Abstract
Epigenetics refers to heritable changes that are not encoded in the DNA sequence itself, but play an important role in the control of gene expression. In mammals, epigenetic mechanisms include changes in DNA methylation, histone modifications and non-coding RNAs. Although epigenetic changes are heritable in somatic cells, these modifications are also potentially reversible, which makes them attractive and promising avenues for tailoring cancer preventive and therapeutic strategies. Burgeoning evidence in the last decade has provided unprecedented clues that diet and environmental factors directly influence epigenetic mechanisms in humans. Dietary polyphenols from green tea, turmeric, soybeans, broccoli and others have shown to possess multiple cell-regulatory activities within cancer cells. More recently, we have begun to understand that some of the dietary polyphenols may exert their chemopreventive effects in part by modulating various components of the epigenetic machinery in humans. In this article, we first discuss the contribution of diet and environmental factors on epigenetic alterations; subsequently, we provide a comprehensive review of literature on the role of various dietary polyphenols. In particular, we summarize the current knowledge on a large number of dietary agents and their effects on DNA methylation, histone modifications and regulation of expression of non-coding miRNAs in various in vitro and in vivo models. We emphasize how increased understanding of the chemopreventive effects of dietary polyphenols on specific epigenetic alterations may provide unique and yet unexplored novel and highly effective chemopreventive strategies for reducing the health burden of cancer and other diseases in humans.
Keywords: Epigenetics, polyphenols, histone modifications, microRNA, DNA methylation, diet, dietary compounds, cancer
1. INTRODUCTION
1.1 Epigenetics and Cancer
Cancer is widely perceived as a heterogeneous group of disorders, which is caused by a series of clonally selected ‘genetic’ changes in key tumor suppressor genes and oncogenes. However, accumulating evidence in the recent years indicate that tumor cell heterogeneity is in part due to significant contribution of ‘epigenetic’ alterations in cancer cells. Consequently, it is now becoming apparent that epigenetic plasticity together with genetic lesions drives tumor progression, and that cancer is the manifestation of both genetic and epigenetic modifications (1–4). Although a small proportion of tumors can be inherited, it is believed that majority of cancers result from changes that accumulate throughout the life because of exposure to various endogenous factors such as nutrients, infections, physical activity, social behavior and other environmental factors. Even when cancer initiation and progression is driven by acquired genetic alterations, epigenetic disruption of gene expression plays an equally important role in development of disease (5), and arguably diet and environment-mediated epigenetic perturbations play a crucial role in cancer progression in humans (6–8).
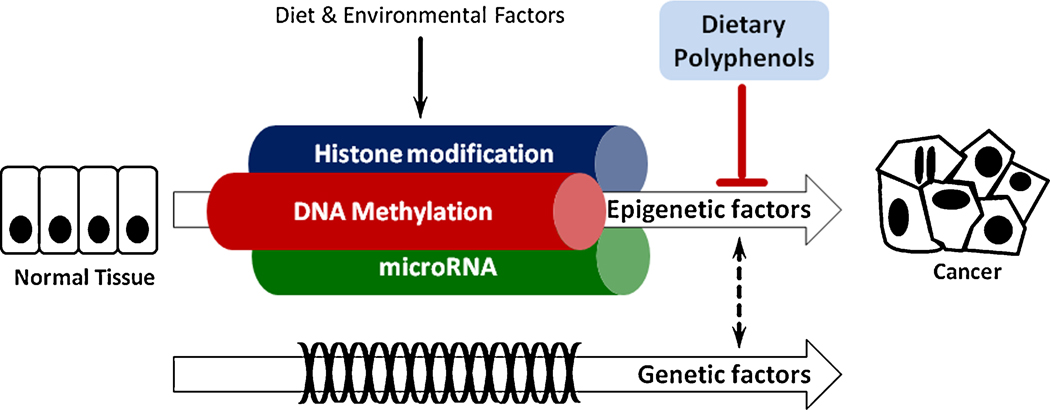
The term ‘epigenetics’, which was first coined by the developmental biologist Conrad H. Waddington in 1942, is defined as reversible heritable changes in gene expression that occur without alteration in DNA sequence, but changes that are sufficiently powerful to regulate the dynamics of gene expression (9). Three distinct and intertwined mechanisms are known to be part of the “epigenome”, which includes DNA methylation, histone modifications, and post transcriptional gene regulation by non-coding microRNAs (miRNAs) (2). These processes affect transcript stability, DNA folding, nucleosome positioning, chromatin compaction, and complete nuclear organization of the genetic material (Figure 1). Synergistically and cooperatively they determine whether a gene is silenced or expressed, as well as the timing and tissue-specificity of the expression of these genes. Disruption of the epigenome certainly underlies disease development. Therefore, disease susceptibility is clearly a result of complex interplay between one’s genetic endowment and epigenetic marks imprinted on one’s genome by endogenous and exogenous factors (10).
Figure 1. Epigenetic mechanisms involved in carcinogenesis.
Carcinogenesis is a long-term process and both genetic and epigenetic factors contribute to cancer development. Epigenetic changes, such as DNA methylation, histone modifications and microRNAs are easily influenced by dietary and environmental factors. Dietary polyphenols can potentially impact all three epigenetic modifications, which in turn contributes towards their chemopreventive potential.
From a clinical point of view, epigenetics offers a very promising and attractive avenue. This is because, unlike genetic changes (mutations, gene deletions etc), epigenetic alterations are potentially reversible. What this means is that unlike mutations, which exist for the lifetime, epigenetically modified genes can be restored; methylation silenced genes can be demethylated, and histone complexes can be rendered transcriptionally active by modification of acetylation and methylation of various histones via nutrients, drugs and other dietary interventions. This is really fascinating, as this provides a perfect opportunity for designing optimal chemopreventive and therapeutic strategies. The mechanism of interaction between various epigenetic factors and regulation of chromatin structure, dynamics, and ultimately gene expression is an active area of research, and recent understanding of these epigenetic mechanisms is highlighted in the sections below.
1.1.1. DNA methylation
DNA methylation of cytosines at CpG dinucleotides is perhaps the most extensively studied epigenetic modification in mammals. DNA methylation, in association with histone modifications is an essential component of the epigenetic machinery, which regulates gene expression and chromatin architecture (11). In mammalian cells, DNA methylation occurs at the 5’ position of the cytosine residues within CpG dinucleotides by the addition of a methyl group to form 5- methylcytosine (12). CpG dinucleotides are not uniformly distributed throughout the human genome, but are often enriched in the promoter regions of genes, as well as regions of large repetitive sequences (e.g. centromeric repeats, LINE and ALU retrotransposon elements) (13). Short CpG-rich regions are also called as “CpG islands”, and these are present in more than 50% of human gene promoters (14). Whilst most of the CpG dinucleotides in the genome are methylated, the majority of CpG islands usually remain unmethylated during development and in undifferentiated normal cells (15). Hyper-methylation of CpG islands within gene promoters can result in gene silencing, while promoters of transcriptionally active genes typically remain hypo-methylated (15). DNA methylation can lead to gene silencing by either preventing or promoting the recruitment of regulatory proteins to DNA. For example, it can inhibit transcriptional activation by blocking transcription factors from accessing target-binding sites e.g. c-myc (16). In other instances, it can provide binding sites for methyl-binding (sequestering) domain proteins, which can orchestrate gene repression through interaction with histone modifying enzymes (17). Thus DNA methylation uses a variety of mechanisms to silence genes, and a direct association between DNA methylation and the phenotype of a cell can be postulated.
The modification at 5-methylcytosine is catalyzed by various DNA methyltransferases (DNMTs). There are three main DNA methyltransferases; DNMT1, which is the major maintenance enzyme that preserves existing methylation patterns following DNA replication by adding methyl groups to the hemi-methylated (partially-methylated) CpG sites (18;19); DNMT3a and DNMT3b on the other hand serve as de novo methyltransferases, which act independent of replication and show equal preference for both unmethylated and hemi-methylated DNA (20;21). The role of DNA methylation-induced transcriptional silencing of genes is now well-established in multiple human malignancies (22). In fact, analogous to mutations and deletions, DNA methylation of genes in most human cancers is now believed to be a most frequent mechanism for the transcriptional silencing of tumor suppressor genes (23). Several detailed and informative reviews on the association between DNA methylation and cancer are available, but these are beyond the scope of this review (11;19;24–27).
1.1.2. Histone modifications
In addition to direct methylation of DNA, chromatin structure is frequently influenced by diverse histone modifications, which also play an important role in gene regulation and tumorigenesis (3;4). Chromatin proteins serve as building blocks to package eukaryotic DNA into higher order chromatin fibers. Each nucleosome encompasses ~146 bp of DNA wrapped around an octamer of histone proteins. These octamers consist of double subunits of H2A, H2B, H3 and H4 core histone proteins (28). The histone proteins coordinate the changes between tightly packed DNA (or heterochromatin), which is inaccessible to transcription, and lightly packed DNA (or euchromatin), which is available for active transcription through binding of transcription factors (29). These changes typically occur in the ‘histone tails’, which extend from the core octamer. The histone tails comprise of a globular C-terminal domain and an unstructured N-terminal tail (30). The N-terminal histone tails are the major sites for post-translational modifications including methylation, acetylation, phosphorylation, ribosylation, ubiquitination, sumoylation and biotinylation (31). The majority of these modifications take place at lysine, arginine and serine residues within the histone tails and regulate key cellular processes such as transcription, replication and repair (31). Unlike DNA methylation, histone modifications can lead to either activation or repression depending upon which residues are involved, and the type of modification present. For instance, lysine acetylation associates with transcriptional activation, while its methylation leads to transcriptional activation or repression depending upon which specific lysine is modified. For instance, tri-methylation of lysine 4 on histone H3 (H3K4me3) is enriched at transcriptionally active gene promoters (32), whereas tri-methylation of H3K9 (H3K9me3) and H3K27(H3K27me3) is present at transcriptionally repressed promoters (31). H3K9me3 and H3K27me3 histone modifications together constitute the two main silencing mechanisms in mammalian cells.
Similar to DNA methylation changes, various histone modifications are potentially reversible, and are dynamically regulated by groups of enzymes that add or remove covalent modifications to histone proteins (3;33). Histone acetyltransferases (HATs) and histone methyltransferases (HMTs) add acetyl and methyl groups, respectively, whereas histone deacetylases (HDACs) and histone demethylases (HDMs) remove acetyl and methyl groups, respectively, from histone proteins (34–36). A number of histone-modifying enzymes including various HATs, HMTs, HDACs, and HDMs have been identified in the recent years, including a large number of dietary polyphenols enumerated in the later sections of this review.
