiotemporal 3D domain by introducing an active surface model. A novel (s, t) domain is created in which a 2D active contour deforms to locate filament ends accurately. Because it utilizes temporal constraints on filament body and tips from the entire sequence, it compares favorable to previous methods that use partial temporal information.
Acknowledgments
This work was supported by NIH grant R21GM083928.
References
- 1.Fujiwara I, Vavylonis D, Pollard TD. Polymerization kinetics of ADP- and ADP-Pi-actin determined by fluorescence microscopy. Proc. Natl. Acad. Sci. USA. 2007;104:8827–8832. doi: 10.1073/pnas.0702510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 2005;88:1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Shen T, Smith M, Fujiwara I, Vavylonis D, Huang X. Automated actin filament segmentation, tracking and tip elongation measurements based on open active contour models. Proc. ISBI. 2009 doi: 10.1109/ISBI.2009.5193303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hadjidemetriou S, Toomre D, Duncan J. Motion tracking of the outer tips of microtubules. Medical Image Analysis. 2008;12:689–702. doi: 10.1016/j.media.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Saban M, Altinok A, Peck A, Kenney C, Feinstein S, Wilson L, Rose K, Manjunath B. Automated tracking and modeling of microtubule dynamics. Proc. ISBI. 2006;1:1032–1035. [Google Scholar]
- 6.Sargin ME, Altinok A, Kiris E, Feinstein SC, Wilson L, Rose K, Manjunath BS. Tracing microtubules in live cell images. Proc. ISBI. 2007 doi: 10.1186/1471-2121-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smal I, Draegestein K, Galjart N, Niessen W, Meijering E. Particle filtering for multiple object tracking in dynamic fluorescence microscopy images: Application to microtubule growth analysis. IEEE Trans. on Medical Imaging. 2008;27:789–804. doi: 10.1109/TMI.2008.916964. [DOI] [PubMed] [Google Scholar]
- 8.Kong K, Marcus A, Giannakakou P, Wang M. Using particle filter to track and model microtubule dynamics. Proc. ICIP. 2007;5:517–520. [Google Scholar]
- 9.Li H, Shen T, Vavylonis D, Huang X. Actin filament tracking based on particle filters and stretching open active contour models. Proc. MICCAI. 2009;2:673–681. doi: 10.1007/978-3-642-04271-3_82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen LD, Cohen I. Finite element methods for active contour models and balloons for 2d and 3d images. IEEE Trans. on Pattern Analysis and Machine Intelligence. 1991;15:1131–1147. [Google Scholar]
- 11.Besl P, McKay H. A method for registration of 3-D shapes. IEEE Trans. Pattern Analysis and Machine Intelligence. 1992;14:239–256. [Google Scholar]