Abstract
An IL28B haplotype strongly determines the outcome of natural and interferon-α treated hepatitis C virus (HCV) infection. To assess whether the polymorphism marking the haplotype (rs12979860) also affects other interferon-α responsive chronic viral illnesses, namely hepatitis B virus (HBV) and human immunodeficiency virus (HIV) type 1 infections, we genotyped 226 individuals with HBV persistence, 384 with HBV recovery, and 2548 with or at high risk for HIV infection. The C/C genotype of rs12979860 was not associated with HBV recovery (odds ratio, 0.99), resistance to HIV infection (odds ratio, 0.97), or HIV disease progression (P>.05). This IL28B single-nucleotide polymorphism affects the immune response to HCV but not to HBV or HIV.
IL28B (interferon [IFN] l3) produces an antiviral state by triggering a cascade through the JAK-STAT pathway that up-regulates the IFN-stimulated genes (ISGs). The effects of IL28B are similar to those of IFN-α and -β; however, IL28B binds to a distinct receptor that may up-regulate a different set of ISGs [1]. This cytokine has been identified as a key modulator of the immune response to hepatitis C virus (HCV), because a single-nucleotide polymorphism (SNP) (rs12979860) upstream of the IL28B gene was associated with spontaneous and treatment-induced HCV clearance [2, 3]. The mechanism of how the SNP affects IL28B function has not been elucidated. One clue to the mechanism may come from determining whether the SNP affects the outcome of other chronic viral infections wherein IFN-α and ISGs are important in the host response, such as hepatitis B virus (HBV) and human immunodeficiency virus (HIV) type 1 infection.
After an HBV infection, 90%-95% of adults mount an effective immune response that leads to recovery from infection and development of hepatitis B surface antibodies (anti-HBs). For the remaining 5%-10%, chronic hepatitis B is established, which predisposes those infected to cirrhosis and liver cancer. As with HCV, IFN-α and ISGs are thought to be important in the immune response to HBV, and pegylated IFN-α is used to treat chronic hepatitis B infection. Thus, IL28B may be important in recovery from an HBV infection.
Despite repeated exposure to HIV, a small percentage of persons resist infection, allowing study of the host genetic basis of such protection. Among those who become infected with HIV, the rate of progression to AIDS varies markedly. As with HBV and HCV, IFN-α is active against HIV, probably through stimulation of ISGs [4]. Furthermore, IL28A (IFN-λ2) and IL29 (IFN-λ1), which bind the same receptor as IL28B, inhibit HIV1 infection of macrophages [5]. Thus, IL28B may have a role in the pathogenesis of HIV infection. Given the varied outcomes after both HBV and HIV infections and the importance of IFN-α and ISGs in these infections, we tested whether the rs12979860 SNP is associated with HBV recovery, resistance to HIV infection, or HIV disease progression.
Methods. The subjects in the HBV cohort were participants in one of the following ongoing parent cohorts: (1) Multicenter AIDS Cohort Study, a study of 5622 men who have sex with men in the United States; (2) AIDS Link to Intravenous Experience, a study of 2921 injection drug users in Baltimore, Maryland; (3) Hemophilia Growth and Development Study, a study of 333 children and adolescents with hemophilia; and (4) Multicenter Hemophilia Cohort Study, a prospectively followed cohort of patients with coagulation disorder from 16 hemophilia treatment centers (for references with cohort descriptions, see [6]).
A nested case-control design was used, in which individuals with a persistent HBV infection were matched to 2 persons from the same cohort with HBV recovery, who were otherwise similar with regard to nongenetic factors, as described elsewhere [6]. Subjects were considered persistently HBV infected if they had hepatitis B surface antigen (HBsAg) at 2 visits separated by ⩾6 months. Individuals with HBV recovery were positive for hepatitis B core antibody (anti-HBc) and anti-HBs without HBsAg at 2 time points separated by ⩾6 months.