1.1.3 microRNAs
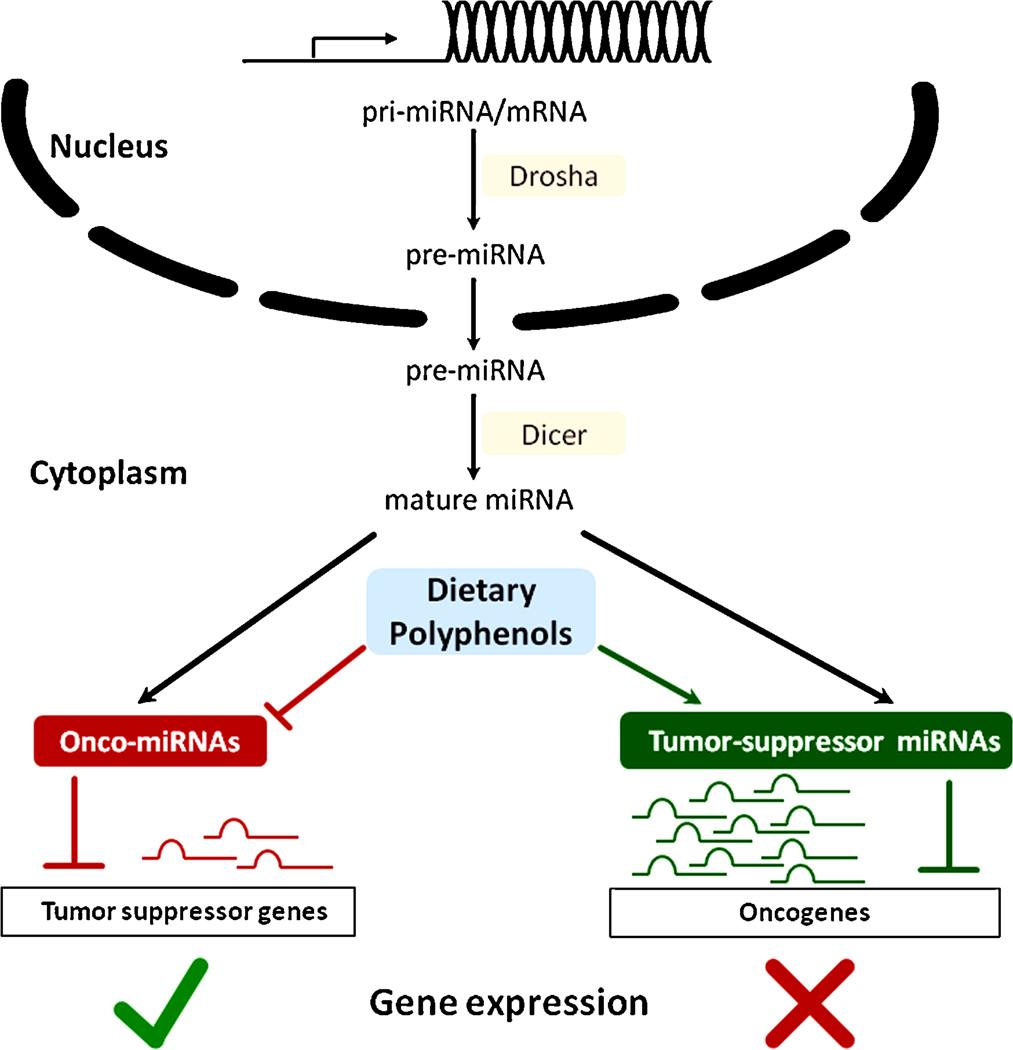
Besides DNA methylation and histone modifications, miRNAs are emerging as key mediators of epigenetic gene regulation in mammals. Non-coding RNAs, including miRNAs, were initially noted to perform catalytic functions in facilitating RNA splicing. In recent years it has been recognized that they participate in the post-transcriptional gene regulation (37;38). miRNAs are small single-stranded RNAs, ~19–24 nucleotides in length, that regulate gene expression through post-transcriptional silencing of the target genes. Sequence specific base pairing of miRNAs with 3’ untranslated regions of the target messenger RNA (mRNA) results in degradation or translational inhibition (39). miRNAs are expressed in a tissue-specific manner and control a wide spectrum of biological processes including cell proliferation, apoptosis and differentiation. Although miRNA are vital to normal cell physiology, aberrant expression of these small non-coding RNAs has been linked to carcinogenesis. In fact, miRNA profiles are now being used to classify human cancers (40–42). One of the interesting features of miRNAs is that similar to regular genes, their own expression can be regulated by other epigenetic mechanisms, such as DNA methylation (43). The influence of miRNA on the epigenetic machinery and the reciprocal epigenetic regulation of miRNA expression suggest that its deregulation during carcinogenesis has an important implication for global regulation of epigenetics and cancer. Additionally, interaction among various components of the epigenetic machinery re-emphasizes the integrated nature of epigenetic mechanisms involved in the maintenance of global gene expression patterns in mammals. Several detailed and informative reviews of the association between miRNA and cancer have been published previously (37;41;44–46).
2. Dietary and Environmental factors and their influence on epigenetics
Perhaps one of the most interesting and important features of epigenetics and its role in disease development is the fact that, unlike the genetic changes, epigenetic marks can be modified by the environment, diet or pharmacological intervention. This feature of epigenetic modifications has fueled enthusiasm for developing therapeutic strategies by targeting the activity of various epigenetic factors, such as DNMTs and HDACs, in order to prevent or treat various disease including human cancers (47;48). Next, we will briefly focus on the historical and current evidence for interactions between the environment, nutrition and the “epigenome”, the rationale for chemoprevention and therapy using dietary factors.
2.1 Nutrient deficiency and human cancer
Nutrients like folic acid, B vitamins and SAM (S-adenosyl methionine) are key components of the methyl-metabolism pathway, and methyl-donating nutrient-rich diet can rapidly alter gene expression, especially during early development when the epigenome is first being established. As a result, diet can influence the degree of methylation by influencing the availability of methyl donors, including folate, choline, and methionine, as well as DNMT activity (49–53). A classic example of the dietary influence in DNA methylation and cancer is the finding that dietary methyl deficiency (of folate, choline, and methionine) in an animal model was shown to alter hepatic DNA methylation patterns and induce liver cancer in the absence of a carcinogen (54). A more recent study revealed that only with early re-feeding of a methyl-sufficient diet during methyl-deficiency-induced hepatocarcinoma in mice can help mitigate aberrant DNA methylation defects, emphasizing that timing need to be considered in any intervention (55).
Selenium is another nutrient that has been linked with DNA methylation, both in cultured cell studies and animal experiments. It was demonstrated that rats fed with selenium-rich diets, both liver and colon DNA were significantly hypomethylated, thus providing a rationale for their potential chemopreventive efficacy (52;56). These effects of selenium were linked to its ability to inhibit DNMT1 activity and decreased DNMT1 protein expression (56).
2.2 Agouti mice
Classical experiments in mice show just how important a mother's diet is in shaping the epigenome of her offspring. At present, the best evidence to demonstrate that nutrition can modulate the epigenetic status of mammals comes from studies with mice carrying the agouti viable yellow (Avy) gene. The normal function of the Avy gene is to confer a wild-type coat color but dominant mutations at the agouti locus cause a pleiotropic syndrome, which confers excessive amounts of yellow pigment on the coat, together with systemic effects including obesity, a non-insulin-dependent diabetic-like condition, and of vulnerability to various types of cancer (57). In the Avy mouse, the expression of the allele varies with its methylation status, and when methylated, the gene behaves like a wild type allele and is expressed only in the hair follicle. When unmethylated the gene is expressed ubiquitously, causing the full agouti syndrome, but intermediate levels of methylation cause a mottled appearance, so that the coat color and other aspects of the agouti phenotype provide a direct readout of the methylation status of the allele. Using this model, Wolff and colleagues showed that by feeding diets supplemented with high levels of folic acid (as methyl donor) to pregnant dams it was possible to modify the expression of the agouti gene in the offspring (58). A higher proportion of offspring with wild type coat color were obtained from supplemented dams, which was consistent with higher levels of DNA methylation of the agouti gene (59;60). One of the most remarkable features of the Avy mouse model is that there is good evidence that the epigenetic marks established by dietary supplementation with methyl donors can be passed to a successive generation via the female germline and that these effects are mediated by polycomb group proteins (61). These results indicate that an individual's adult health is heavily influenced by early prenatal factors, and that our health is not only determined by what we eat, but also what our parents ate.
2.3 Maternal behavior and epigenetic changes in animals
Until date, there are not too many well-authenticated examples of direct effects of the environment on epigenetic status in mammals, but one of the remarkable examples is the effect of maternal care behavior on the offspring of rodents. In a very interesting study, it was demonstrated that high levels of pup-licking, grooming and “arched-back nursing” by rat mothers modified the levels of DNA methylation at a glucocorticoid receptor (GR) gene promoter in the hippocampus of the offspring, leading to altered histone acetylation and binding of a transcription factor (NGFI-A) to the GR promoter (62). Remarkably, central infusion of a histone deacetylase (HDAC) inhibitor abolished the group differences in histone acetylation, DNA methylation, NGFI-A binding, expression of the GR and hypothalamic–pituitary–adrenal responses to stress (63). These researchers subsequently showed that differences in maternal care modify the expression of more than 900 genes in the offspring. The probable involvement of epigenetic reprogramming in these effects was strongly implied by the observation that a proportion of these changes could be modified by treatment with a HDAC inhibitor or with the methyl donor methionine (64). Thus an epigenetic determinant of maternal behavior may be transmitted across generations.
2.4 Vernalization in plants
In addition to mammals, the contributions of environmental and epigenetic factors have also been studied in the plants. One of the best examples of environmental effects influencing the physiology of an organism by modifying its epigenetic status is the phenomenon of vernalization, in which exposure of a plant to low temperatures induces earlier flowering (65). A protein encoded by flowering locus C (FLC), which acts as a repressor of flowering in Arabidopsis was identified (66). Exposure to low-temperatures down-regulates FLC activity and induces earlier flowering, but interestingly FLC activity together with late flowering are restored in each plant generation. The suppression of FLC is associated with a reduction of histone H3 trimethyl-lysine 4 (H3K4), and acetylation of both histones H3 and H4, around the promoter-translation start of FLC.
2.5 Other nutritional factors and their effect on epigenetics
Nutrition is a major environmental aspect that may influence epigenetic mechanisms in multitude of ways. A number of biologically active food constituents have been shown to affect the metabolic processes associated with energy metabolism through changes in DNA methylation status of genes directly or indirectly. In obese individuals, excess adipose tissue accumulates over time as a consequence of energy intake exceeding expenditure. Adipocytes are a rich source of endocrine factors and other pro-inflammatory cytokines (such as TNF-α and IL-6). These pro-inflammatory cytokines are notorious for their adverse effects in causing increased inflammation, which is strongly associated with carcinogenesis (67). Indeed, inflammatory bowel disease has been shown to be a driver of aberrant DNA methylation in the colon (12;68). One potential mechanism for this effect is through the activity of IL-6, which has been shown to support the aberrant methylation of the p53 promoter via up-regulation of DNMT1 gene expression (69;70).
3. EPIGENETIC THERAPY
Epigenetic therapy, the use of drugs to correct epigenetic defects, is currently a new and fascinating area for drug development in the field of cancer prevention and therapy. Epigenetic therapy is a potentially very useful form of therapy because epigenetic defects, in contrast to genetic defects, are reversible (71;72). Besides their promise as therapeutic agents, epigenetic drugs may also be used for prevention of various diseases, including cancer chemoprevention (73). Additionally, there is growing enthusiasm that epigenetic drugs alone or in combination with conventional anticancer drugs may prove to be a significant advance over the conventional anticancer drugs, which inherently tend to be very toxic by themselves (74). Given the fact that epigenetic alterations underpin a broad range of human diseases, the scope of epigenetic therapy is tremendous and likely to expand in the coming years.
The current generation of epigenetic drugs primarily target to inhibit the activity and expression of DNMTs and HDACs. However, since many other molecules are also involved in epigenetic mechanisms that regulate gene expression, there are other potential targets, which yet remain undiscovered. Nonetheless, epigenetic drugs presently under evaluation in various pre-clinical and clinical trials can be classified into two groups, depending on whether they inhibit DNMTs or HDACs. Among the DNMT inhibitors, nucleoside inhibitors, such as 5-azacytidine (5-Aza-CR, or commercially available as Vidaza) and 5-aza-2-deoxycytidine (5-Aza-CdR, or commercially sold as Decitabine) are the most important and widely studied epigenetic drugs (75). In addition to this, certain non-nucleoside inhibitors such as procainamide, procaine and EGCG have also shown certain potential for inhibiting DNMT activity in various experimental and clinical studies (76–81). With regards to HDAC inhibitors, trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA), valproic acid and phenyl butyrate, have been widely used with some success in various studies (82–85). Several of these potentially useful epigenetic drugs are undergoing preclinical and clinical drug trials. Most of these trials have involved various types of cancers such as solid tumors and hematological malignancies (73;86).
Although the current generations of epigenetic drugs have provided the proof of principle in its favor, epigenetic therapy has its limitations. Some of these shortcomings include that both DNMT and HDAC inhibitors may activate oncogenes due to lack of specificity, resulting in accelerated tumor progression (87). Moreover, epigenetic states, once corrected, may revert to the original state because of the reversible nature of DNA methylation patterns (88). Taken together, the lack of specificity and the associated high levels of toxicity profiles with the synthetic epigenetic drugs have prompted the dire need for the discovery and development of safe and more specific epigenetic chemopreventive and therapeutic drugs, which can translate into clinics in the near future. Epidemiological and experimental data in recent years have clearly provided evidence that diet and diet-derived plant polyphenols have potent anti-cancer properties, and some of these effects are orchestrated via modulation of epigenetic machinery within cancer cells.
4. DIETARY POLYPHENOLS AND CANCER CHEMOPREVENTION
Polyphenols constitute one of the largest and ubiquitous group of phytochemicals. One of the primary functions of these plant-derived polyphenols is to protect plants from photosynthetic stress, reactive oxygen species, and consumption by herbivores. Polyphenols are also an essential part of the human diet, with flavonoids and phenolic acids being the most common ones in food. Not surprisingly, there is a growing realization that lower incidence of cancer in certain populations may probably be due to consumption of certain nutrients, and especially polyphenol rich diets. Consequently, a systematic dissection of the chemopreventive potential of polyphenolic compounds in the recent years has clearly supported their health benefits, including anti-cancer properties. Given the challenges of cancer therapy, ‘chemoprevention’-which uses pharmacological or natural agents to impede, arrest or reverse carcinogenesis at its earliest stages’ remains the most practical and promising approach for the management of cancer patients (89).
Till date, a substantial number of studies in cultured cells, animal models and human clinical trials have illustrated a protective role of dietary polyphenols against different types of cancers (90–93). Polyphenols are present in fruits, vegetables, and other dietary botanicals and some of these are depicted in Figure 2. Some estimates suggest that more than 8000 different dietary polyphenols exist, and these can be divided into ten different general classes based on their chemical structure (94). Phenolic acids, flavonoids, stilbenes and lignans are the most abundantly occurring polyphenols that are also an integral part of everyday nutrition in populations worldwide. Some of the common examples of the most studied and promising cancer chemopreventive polyphenols include EGCG (from green tea), curcumin (from curry) and resveratrol (from grapes and berries). Significant gains have been made in understanding the molecular mechanisms underpinning the chemopreventive effects of polyphenols, and consequently, a wide range of mechanisms and gene targets have been identified for individual compounds. Various mechanistic explanations for their chemopreventive efficacy include their ability to interrupt or reverse the carcinogenesis process by acting on intracellular signaling network molecules involved in the initiation and/or promotion of cancer, or their potential to arrest or reverse the progression stage of cancer (95;96). Polyphenolic compounds may also trigger apoptosis in cancer cells through the modulation of a number of key elements in cellular signal transduction pathways linked to apoptosis (caspases, bcl-2 genes) (90;95;96). Several elegant reviews have described in detail specific genetic and signaling mechanisms that are targeted by different polyphenols, and this is beyond the scope of this review article (97–99). However, recent research has suggested that some of the chemopreventive potential of dietary polyphenols may in part be due to their ability to modulate epigenetic alterations in cancer cells. This is of interest, as epigenetic modifications occur early and are potentially reversible, making dietary polyphenol-induced chemoprevention of various human cancers an attractive possibility from a clinical standpoint. However, the mechanism how flavonoids do regulate and effect various epigenetic modifications in cancer cells is a topic that is still in its infancy. Nevertheless, increasing number of reports have repeatedly shown the promise of epigenetic prevention and possibly therapy by dietary polyphenols. This review, which is first of its kind, provides a comprehensive review of the chemopreventive effects of various dietary polyphenols in regulating specific epigenetic alterations in human cancers. In particular, in the following sections of this review, we will summarize the existing data on the role of a large number of dietary agents on DNA methylation, histone modifications and regulation of expression of non-coding miRNAs in various in vitro and in vivo models of human cancers.
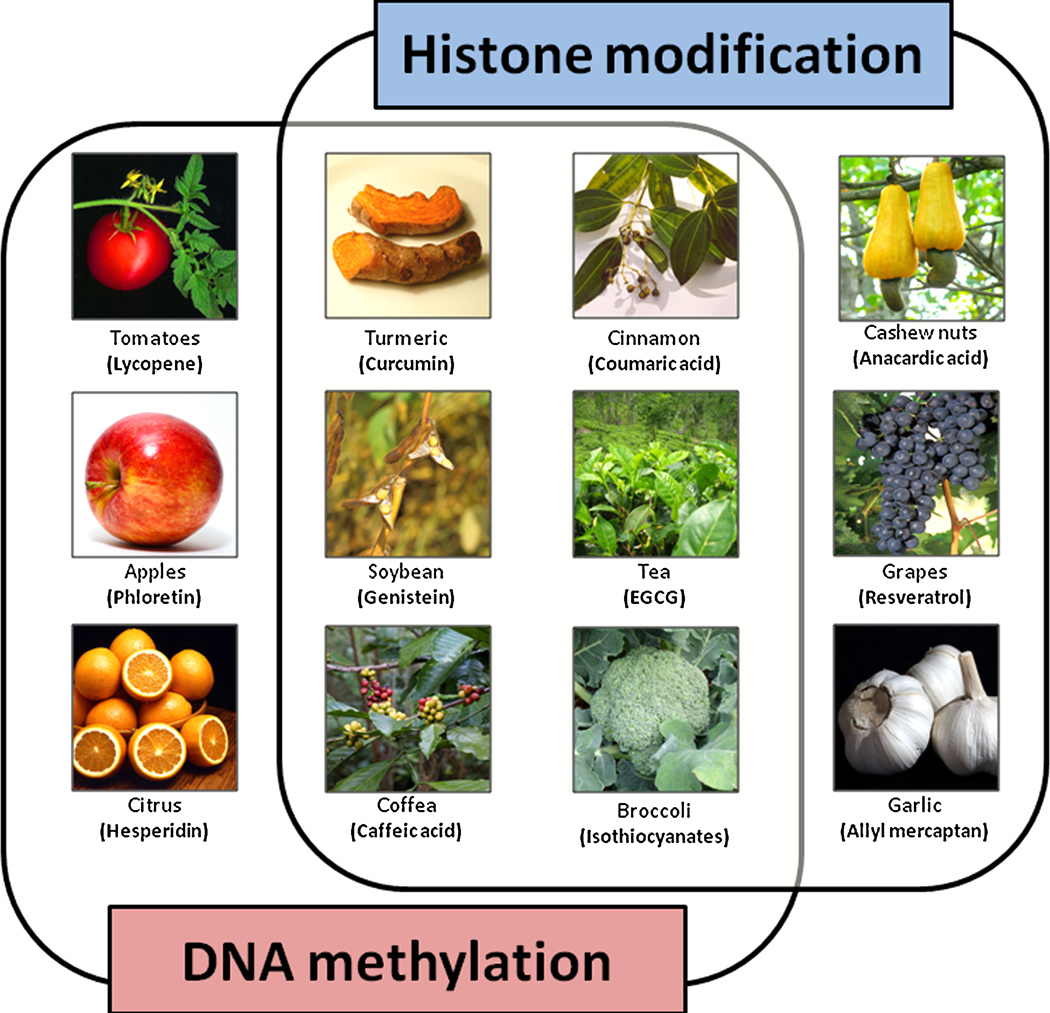
Figure 2. Illustration depicting major plants with evidence for epigenetic modifications.
The figure illustrates photographs from major plants with demonstrated evidence for epigenetic alterations in cancer cells. The ‘active principles’ for each of the plants are shown within parenthesis. These images were borrowed from various websites for illustration purposes only.