The HIV cohort came from participants in the same parent cohorts as the HBV study, with the addition of the DC gay cohort, which includes men who have sex with men from Washington, DC, and the San Francisco City Clinic Cohort, which includes men who have sex with men from the San Francisco sexually transmitted disease clinic (for references with cohort descriptions, see [7]). Subjects who were persistently HIV negative despite ongoing high-risk behavior are considered high-risk seronegative (HRSN), as described elsewhere [7]. Subjects who were HIV positive at entry into the parent cohort are seroprevalent subjects, and those who became HIV positive while in the parent study are HIV seroconverters. Informed consent was obtained from all participants. This study was approved by the institutional review boards at participating institutions.
All serum specimens were stored at -700 C before testing. HIV antibody was determined by enzyme immunoassay with Western blot confirmation. HBsAg, anti-HBs, and anti-HBc were measured using commercially available kits according to the manufacturer's specifications (Auszyme, Ausab, and Corzyme, respectively; Abbott Laboratories). HCV antibody and HCV RNA were assayed using commercially available kits according to the manufacturer's specifications, as described elsewhere [2]. HCV infection was defined as positive results for HCV antibody and HCV RNA. Samples were genotyped for the C or T allele at rs12979860 using a predeveloped TaqMan allelic discrimination assay (Applied Biosystems), as described elsewhere [3].
The SNP was in Hardy-Weinberg equilibrium in the HBV and HIV cohorts, as determined by a χ2test using 1 degree of freedom. For the HBV study, the Cochran-Mantel-Haenszel test was used for the allelic analysis. For dominant and recessive models and for comparing C/C and T/T, conditional logistic regression analysis was used to adjust for cohort, HCV status, and the presence of a 32-base pair deletion in CCR5 (CCR5Δ32), which is associated with HBV recovery [6]. All analyses were stratified by HIV status and ethnicity.
To determine whether the SNP was associated with resistance to HIV infection, we compared the allele frequency between the HRSN and the HIV-infected subjects (HIV seroconverter and seroprevalent subjects combined), using the Cochran-Mantel-Haenszel test. We tested dominant and recessive models, in addition to comparing C/C with T/T, using logistic regression analysis to adjust for cohort, ethnicity, and HCV status. To determine whether the SNP was associated with HIV disease progression in the HIV seroconverters, Cox proportional hazards models were used to test the following AIDS-related outcomes: CD4 T cell count <200 cells/μL, progression to AIDS according to the 1987 and 1993 Centers for Disease Control definitions [8, 9], and death from an AIDS-related cause.
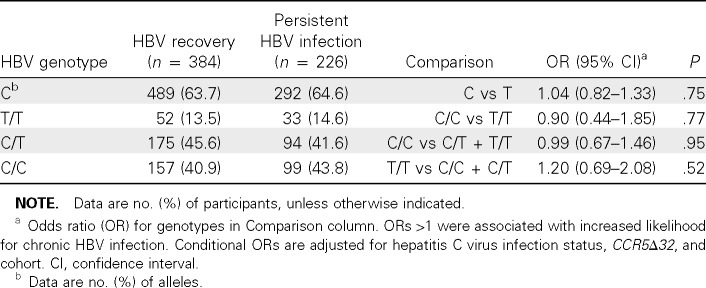
Results. In the HBV cohort, there were 226 individuals with HBV persistence and 384 with HBV recovery; thus, only 1 matched control was available for 68 subjects. No significant differences were detected between these HBV groups in terms of chronic HCV prevalence (21.5% vs 16.9%). The matched nongenetic factors were also similar (median age, 31 years; 75% white and 20% black; 98% male; 69% HIV positive).
The C allele frequency was significantly higher in white subjects (69.1%) than in black subjects (44.4%) (P< .001) and was similar to population frequencies [2]. However, there was no significant difference in C allele frequency between subjects with HBV persistence and those with HBV recovery (64.6% and 63.7%, respectively; P = .76) (Table 1). Stratification by ethnicity (P = .51 for both blacks and whites) or HIV status (P = .94 for HIV-positive, P = .74 for HIV-negative) also did not reveal an association with HBV outcome.