5. POLYPHENOLS AND DNA METHYLATION
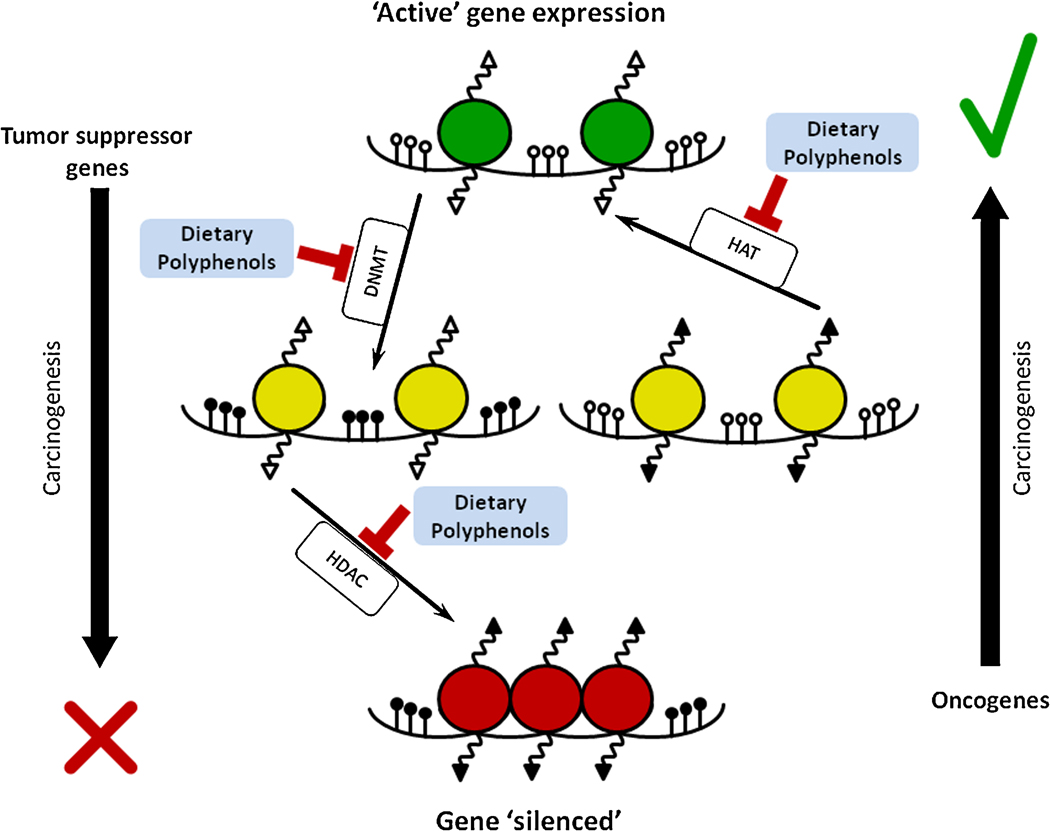
As mentioned earlier, hyper-methylation induced transcriptional silencing of tumor suppressor genes constitutes a frequent epigenetic defect in many human cancers. Reversal of gene hypermethylation, which may in part be achieved by inhibiting DNMT activity in cancer cells, is a plausible and promising avenue for developing epigenetic drugs. In this regard, in spite of the promising effects shown by synthetic DNMT inhibitors in clinical studies, their usefulness has been limited due to lack of specificity and resultant toxicity. Several dietary polyphenols have shown potential as DNMT inhibitors and in their ability to reverse methylation-induced silencing and restore the expression of various tumor suppressor genes (Figure 3). Herein, we will have summarized in detail, experimental data demonstrating the effect of various dietary polyphenols on DNA methylation changes in various models of human cancers (Table 1).
Figure 3. Effects of dietary polyphenols on the DNA methylation and histone modifications.
Simplified scheme demonstrates a number of epigenetic changes that occur during carcinogenesis. In cancers, tumor suppressor genes become “inactivated” (shown as red circles) while oncogenes are “activated” (green circles). Epigenetic gene expression regulation is a complex process and several key enzymes play crucial roles. DNA methyltransferase (DNMT) is responsible for transfer of methyl group to 5′-cytosine. Histone acetylases (HAT) and histone deacetylases (HDAC) are responsible for the acetylation and de-acetylation of lysine residues within histone tails, respectively. Because of these histone modifications, conformational changes in chromatin structure lead to changes in DNA accessibility for transcription regulators and polymerases. Polyphenols can impact these enzymes in specific ways induces reversibility of epigenetic dysregulation in cancer cells.
Table 1.
Polyphenols and DNA methylation.
| Dietary agent | Plant source | Molecular mechanism | Validated gene target(s) | In vitro model | In vivo model | Concentration | Treatment duration |
References |
|---|---|---|---|---|---|---|---|---|
| Apigenin | Parsley, celery | DNMT inhibitor | Esophageal | 20–50 µM | * | (103) | ||
| Baicalein | Indian Trumpet | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
| Betanin | Beetroot red | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
| Biochanin A | Soy | DNMT inhibitor | Esophageal; Prostate | Daphnids | 20–100 µM | 6 days | (103;115;230) | |
| Caffeic acid | Coffea | DNMT inhibitor | RARβ, CDKN2A | Breast | 1–50 µM | 8 days | (126) | |
| Catechin | Green tea | DNMT inhibitor | RARβ | Breast | 5–50 µM | 3–6 days | (101) | |
| Chlorogenic acid | Coffea | DNMT inhibitor | RARβ, CDKN2A | Breast | 1–50 µM | 8 days | (126) | |
|
Coumaric/Hydroxycinn amic acid |
Cinnamon | DNMT inhibitor | Esophageal | 20–50 µM | * | (103) | ||
| Curcumin | Turmeric | DNMT inhibitor | Esophageal; Leukemia; | 3–50 µM | 3 days | (103;130;131) | ||
| Cyanidin | Berries, grapes | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
| Daidzein | Soy | DNMT inhibitor | Esophageal; Prostate; Mice | 20–100 µM | 6 days | (103;115;121) | ||
| Ellagic Acids | Berries | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
| Epicatechin | Green tea | DNMT inhibitor | Esophageal; Breast | 50 µM | * | (100;101) | ||
| Epicatechin gallate | Green tea | DNMT inhibitor | Esophageal | 20–50 µM | * | (100) | ||
| Epigallocatechin | Green tea | DNMT inhibitor | Esophageal | 20–50 µM | * | (100) | ||
|
Epigalocatechin-3- gallate |
Green tea | DNMT inhibitor |
RARβ, MGMT, MLH1, CDKN2A, RECK, TERT, RXRα, CDX2, GSTP1, WIF1 |
Esophageal; Oral; Prostate; Urinary; Lung; Colon; Leukemia; Lymphoma; |
Agouti mouse Mouse models: skin, prostate, colon and uterine cancer Human: gastric and oral cancers, premenopausal women |
10 µg/ml; 20– 100 µM; 0.3– 0.6% |
2–6 days | (78;80;100– 105;108– 111;113;114;114;192;231) |
| Fisetin | Poison ivy | DNMT inhibitor | Esophageal; Breast | 5–20 µM | (101;103) | |||
| Galangin | Galangal root, propolis |
DNMT inhibitor | Breast | 20 µM | 3 days | (132) | ||
| Garcinol | Garcinia | DNMT inhibitor | Esophageal | 20–50 µM | * | (103) | ||
| Genistein | Soy | DNMT inhibitor ↓ DNMTs, MBD1, MBD4 MeCP2 expression |
RARβ, MGMT, CDKN2A, GSTP1, HMGN5, BTG3, TERT |
Esophageal; Prostate | Daphnids | 3.75–100 µM; 50–300 mg/kg/d |
3–6 days | (7;8;103;115– 123;192;230;232) |
| Hesperidin | Citrus | DNMT inhibitor | Esophageal | 20–50 µM | * | (103) | ||
| Isothiocyanates | Broccoli, broccoli sprouts |
Unknown | GSTP1 | Esophageal; Prostate | 2.5 µM | 5 days | (128;129) | |
| Luteolin | Parsley, celery | DNMT inhibitor | Esophageal | 20–50 µM | * | (103) | ||
| Lycopene | Tomatoes | Unknown | GSTP1, RARβ, HIN-1 | Breast | 2 µM | 1–2 weeks | (117) | |
| Myricetin | Berries | DNMT inhibitor | Esophageal; Breast | 5–25 µM | 3 days | (101;103;132) | ||
| Naringenin | Citrus | DNMT inhibitor | Esophageal | 20–50 µM | * | (103) | ||
| Phloretin | Apples | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
|
Piceatannol (Resveratrol metabolite) |
Grapes, blueberries | DNMT inhibitor | Breast | 20–40 µM | 3 day | (132) | ||
| Protocatechuric acid | Olives | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
| Quercetin | Citrus | DNMT inhibitor | CDKN2A | Esophageal; Breast; Colon | 5–20 µM | 5 days | (101;103;233) | |
| Resveratrol | Grapes, wines, eucalyptus |
DNMT inhibitor | Breast; Lung | 20–40 µM | 1 day | {Paluszczak, 2009 213 /id;Panayiotidi s, 2006 90 /id;Stefanska, 2010 340/id} |
||
|
Rosmarinic acid /Rosmarinic |
Rosemary | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
| Sinapic acid | Sinapis (mustard) | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) | ||
| Sulforaphane | Broccoli | ↓ DNMTs expression | Esophageal; Colon | 50 µM | * | (127;192) | ||
| Syringic Acid | Red grapes | DNMT inhibitor | Breast | 20–40 µM | 3 days | (132) |
DNA methyltransferase activity assay using only nuclear extracts.
5.1 Epigallocatechin-3-gallate (EGCG)
EGCG, the major polyphenol in green tea, has been extensively studied as a potential demethylating agent. EGCG is methylated by catechol-O-methyltransferase (COMT), the enzyme responsible of the inactivation of catechol molecules, such as dietary polyphenols. This enzyme introduces a methyl group to the catecholamine group, which is donated by S-adenosyl methionine (SAM). Demethylation of SAM results in the formation of S-adenosyl-L-homocysteine (SAH), and as SAH is a potent inhibitor of DNMT. Generation of SAH has been hypothesized as one of the mechanisms for the demethylating properties of this compound. On the other hand, EGCG can form hydrogen bonds with different residues in the catalytic pocket of DNMT, thus acting as a direct inhibitor of DNMT1 (100;101). The inhibition of DNMT may prevent the methylation of the newly synthesized DNA strand, resulting in the reversal of the hypermethylation and the re-expression of the silenced genes. Finally, it has been shown that EGCG is also an efficient inhibitor of human dihydrofolate reductase. Like other antifolate compounds, EGCG acts through interaction with folic acid metabolism in cells, causing the inhibition of DNA and RNA synthesis and altering DNA methylation (102).
In a seminal paper in 2003, Fang et al. showed that treatment of human esophageal cancer cells with EGCG caused a concentration- and time dependent reversal of hypermethylation of several known tumor suppressor genes such as p16, RAR, MGMT, and MLH1 genes (103). In the same work, reactivation of some methylation-silenced genes by EGCG was also demonstrated in human colon cancers and prostate cancer cells. Since then, several groups found similar in vitro results. Partial demethylation of hypermethylated RARβ by EGCG was demonstrated in breast cancer cell lines MCF-7 and MDA-MB-231 cells. Kato et al. showed that treatment of oral cancer cells with EGCG partially reversed the hypermethylation status of the RECK gene and significantly enhanced the expression level of RECK mRNA (104). Pandey et al. demonstrated that exposure of human prostate cancer LNCaP cells to green tea polyphenols caused a concentration- and time dependent re-expression of a known precursor to the genesis of prostate cancer (GSTP1), and methylation analysis revealed extensive demethylation in the GSTP1 promoter region (105). Berletch et al. showed that treatment of MCF-7 breast cancer cells with EGCG resulted in a time-dependent decrease in hTERT promoter methylation (80;106;107).
On the other hand, significant demethylation and activation of several genes by EGCG were not observed by other authors. Chuang et al. examined a total of six different genes/repetitive elements (p16, RARβ, MAGE-A1, MAGE-B2 and Alu) in three separate cell lines (T24, HT29, and PC3) for their DNA methylation levels and their mRNA expression levels using several demethylating agents (108). Treatment with EGCG did induce neither DNA demethylation nor reexpression of the analyzed genes. In addition, Stresemann et al. also performed a comparative analysis of compounds that had been previously reported to inhibit DNA methyltransferase activity in cancer cell lines, including EGCG (109). These authors determined the cytosine methylation level and the methylation status of TIMP3 in different cell lines and found that EGCG did not inhibit DNA methylation. There are many potential reasons for the discrepancies between studies, including different methods of analysis, possible gene-specificity or cell line–specificity of EGCG, or that treatment method might had been ineffective to show efficacy. Based on their results, Stresemann et al. argued that cellular effects induced by EGCG could probably be attributed to the oxidative stress induced by this compound.
Whether EGCG can reverse DNA hypermethylation and reactivate methylation-silenced genes in vivo still remain to be determined. Mittal et al. showed that topical treatment of EGCG in hydrophilic cream inhibits UVB induced global DNA hypomethylation pattern in chronically UVB-exposed mice (110). Immunohistochemical detection of DNA methylation pattern was performed using anti–5-methylcytosine monoclonal antibody. Since global DNA hypomethylation is a phenomenon usually associated with hypermethylation and inactivation of specific genes during carcinogenesis, the authors hypothesized that their observation was not contradictory to the concept that EGCG can prevent or reverse the hypermethylation of certain specific genes. Kinney et al. recently tested whether oral consumption of green tea polyphenols (GTP) could affect normal or cancer-specific DNA methylation in vivo, using a mice model (111). Wild-type and transgenic adenocarcinoma of mouse prostate (TRAMP) mice were given 0.3% GTPs in drinking water beginning at 4 weeks of age. To monitor DNA methylation, the authors measured 5-methyl-deoxycytidine (5mdC) levels, methylation of the B1 repetitive element, and methylation of the Mage-a8 gene. GTP treatment did not inhibit tumor progression in TRAMP mice and no dose-dependent alterations in DNA methylation status were observed. Yuasa et al. performed a retrospective analysis examining the methylation status of several genes in primary gastric carcinomas in relation to past lifestyle of the patients, including dietary habits (112;113). Methylation of CDX2 and BMP-2, measured by methylation specific PCR, correlated with the decreased intake of green tea and cruciferous vegetables.
Finally, Tsao et al. recently published a phase II randomized, placebo-controlled trial of green tea extract (GTE) in patients with high-risk oral premalignant lesions (OPL) (114). The OPL clinical response rate was higher in all GTE arms at different doses (n = 28; 50%) versus placebo (n = 11; 18.2%; P = 0.09) but did not reach statistical significance. Only two patients in the GTE arm had baseline p16 promoter methylation that could be evaluated following treatment, which did not reverse methylation status in either patient.
Although most of the evidence about the epigenetic properties of tea natural compounds has focused on EGCG, also other catechins such as catechin, epicatechin, epicatechin gallate and apigallocatechin have also been found to share similar features, although with much less DNMT inhibitory activity compared to EGCG (100;101;103).
5.2 Genistein
Genistein, one of the many phytoestrogens contained in soybeans, has been lately studied as a demethylating agent. Stronger than other soy isoflavones (Biochanin A or diadzein), genistein induces a dose-dependent inhibition of the DNMT activity, showing competitive and noncompetitive inhibition with respect to the substrate poly(dI-dC).poly(dI-dC) and noncompetitive inhibition with respect to SAM (115;116).
Treatment of KYSE 510 esophageal squamous cell carcinoma cells with genistein partially reversed DNA hypermethylation and reactivated p16, RARβ and MGMT. This was demonstrated by the appearance of unmethylation-specific bands by methylation-specific-PCR as well as by the increased mRNA levels determined by RT-PCR. Partial reversal of DNA hypermethylation and reactivation of RARβ were also observed in KYSE 150 cells and prostate cancer LNCaP and PC3 cells. In the same work, genistein in combination with other DNA methyltransferase or HDAC inhibitors (such as TSA) enhanced the reactivation of methylation-silenced genes (115).
King-Batoon et al. found that a low, nontoxic concentration of genistein (3.125 µM) partially demethylated the promoter of the GSTP1 tumor suppressor gene in MDA-MB-468 breast cancer cells (117). RT-PCR confirmed a lack of GSTP1 expression in untreated MDA-MB-468, with restoration of GSTP1 expression after genistein treatment. Similarly, treatment of renal and prostate cancer cells with genistein induced reversal of hypermethylation and reactivation of B-cell translocation gene 3 (BTG3), a known tumor suppressor gene in some malignancies (118;119).
Although in vitro studies have shown that genistein apparently induces DNA demethylation through DNMT inhibition, three animal studies have observed rather increased DNA methylation following the treatment. Day et al. treated male mice with different diet schemes including genistein for 2–4 weeks and control diet, and analyzed methylation changes in different tissues using a technique called mouse differential methylation hybridization (DMH) array (120). In this assay, the presence or absence of a spot would indicate hypermethylation or hypomethylation, respectively, relative to the control, including 300 spots covering 900 CpG islands. Interestingly, DNA from prostate showed a particular spot on the 4-wk genistein treatment membrane significantly darker than the equivalent spots on any other membrane, thus suggesting hypermethylation caused by genistein. Dolinoy et al. analyzed the effect of genistein exposure during gestation on DNA methylation in the offspring, due to the fact that this is when the epigenome is most susceptible to environmentally induced dysregulation (7). To determine if maternal genistein affects offspring by altering the epigenome in utero, the authors assessed coat color, DNA methylation, and body weight in genetically identical heterozygous yellow agouti (Avy/a) offspring. The expression of the Avy allele usually leads to yellow fur, obesity and tumorigenesis, while CpG methylation in an intracisternal A particle retrotransposon upstream of the Agouti gene correlates inversely with ectopic Agouti expression. The results showed that genistein induced CpG hypermethylation of six CpG sites in this region, shifting coat-color distribution toward pseudoagouti (brown), thereby decreasing the incidence of adult-onset obesity in Avy/a offspring. Guerrero-Bosagna et al. evaluated the sexual maturity, morphometric parameters and DNA methylation status in mice treated with soy isoflavones (genistein and daidzein) and found that this diet can result in an advancement of sexual maturation in female pups as well suppress normal gender differences in the DNA methylation pattern of a tissue specific methylated gene such as Acta1, inducing hypermethylation of this gene only in females (121). Conversely, Tang et al. showed that treatment of neonatal mice with genistein prevented the hypermethylation of nucleosomal binding protein 1 (Nsbp1) in the uterus throughout life, inducing uterine adenocarcinoma in aging animals (122).
The effect of genistein on methylation in humans has been recently tested. Qin et al. performed a double-blind, randomized trial in 34 healthy premenopausal women receiving 40 mg or 140 mg isoflavones daily (including genistein, daidzein, and glycitein) through one menstrual cycle (123). Methylation assessment of 5 cancer related genes known to be methylated in breast cancer (p16, RASSF1A, RARβ2, ER, and CCND2) was performed on intraductal specimens. In agreement with the animal data, the results showed that RARβ2 and CCND2 hypermethylation was increased after treatment and correlated with the genistein level.
5.3 Lycopene
Lycopene is a bright red carotene and carotenoid pigment found in tomatoes and other red fruits and vegetables. Lycopene can modulate the expression of numerous genes relevant to cell cycle control, DNA repair, and apoptosis in breast cancer cells as shown in gene array studies (124;125). King-Batton et al. showed that treatment of MDA-MB-468 breast cancer cells with a single dose of lycopene partially demethylated the promoter of the GSTP1 tumor suppressor gene, with a concomitant increase in expression (117).
5.4 Coffee polyphenols
Caffeic acid and chlorogenic acid are catechol-containing coffee polyphenols that, in a similar way to the tea polyphenols, have shown to be demethylating agents. Lee et al. studied the modulating effects of these two compounds on the in vitro methylation of synthetic DNA substrates and also on the methylation status of the promoter region of RARβ in two human breast cancer cells lines (126). The presence of caffeic acid or chlorogenic acid inhibited in a concentration-dependent manner the DNA methylation catalyzed by DNMT1, predominantly through a non-competitive mechanism. This inhibition, similar to other dietary polyphenols, was largely due to the increased formation of SAH. Treatment of MCF-7 and MAD-MB-231 human breast cancer cells with these two compounds partially inhibited the methylation of the promoter region of RARβ.
5.5 Sulforaphane
Sulforaphane, a dietary phytochemical obtained from broccoli, has been implicated in several physiological processes consistent with anticarcinogenic activity, including enhanced xenobiotic metabolism, cell cycle arrest, and apoptosis. Although the effect of sulforaphane as a demethylating agent has not been specifically studied, this compound was found to downregulate DNMT1 in CaCo-2 colon cancer cells (127).