Table 1.
rs12979860 and Likelihood of Developing Chronic Hepatitis B Virus (HBV) Infection
Because the homozygous C/C genotype was most strongly protective in HCV, we next compared the frequency of C/C with that of C/T or T/T (dominant model) and the frequency of T/T with that of C/C or C/T (recessive model) in subjects with HBV recovery or persistence (Table 1). In the adjusted multivariate analysis, neither model showed an association with HBV recovery, nor did a comparison between subjects with C/ C and those with T/T genotype. Stratification of these analyses by race or HIV status (data not shown) did not demonstrate a difference.
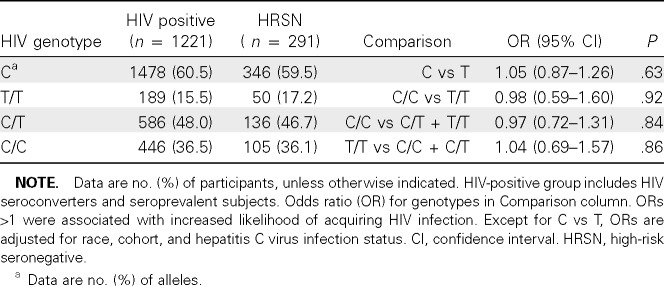
The HIV portion of the study included 1512 individuals of whom 291 were HRSN, 1036 were HIV seroconverters, and 185 were HIV seroprevalent. The cohort was 68% white, 26% black, and 6% other ethnicities. The HCV infection status was known for 1323 (87.5%) of the cohort; of those, 46% were HCV infected.
As in the HBV study, the C allele was more frequent in whites than in blacks (P < .001), and the frequencies were similar to population frequencies [2]. The C allele frequency did not differ between the HIV-infected and HRSN subjects (Table 2). Neither the C/C genotype nor the adjusted genotype models showed an association with resistance to HIV infection.
Table 2.
rs12979860 and Likelihood of Acquiring Human Immunodeficiency Virus (HIV) Infection
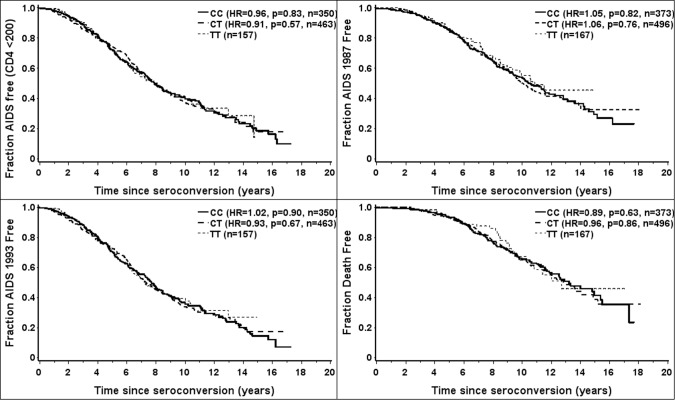
The influence of rs12979860 on the rate of progression to AIDS was determined in 1036 HIV seroconverters, as described in Methods. The adjusted genotype models did not demonstrate an association of HIV disease progression to the outcomes when examined across ethnic groups (Figure 1) or stratified by ethnic group (data not shown).
Figure 1.
Kaplan-Meier survival curves combining all ethnic groups demonstrate no difference in the time to various outcomes of human immunodeficiency virus (HIV) infection based on rs12979860 genotypes C/C, C/T, T/T in 1036 HIV seroconverters. HR, hazard ratio.
Discussion. This study clearly demonstrates that the SNP 4 kilobases upstream of IL28B (rs12979860) was not associated with outcomes of 2 common chronic viral infections, namely, HBV recovery and HIV infection and disease progression. Prior studies have shown that this SNP is associated with spontaneous and treatment-induced clearance of HCV and is one of the strongest known genetic associations with any chronic viral infection to date. Thus, even though both IFN-α and IL28B signal through the JAK-STAT pathway, this IL28B SNP apparently has a distinct effect on the immune response to HCV.
It is somewhat surprising that there was no association, given that IL28B stimulates ISGs, which play an important role in the immune response to HIV and HBV infections. In HBV transgenic mice, ISGs are a major mechanism of noncytolytic inhibition of HBV replication [10]. Furthermore, in an immortalized murine hepatocyte cell line that replicates HBV infection, exogenous murine IFN-λ2 (IL28A) inhibited HBV replication by >90%, inhibition that was thought to be mediated by up-regulation of ISGs [11]. Studies of nonhuman primate models of HIV infection also demonstrate that ISGs are up-regulated in blood and lymph nodes during acute infection [12].