5.6 Isothiocyanates
Isothiocyanates comprise another class of dietary compounds known to affect the epigenome. Isothiocyanates are metabolites of glucosinolates present in a wide variety of cruciferous vegetables and demonstrated to have anticancer properties. Treatment of prostate cancer cells with phenethyl isothiocyanate, a metabolite of gluconasturtin from watercress, was shown to lead to demethylation and re-expression of GSTP1 (128). On the other hand, treatment with different isothiocyanates prevented the esophageous tumorigenesis induced by the methylating agent N-nitrosomethylbenzylamine (NMBA) in male rats (129).
5.7 Curcumin
Curcumin, the major component of tumeric, has shown strong anti-inflammatory, anti-angiogenic and antioxidant, wound healing and anticancer effects for various diseases. It has recently shown that curcumin and one of its major metabolites, tetrahydrocurcumin can inhibit M.SssI, an DNMT1 analog, activity and its inhibitory activity may arisen from a potential covalent blocking of a catalytic group in DNMT1. This inhibition seems to be lower than other compound such as EGCG (103;130;131). More interestingly, curcumin exposure to genomic DNA of MV4-11 leukemia cell line induced a decrease in global DNA methylation comparable to decitabine (130). Our group has recently performed genome-wide methylation analysis in several colon cancer cell lines treated with curcumin, and have discovered that curcumin induces global methylation alterations in all cell lines in the time dependent manner (unpublished data).
5.8 Rosmarinic acid
Rosmarinic acid is a natural polyphenol antioxidant carboxylic acid found in many Lamiaceae herbs used commonly as culinary herbs such as lemon balm, rosemary, oregano, sage, thyme and peppermint. Rosmarinic acid has been recently shown to be a potent inhibitor of DNMT1 activity in nuclear extracts from MCF7 breast cancer cells and decrease the protein levels of DNMT1. However, this compound was unable to demethylate and reactivate known hypermethylated genes such as RASSF1A, GSTP1 and HIN-1 in this cell line (132).
5.9 Resveratrol
Resveratrol, a phytoalexin made naturally by several plants, has been produced by chemical synthesis because of its potential anti-cancer, anti-inflammatory, blood-sugar-lowering and other beneficial cardiovascular effects. There is limited evidence about the potential demethylating activity of this compound. Resveratrol has shown to be a weak DNMT activity inhibitor in nuclear extracts from MCF7 cells, and as rosmarinic acid, was unable to reverse the methylation of several tumor suppressor genes (132). In MCF-7 cells, resveratrol improved the action of adenosine analogues to inhibit methylation and to increase expression of RARβ2, although without significant effect on its own (133).
5.12 Other compounds with effects on DNA methylation
In addition to various dietary polyphenols described above, further compounds exist for which the evidence to modulate DNA methylation is less robust. Several of these additional compounds are not described here, but are listed in the Table 1.
6. POLYPHENOLS INDUCED HISTONE MODIFICATIONS
In addition to their ability to induced changes in DNA methylation, evidence indicates that dietary polyphenols can also regulate gene expression through changes in histone modifications (Figure 3). In this regard, several polyphenols are known to possess potent HAT and HDAC inhibitory activities. The text below and Table 2 systematically summarize the current understanding on the effects of dietary polyphenols on histone modifications, which may play a significant role in the chemopreventive potential of these compounds.
Table 2.
Polyphenols and histone modifications
| Dietary Agent | Plant source | Molecular mechanism |
Histones and Gene targets |
In vitro model | In vivo model | Concentration | References |
|---|---|---|---|---|---|---|---|
| 3,3-diindolylmethane | Broccoli | ↓ HDAC expression |
COX-2 | Colon; Breast | Mouse: colon cancer | 10–60 µM | (196–198) |
|
6-methoxy-2E,9E- humuladien-8-one |
Ginger | HDAC inhibitor | Breast | 1.25 µM | (234) | ||
| Allicin | Garlic | Unknown | H4 acetylation | Erythroleukemia | 2–200 µM | (176) | |
| Allyl mercaptan | Garlic | HDAC inhibitor | H3/H4 acetylation p21WAF1 |
Erythroleukemia; Liver; Colon |
Rat: liver | 2–500 µM (100mg/kg) |
(173–175;177;178) |
| Anacardic acid | Cashew nuts | HAT inhibitor | H3K9 and H3K14 deacetylation NF-kB activation |
Leukemia; Plasmodium; Cervix; Embryonic kidney; Breast; Lymphoma; Prostate; Lung; Esophageal; Skin |
3–200 µM | (138;139;153–160) | |
| Biochanin A | Soy | HDAC inhibitor | RARβ | Esophageal; Prostate | 20–100 µM | (115) | |
| Butein | Varnish Tree | Induction of SIRT1 activity |
Cervix | Drosophila | 100 µM | (203;204) | |
| Caffeic acid | Coffea | HDAC inhibitor | Cervix; Colon | 1–2.54 mM | (152;235) | ||
| Catechin | Green tea | HAT inhibitor | Lymphocytes | 100 µM | (165) | ||
| Chlorogenic acid | Coffea | HDAC inhibitor | Cervix | 0.375 mM | (152) | ||
| Cinnamic acid | Cinnamon | HDAC inhibitor | Cervix; Colon | 1–2 mM | (152;235) | ||
|
Coumaric/ Hydroxycinnamic acid |
Cinnamon | HDAC inhibitor | Cervix; Colon | 1–2 mM | (235) | ||
| Curcumin | Turmeric | HAT and HDAC inhibitor |
H3/H4 deacetylation GATA4, EOMES, GZMB, PRF1 |
Cervix; HIV; Hepatoma; Leukemia; Prostate; Brain; Lymphoma; Lymphocytes; |
Plasmodium falciparum; Herpes virus Mouse: epilepsy Rat: diabetes, heart failure |
6.25–135 µM (0.3–75 mg/kg) |
(134–152) |
| Daidzein | Soy | HDAC inhibitor | Histones acetylation RARβ |
Esophageal; Prostate | 12.8–100 µM | (115;199) | |
| Diallyl disulfide | Garlic | HDAC inhibitor | H3/H4 acetylation p21WAF |
Erythroleukemia; Leukemia; Liver; Colon; Prostate; Fibroblasts |
Rat: colon cancer | 20–200 µM, (42–200 mg/kg) |
(173;175;177;179– 183;236) |
| Dihydrocoumarin | Yellow Sweet Clover | SIRT1 and SIRT2 inhibitor |
p53 acetylation | Lymphoblastoid cell line | 0.75–50 mM | (220) | |
| Epicatechin | Green tea | HAT inhibitor | Lymphocytes | 100 µM | (165) | ||
| Epicatechin gallate | Green tea | HAT inhibitor | Lymphocytes | 100 µM | (165) | ||
| Epigallocatechin | Green tea | HAT inhibitor | Lymphocytes; Colon | 100 µM | (165;166) | ||
|
Epigalocatechin-3- gallate |
Green tea | HAT inhibitor HMT inhibitor |
H3/H4 acetylation H3K27 tri-methylation NF-κB, IL-6, BMI-1, EZH2, SUZ12 |
Lymphocytes; Colon; Keratinocytes; Prostate |
5–100 µM | (105;165–167) | |
| Equol | Soy | HDAC inhibitor | H2A/H2B/H3/H4 acetylation | Drosophila | 12.8 µM | (199) | |
| Fisetin | Poison ivy | SIRT1 activator | Cervix; Drosophila | 100 µM | (203;204) | ||
| Flavone | Feijoa | HDAC inhibitor | Histones acetylation p16, p21, TRAIL |
Myeloid leukemia | 170–340 µM | (237) | |
| Garcinol | Garcinia | HAT inhibitor | Global gene expression down- regulation |
Leukemia; Cervix; Lymphocytes; HIV |
5–100 µM | (139;156;161– 164;238) |
|
| Genistein | Soy | HAT activator HDAC inhibitor |
H2A/H2B/H3/H4 acetylation p21, p16, PTEN, CCLD, p53, FOXA3, SIRT1, BTG3 hTERT, RARβ |
Esophageal; Prostate; Breast; Renal |
5–100 µM | (115;116;118;119;199–201) | |
| Isoliquiritigenin | Liquorice | SIRT1 activator | Cervix; Drosophila | 100 µM | (203;204) | ||
| Isothiocyanates | Broccoli, wasabi | HDAC inhibitor | H3/H4 acetylation, p21, GSTP1 |
Prostate; Erythroleukemia; Leukemia; Prostate |
20–100 µM | (128;174;193–195) | |
| Luteolin | Parsley, celery | SIRT1 activator | Cervix | 100 µM | (203) | ||
| Piceatannol | Grapes, blueberries | SIRT1 activator | Cervix; Drosophila | 100 µM | (203;204) | ||
| Polyphenon B | Black and green tea | ↑ HDAC1 expression |
Rat: liver cancer | 0.05 % | (168) | ||
| Quercetin | Citrus, apple, berries | SIRT1 activator HAT inhibitor |
IP-10, MIP-2 | Cervix; Drosophila; Small intestine; |
Mouse: bowel inflammation |
100 µM | (203;204;219) |
| Resveratrol* | Grapes, wine, eucalyptus |
SIRT1 activator | TNFα, IL-8, RBP | Cervix; Endothelial; Embryonic kidney; Macrophages; Lung; Liver; Cardiomyocytes; |
Yeast; Drosophila; Mouse: colon cancer; Rat: lung cancer |
10–200 µM | (203–212;239–242) |
| S-allylmercaptocysteine | Garlic | HDAC inhibitor | H3/H4 acetylation | Erythroleukemia; Colon | 20–250 µM | (176;178) | |
| Sanguinarine | Opium poppy | Histone methylation inhibitor HAT inhibitor |
H3K9/H3K4 demethylation H3/H4 deacetylation |
Liver; Cervix | 5–75 µM | (221) | |
| Silibinin | Milk thistle | ↑ histone acetylation |
H3/H4 acetylation p21, p27, CASP3, CASP9. |
Hepatoma | 120–240 µM | (243) | |
| Sulforaphane | Broccoli | HDAC inhibitor | H3/H4 acetylation RARβ, HBD-2, p21,BAX |
Esophageal; Prostate; Colon; Kidney; Breast |
Mouse: colon cancer; prostate cancer xenografts Human: blood |
15–25 µM (443 µg/kg, 68g) |
(115;166;185–191) |
| Theophylline | Black and green tea | HDAC activator | Alveolar macrophages; Epithelial cells; Blood monocytes |
10 µM | (169–172) | ||
| Ursolic Acid | Basil | HDAC inhibitor | Histone acetylation | Leukemia | 5–20 µg/ml | (244) |
This table only provides selected publications for resveratrol as a SIRT1 inhibitor.
6.1 Curcumin
Strong evidence from in vitro and in vivo experiments suggests that curcumin functions as a potent histone modifying compound, especially as a HAT inhibitor. In one of the earliest studies using computational screening algorithms, curcumin was shown to bind to HAT enzymes in a covalent manner (134;135). Subsequently, several independent research groups showed that curcumin strongly inhibits p300/CBP activity in cell extracts from multiple cancers including cervix, hepatoma and leukemia at a concentration of 20µM or higher (136–139). Using prostate PC3-M cells and peripheral blood lymphocytes, Marcu et al. showed that curcumin selectively promotes proteasome-dependent degradation of p300/CBP without affecting other HATs such as PCAF or GCN5 (135). Inhibition of p300/CBP was shown to be associated with repression of histones H3/H4 and non-histone proteins such as p53, HIV-Tat protein, as well as HAT-dependent chromatin transcription (136). Moreover, curcumin was shown to effectively prevent histone hyperacetylation induced by the histone deacetylase (HDAC) inhibitor MS-275 in both cancer cells (PC3-M and HeLa) and peripheral blood lymphocytes (135;140).
In addition to its effect in cancer cells, curcumin has further been shown to modulate the immunologic memory of CD8+ T-lymphocytes partially through deacetylation of H3K9 at the promoter region of several key transcription factors such as Eomesodermin (EOMES) and its targets perforin (PRF) and granzyme B (GZMB) (141). In brain cancer cells and brain-derived neural cells, curcumin effectively induced histone H3/H4 hypoacetylation, and this effect was associated with neuronal differentiation, synaptogenesis, progenitor cell migration and neurogenesis both in vitro and in vivo, suggesting its importance in neural stem cell fate controlling (142). Besides human cells, Cui et al. showed that curcumin strongly inhibits one of the Plasmodium falciparum HAT’s nuclear activity (P.falciparum general control nonderepressed 5 (PfGCN5)) which induced hypoacetylation of H3K9 and -K14 (143). The same group further demonstrated that curcumin-related H3K9 hypoacetylation at the promoter region of the certain genes was associated with gene silencing (144).
Building upon the evidence gathered from in vitro studies, several groups have corroborated the HAT inhibitory effect of curcumin in animal models. In the kainate-induced status epilepticus mice model, that associates with H3S10 phosphorylation, H4 acetylation and CBP activation, pretreatment with curcumin (30 mg/kg) attenuated all histone modifications and the severity of the status epilepticus (145). Several studies have shown the beneficial effects of curcumin on the progression of streptozotocin-induced diabetes nephropathy in male Sprague-Dawley rats. In this animal model, curcumin treatment was associated with inhibition of p300, NF-kB, H3S10 phosphorylation and H3 hyperacetylation (146;147). Furthermore, two independent groups have shown that curcumin acts as a protective agent against cardiac hypertrophy, inflammation and fibrosis in animal models through both suppression of HAT activity (p300) and downregulation of GATA4, NF-κB and TGFβ/Smad signaling pathways (148;149). In these studies, curcumin abrogated H3/H4 acetylation, GATA4 acetylation levels and relative levels of p300/GATA4 complex, which is otherwise markedly increased in the hypertensive hearts of these rats (148;149).
In addition to HAT-inhibitory effect of curcumin, a few recent studies have inconclusively suggested a possible HDAC-inhibitory effect as well (137). Using Burkitt-lymphoma Raji cells, Liu et al. and Chen et al. showed that curcumin treatment was associated with down-regulation of HDAC1, HDAC3 and HDAC8 proteins, whereas H4 protein expression was up-regulated (150;151). Although these results require further experimental confirmation, another recent study has supported the HDAC–inhibitory effects of curcumin (152).
6.2 Anacardic Acid
Anacardic acid (AA) is an active compound of cashew nuts that has been shown to be a specific HAT inhibitor. AA has been shown to be a potent inhibitor of p300, PCAF and Tip60 HAT factors (153;154). Although AA does not affect DNA transcription directly, HAT-dependent transcription was strongly inhibited and was associated with simultaneous histone H3 and/or H4 hypoacetylation in HeLa and MCF7 cancer cells (153;155). In fact, on the basis of knowledge gained from its HAT inhibitory activity, AA chemical formula has been widely used for the development of new synthetic HAT inhibitors and activators (138;139;153;155–157).
Only very few studies have evaluated the molecular biological relevance of AA-related HAT inhibitory activity. Sung et al. showed that AA inhibits both inducible and constitutive NF-κB activation, and suppresses activation of IκBα kinase which leads to abrogation of its phosphorylation and eventual degradation in multiple cancer cells (myeloid and T-cell lymphoma cells, human embryonic cells, lung, prostate and esophageal cancer cells) (158). In the same study, it was demonstrated that AA inhibits acetylation and nuclear translocation of p65, and suppresses NF-κB-dependent reporter gene expression, which is dependent upon p300 HAT expression/activity (158). In addition, using human dermal fibroblasts Kim et al. demonstrated that AA effectively inhibits UV-induced cancer formation and premature skin aging by inhibiting UV-enhanced levels of c-H2AX, p53, and acetylation of H3 (159). In P. falciparum, similar to curcumin, AA has also been shown to inhibit PfGCN5 activity in nuclear extracts (160). In this elegant study, treatment with AA induced hypoacetylation of H3K9 and -K14, and resulted in down-regulation of 207 genes that were partially enriched for H3K9 acetylation. These data provide support for the HAT-inhibitory effects of AA not only in mammalian cells, but also in malaria parasites as well (160).
6.3 Garcinol
Garcinol is a highly cytotoxic polyisoprenylated benzophenone derivative from garcinia fruit rinds. Multiple studies have shown that garcinol is a potent inhibitor of different HATs, such as, p300 and PCAF (139;156;161). A recent mechanistic work using fluorescence, docking and mutational studies, has revealed that garcinol induces alteration in the secondary structure of the HAT proteins (162). Analogous to curcumin and AA, garcinol also possesses significant histone H3 and H4 deacetylating activities (161;163). In addition, garcinol also has the ability to inhibit autoacetylation of p300, which is one of the key regulatory mechanism for its catalytic activity in HeLa cor histones (163). Although none of the studies have specifically evaluated the effects of garcinol on gene-specific histone modifications, existing data indicates that garcinol and/or its synthetic derivate LTK14, down-regulates the expression of multiple genes in cervical cancer cells and T-lymphocytes, supporting its HAT inhibitory activity (161;164).
6.4 EGCG and Green tea polyphenols
EGCG is one of the first compounds that was recognized as an epigenetic modulator in cultured cancer cells. Although effects of EGCG have mainly been studied in the context of DNA methylation, recent data suggest that EGCG also acts as a histone modifier. Of all the catechins present in green tea, EGCG has shown to be is the most promising and potent modulator of histone marks in cancer cells (165). In a recent study by Choi et al., several green tea polyphenols were screened for their HAT activity in various cell extracts from B-lymphocytes. The authors discovered that among the studied tea polyphenols, EGCG was the most potent HAT inhibitor, and possessed global specificity towards various HAT enzymes in the following order p300>CBP>Tip60>PCAF (165). EGCG treatment inhibited the acetylation of p65 and the expression of NF-κB target genes in response to diverse stimuli, thus having great potential as a chemopreventive agent of chronic inflammation (165). The activity of EGCG toward other histone modifying enzymes such as HDACs, SIRTs and HMTs remains controversial. While Choi et al. and Nair et al. found no activity toward these enzymes (165;166), Pandey et al. demonstrated that green tea polyphenols show both inhibition of HDAC activity and reduction in mRNA expression of HDAC1, 2 and 3 in prostate cancer cells. These changes were subsequently associated with time-dependent increase in the acetylation of H3 and H4 (105).
In addition to the effect on HAT and HDAC activities, EGCG has recently shown to affect Polycomb Group (PcG) protein complexes PRC2 (EED) and PRC1 (BMI-1) in immortalized keratinocytes and skin cancer cells (167). Both PRC1 and PRC2 actively participate in epigenetic regulation of gene expression by increasing histone methylation and reducing acetylation, which leads to chromatin compaction and transcriptional silencing of genes in cancer cells (167). Treatment of skin cancer cells or immortalized keratinocytes with 60µM EGCG reduced the expression of BMI-1 and EZH2, which was associated with reduction in survival and global reduction in histone H3K27me3, a hallmark of PRC2 complex action (167).
6.5 Other tea polyphenols: Polyphenon B and Theophylline
There is limited evidence regarding the epigenetic properties of polyphenon B (black tea polyphenol), in cultures cancer cells. In a single study on DAB-induced liver cancer animal model, polyphenon B (0.05%) induced a significant decrease in HDAC1 expression in male Sprague-Dawley rats, compared to the untreated controls (168).
Theophylline, also known as dimethylxanthine, shows a structural similarity to caffeine and is present in low concentrations in tea. Cosio et al. evaluated the HDAC modulating effect of theophylline in smokers and chronic obstructive pulmonary disease (COPD) patients, a situation known to have decreased HDAC activity (169). Interestingly, theophylline treatment was associated with down-regulation of the inflammatory response through modulation of HAT, HDAC activity, and NF-κB activation. Following studies from the same group demonstrated that low-doses of theophylline increased HDAC activity in epithelial cells and macrophages, and further reduced IL-8 and TNFα concentrations (170–172). This mechanism occurred at therapeutic concentrations and independently of its phosphodiesterase inhibition, improving the anti-inflammatory effects of steroids (172).