It is not known how rs1297860 affects the function of IL28B, but presumably it alters the immune function to HCV but not to HBV or HIV. IL28B has interesting characteristics that may explain this finding. In addition to inducing ISG expression, IL28B may activate alternate antiviral pathways, such as the adaptive immune response, which may be more important in HCV. This is supported by a study in which IL28B, when used as a vaccine adjuvant, significantly decreased splenic regulatory T cells, increased splenic and peripheral blood CD8+ T cells, and led to increased antigen-specific perforin induction and degranulation [13]. IL28B may also lead to a different antiviral state than IFN-α, because these 2 molecules up-regulate different ISGs and do so with different kinetics [1]. It was also shown that phosphorylation of STAT1 and STAT2 by IFN-α is slower but more sustained than phosphorylation by IFN-λ, suggesting differences in signal transduction between these 2 IFNs [1].
It is also possible that the IL28B pathway may have a more dominant response to HCV than to HBV or HIV; therefore, the SNP would have a greater effect in HCV infection. In support of this hypothesis, it was shown that IFN-λ1 (IL29) induced transcription of antiviral genes in 2 hepatoma cell lines, but in 1 cell line there was no effect on HBV replication and in the other there was a marginal decrease [14]. Furthermore, in the HBV transgenic mouse, IFN-λ2 (IL28A) was unable to induce intrahepatic ISG expression or to decrease HBV replication [11]. Additional work is needed to determine whether these findings can be extrapolated to IL28B. It is also plausible that the IL28B and IFN-α pathways are synergistic and that the synergism more strongly enhances the immune response to HCV than to HBV or HIV.
One limitation of this study was that it could not determine whether the IL28B SNP influences the response of chronic hepatitis B treatment to IFN-α. Another limitation is that IL28B may still have a role in the immune response to hepatitis B or HIV that is not affected by this SNP.
In conclusion, the SNP upstream of IL28B that has the strongest genetic association with HCV recovery to date has no effect on HBV recovery, HIV infection, or HIV disease progression. These data are consistent with HIV genome-wide association studies in which the IL28B SNPs were not associated with HIV RNA levels [15]. Thus, the effects of this SNP cannot be generalized to these chronic viral infections in which IFN-α is important. Additional studies are needed to understand the mechanisms underlying the beneficial effect of this SNP in HCV infection but not in HIV or HBV infection.
Acknowledgments
The authors thank Abbott Laboratories for donating HBsAg and anti-HBs kits and the cohort participants for making this study possible.
This work was supported by the Investigators in the Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund (C.L.T.). Data in this study were collected by the Multicenter AIDS Cohort Study (MACS), with centers (principal investigators) at the Johns Hopkins University Bloomberg School of Public Health (Joseph B. Margolick, Lisa Jacobson); Howard Brown Health Center and Northwestern University Medical School (John Phair); University of California, Los Angeles (Roger Detels); and University of Pittsburgh (Charles Rinaldo). The MACS is funded by the National Institute of Allergy and Infectious Diseases, with additional supplemental funding from the National Cancer Institute (NCI) and the National Heart, Lung and Blood Institute (grants UO1-AI-35042, 5-MO1-RR-00722 [GCRC], UO1-AI-35043, UO1-AI-37984, UO1-AI-35039, UO1-AI-35040, UO1-AI-37613, and UO1-AI-35041). Web site: http://www .statepi.jhsph.edu/macs/macs.html.
The Multicenter Hemophilia Cohort Study is supported by the NCI (contract N02-CP-55504 with RTI International). The AIDS Link to Intravenous Experience (ALIVE) study is supported by the National Institute on Drug Abuse (grants R01-DA-04334 and R01-DA-12568). The Hemophilia Growth and Development Study is supported by the Bureau of Maternal and Child Health and Resources Development (MCJ-060570), the National Institute of Child Health and Human Development (NO1HD-4–3200), the Centers for Disease Control and Prevention, and the National Institute of Mental Health.
Additional support has been provided by grants from the National Center for Research Resources of the National Institutes of Health (NIH) to the New York Hospital-Cornell Medical Center Clinical Research Center (grant MO1-RR06020), Mount Sinai General Clinical Research Center, New York (grant MO1-RR00071), University of Iowa Clinical Research Center (MO1RR00059), and University of Texas Health Science Center, Houston (grant MO1-RR02558), and R01-HD-4–1224. This project has been funded in whole or in part with federal funds from the NCI, NIH (contract N01CO-12400) and in part by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Footnotes
Potential conflicts of interest: none reported.
Presented in part: 61st Annual Meeting of the American Association for the Study of Liver Diseases, Boston, Massachusetts, 29 October to 2 November 2010 (abstract 1408).
Financial support: Financial support is provided in the Acknowledgments.
Disclaimer: The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.
References
- 1.Marcello T, Grakoui A, Barba-Spaeth G, et al. Interferons alpha and lambda inhibit hepatitis C virus replication with distinct signal transduction and gene regulation kinetics. Gastroenterology. 2006;131(6):1887–1898. doi: 10.1053/j.gastro.2006.09.052. [DOI] [PubMed] [Google Scholar]
- 2.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461(7265):798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461(7262):399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 4.Lapenta C, Santini SM, Proietti E, et al. Type I interferon is a powerful inhibitor of in vivo HIV-1 infection and preserves human CD4 +T cells from virus-induced depletion in SCID mice transplanted with human cells. Virology. 1999;263(1):78–88. doi: 10.1006/viro.1999.9869. [DOI] [PubMed] [Google Scholar]
- 5.Hou W, Wang X, Ye L, et al. Lambda interferon inhibits human immunodeficiency virus type 1 infection of macrophages. J Virol. 2009;83(8):3834–3842. doi: 10.1128/JVI.01773-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thio CL, Astemborski J, Bashirova A, et al. Genetic protection against hepatitis B virus conferred by CCR5Delta32: evidence that CCR5 contributes to viral persistence. J Virol. 2007;81(2):441–445. doi: 10.1128/JVI.01897-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean M, Carrington M, Winkler C, et al. Genetic restriction of HIV1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention (CDC) Revision of the CDC surveillance case definition for acquired immunodeficiency syndrome: council of state and territorial epidemiologists; AIDS program, Center for Infectious Diseases. MMWR Morb Mortal Wkly Rep. 1987;36(Suppl 1)(Suppl 1):1–15. [PubMed] [Google Scholar]
- 9.1993 Revised classification system for HIV infection and expanded surveillance case definition for AIDS among adolescents and adults. MMWR Recomm Rep. 1992;41:1–19. (RR-17) [PubMed] [Google Scholar]
- 10.Guidotti LG, Morris A, Mendez H, et al. Interferon-regulated pathways that control hepatitis B virus replication in transgenic mice. J Virol. 2002;76(6):2617–2621. doi: 10.1128/JVI.76.6.2617-2621.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robek MD, Boyd BS, Chisari FV. Lambda interferon inhibits hepatitis B and C virus replication. J Virol. 2005;79(6):3851–3854. doi: 10.1128/JVI.79.6.3851-3854.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquelin B, Mayau V, Targat B, et al. Nonpathogenic SIV infection of African green monkeys induces a strong but rapidly controlled type I IFN response. J Clin Invest. 2009;119(12):3544–3555. doi: 10.1172/JCI40093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morrow MP, ankhong P, Laddy DJ, et al. Comparative ability of IL12 and IL-28B to regulate Treg populations and enhance adaptive cellular immunity. Blood. 2009;113(23):5868–5877. doi: 10.1182/blood-2008-11-190520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hong SH, Cho O, im K, Shin HJ, Kotenko SV, Park S. Effect of interferon-lambda on replication of hepatitis B virus in human hepatoma cells. Virus Res. 2007;126:245–249. doi: 10.1016/j.virusres.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Fellay J, Ge D, Shianna KV, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5(12):1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]