6.6 Allyl-derivates
Allyl-derivates from garlic were one of the first compounds that were described to have an impact on histone acetylation, suggesting that these compounds may inhibit HDAC enzyme activity in mouse and human leukemia cells DS19 and K562 (173). Initially it was shown that various allyl-derivates such as allyl mercaptan (AM), diallyl disulfide (DADS), S-allylcysteine(SAC), S-allylmercaptocysteine (SAMC) and allicin induce increased histone acetylation (H3/H4) both in cultured cancer cells such as DS19 and at higher concentrations in liver from rats (173–176). Among various allyl-derivates and precursors, AM is the most potent HDAC inhibitor in colorectal cancer and leukemic cells (173;177;178). Using docking simulation model with human HDAC8 protein, Nair et al. have demonstrated that AM interacts with the enzyme active site, and this was accompanied by a rapid accumulation of H3/H4 histones (166). Similar to other HDAC-inhibitors, AM increased histone H3 acetylation on the CDKN1A gene, with a concomitant increase in binding of transcription factor Sp3 and p53 to the promoter region (178). Although less significant, several studies have shown that DADS treatment in CaCo2 and HT29 colorectal cancer cells also produced increased acetylation of H3 and H4, and up-regulation of CDKN1A (177;179–183). SAMC and DADS treatments were also shown to associate with increased E-cadherin expression, however, its relationship concerning HDAC inhibitory effects has not been evaluated (180;184). In addition to in vitro evidence accumulated thus far, animal studies with ally-derivatives have shown that treatment with AM and/or DADS increases acetylation of histones and causes up-regulation of p21 expression in normal liver and hepatoma cells and in rat colonocytes (175;180;181). Although these findings are very encouraging, there is concern about the high concentrations of allyl-derivatives used in animal studies, which are unlikely to be physiologically achievable in humans if such compounds are considered for clinical intervention (175;180;181). Considering this, it was proposed that SAMC may be a safer and more effective choice, as it was shown to induce growth arrest in mouse erythroleukemia cells at relatively low concentrations (176). However, SAMC is inherently a weaker HDAC inhibitor that AM or DADS, as evidence when it was tested in prostate cancer cell lines (184).
6.7 Isothiocyanates (Sulforaphane and Isothiocyanates derivates)
Isothiocyanates such as phenethyl isothiocyanate (PEITC) and sulforaphane (SFN) are the main compounds found at high levels in broccoli. Until now multiple studies have confirmed the potent HDAC inhibitory activity of isothiocyanates. Using human kidney cells and colorectal cancer cells, Myzak et al. have demonstrated that one of the SFN metabolites (SFN-Cys) acts as a HDAC inhibitor, as predicted by computer modeling (185). Although Nair et al. failed to demonstrate HDAC inhibitory effect when SFN was combine with EGCG in HT29 colon cancer cells (166), several studies have clearly confirmed the HDAC inhibitory activity of SFN (150–25µM) in multiple human cancer cell lines (colon, prostate and breast cancer cells) (185–188). HDAC inhibition in cell lines and animals was associated with increased histone H3/H4 acetylation (185–187;189). Such effects of SFN were associated with increased H4 acetylation in the p21 and Bax promoters, which resulted in significant up-regulation of both gene and protein expression in prostate cancer cells (185–187;189). However, Pledgie-Tracy et al. failed to observe any increase in histone acetylation induced by SFN in breast cancer cells (188).
In a mice model, SFN-containing diet is optimal for achieving SFN tissue concentrations in the 3–30µM range (190). SFN-enriched diets in these animals were shown to suppress tumor development in APCmin/+ mice via increase in overall H3/H4 histone acetylation, and a concomitant up-regulation of p21 expression (189). Not only this, in a pilot study in human volunteers, consumption of 68g broccoli sprouts resulted in a significant inhibition of blood HDAC activity 3 h following intake (187). SFN treatment induced changes in the gene expression of numerous genes in human colon cancer cell lines, although it remains unknown if these changes are consequence of modifications in their corresponding histone marks (127;191). In support of this, squamous esophageal cells treated with SFN showed a marked increase in the RARβ expression in a similar manner as Trichostatin A (TSA) and 5-aza-CdR, two potent HDAC and DNMT inhibitors, respectively (192).
Allyl-isothyocyanate, one of the first compounds isolated from broccoli, was shown to increase acetylation of histones in mouse erythroleukemia cells (174). In this study, this effect was independent of changes in HAT activity, which implicates HDAC inhibition as a possible mechanism for these results (174). This has been confirmed by several other studies in which isothiocyanates have been shown to inhibit HDAC activity inducing histone acetylation and up-regulation of p21/Bax expression in various cancer cell lines (leukemia, esophageal squamous and prostate cancer cells) (128;193–195).
6.8 3,3′-Diindolylmethane
3,3’-Diindolylmethane (DIM) is an active compound derived from the digestion of indole-3-carbinol, which is found in Brassica family of vegetables, such as broccoli or cauliflower. In a study to determine the HDAC inhibitory effects of DIM, Bhatnagar et al. found that this compound significantly inhibits the expression of HDAC1, HDAC2 and HDAC3 in colon cancer cells, which was associated with strong inhibition of anti-apoptotic protein survivin both in colon cancer cells and APCmin/+ mice (196). Li et al. have recently provided insight into the mechanism responsible for HDAC1 inhibition. Using HT29 and SW620 colon cancer cell lines, the authors demonstrated that DIM selectively induces proteasome-mediated degradation of class I histone deacetylases (HDAC1-3 and HDAC 8), which resulted in increased p21 and p27 expression (197). In another study, treatment of MCF-7 breast cancer cells with DIM prevented histone H4 acetylation at the COX-2 gene promoter, thus inhibiting over-expression of this gene (198).
6.9 Isopflavone and soy peptides
Genistein is the major isoflavone present in soybeans. Accumulating evidence indicates that genistein possesses the highest histone modifying activity in comparison to the other isoflavones, biochanin A and diadzein (or its derivate equol). In the few available studies, genistein has shown to increase histone acetylation in esophageal squamous and prostate cancer cells (115;119;199). In further support of this, several studies have revealed an increased activation of HAT after genistein treatment in renal and prostate cancers, and in some of these studies, this was associated with increased mRNA expression of CREBBP, HAT1, PCAF and EP300 (118;119;200).
Three studies have evaluated the HDAC activity of genistein using in nuclear extracts from cancer cells indicating that this compound decreases the HDAC activity in squamous esophageal and renal cancer cell lines, but not in prostate cancer cells (115;118;119). In addition, there is evidence suggesting that genistein inhibits the expression of SIRT1, one of the NAD+ dependent histone deacetylases (201). So, either by HAT activation or HDAC inhibition, genistein is clearly associated with activation of tumor suppressor genes (such as p21, p16, FOXA3a, PTEN etc.) or inhibition of oncogenes (hTERT).
6.10 Resveratrol
Resveratrol is believed to play a significant role in the reduction of cardio-vascular events (202). Multiple studies have shown that resveratrol is associated with activation of NAD+ dependent histone deacetylase sirtuin 1 (SIRT1) and p300 in multiple in vitro and in vivo models (203–206). In one of the key studies, Howity et al. showed that resveratrol intake could extend the lifespan of several yeast (Saccharomyces cerevisiae) and worms strains (Drosophila melanogaster) and have favorable effects against metabolic disorders including obesity and insulin resistance (203–205;207). Although it is debated whether the direct SIRT1 activation-induced by resveratrol might be responsible for such effects (208–211), recent elegant animal studies have demonstrated that cancer preventive effects of resveratrol are significantly dependent on SIRT expression in APCmin/+ mice, suggesting the importance of SIRT1 activating effect as a key mechanism in resveratrol tumor prevention (212). To further illustrate the molecular mechanism underlying how activated SIRT1 triggers cell death, it was demonstrated that SIRT1 negatively regulates expression of Survivin, which encodes an anti-apoptotic protein, by deacetylating H3K9 within the promoter of Survivin (213). Additionally, it was shown that SIRT1 mediated BRCA1 signaling in breast cancer cells by inhibiting tumor growth through the repression of transcription of oncogenes or activity of oncoproteins (205;213). In another recent study, it was demonstrated that resveratrol treatment also enhanced p53 acetylation and apoptosis in prostate cancer cells by inhibiting MTA1/NuRD complex(214). Previously, it was demonstrated that resveratrol-induced SIRT1 activation lead to the modulation of PGC-1α functions in animals (215). The authors suggested that resveratrol treatment ultimately impacted the regulation of energy homeostasis, which might be a crucial feature for its chemopreventive potential (215). Since it is beyond the scope of this article to describe the detailed effects of resveratrol on epigenetics and ageing, we direct the readers to some of the previously published reviews on the topic (216–218).
6.11 Quercetin
Quercetin, a potent anti-tumor dietary polyphenol, is predominantly present in citrus fruits and buckwheat. Quercetin has been shown to activate NAD+ dependent histone deacetylase SIRT1 in yeast, but this effect was less pronounced compared to resveratrol (203). Ruiz et al. evaluated the effect of quercetin on the TNFα mediated expression of interferon-g-inducible protein 10 (IP-10) and macrophage inflammatory protein 2 (MIP-2) genes in murine intestinal epithelial cells (219). Treatment with quercetin was associated with inhibition of the HAT activity on the promoter region of these genes, which resulted in reduced gene expression (219).
6.12 Dihydrocoumarin
Dihydrocoumarin (DHC) is an active compound found in Melitous officinalis (sweet clover) that is widely used in food and cosmetic industries. DHC has recently identified as an inhibitor of the HDAC family of Sirtuins, which have a firmly established role in aging (220). DHC disrupted heterochromatic silencing and inhibited yeast Sir2p and human SIRT1/2 deacetylase activity, which caused p53 acetylation and increase in apoptosis in vitro (220).
6.13 Sanguinarine
Sanguinarine (SGR) is commonly extracted from several plants such as bloodroot (Sanguinaria canadensis) or from the root, stem and leaves of the opium poppy. SGR has been shown to induce conformational changes by interacting with chromatin (221). This compound potently inhibited HAT activity in rat liver and cervix cancer cell lines, and this was associated with dose-dependent decrease in H3/H4 acetylation. In addition, binding of SGR to chromatin inhibited H3K4 and H3R17 methylation, an epigenetic mark associated with transcriptional activation, more efficiently than H3K9 methylation, a marker for silent heterochromatin. Interestingly, SGR treatment was found to modulates global gene expression (221).
6.14 Other compounds with Histone modification properties
In addition to the well-described effects of various dietary polyphenols on histone modifications described above, there are several other such compounds for which limited evidence exists for their ability to modulate histone modifications. Many of these additional compounds are not mentioned here, but are listed in the Table 2.
7. POLYPHENOLS AND miRNA EXPRESSION
MicroRNAs have been recently discovered as key regulators of gene expression. Although there is still limited evidence, recent reports have identified that dietary polyphenols can also modulate gene expression by targeting various oncogenic or tumor suppressive miRNAs. In the last section of this review, we will summarize the current evidence supporting the effect of dietay polyphenols on specific target miRNAs (Table 3 and Figure 4).
Table 3.
Polyphenols and microRNAs
| Dietary agent | Plant source | Analysis Method | Micro RNAs | Target gene | In vitro model | In vivo model | Concentration | Treatment duration |
References |
|---|---|---|---|---|---|---|---|---|---|
| 3,3-diindolylmethane | Broccoli | Microarray; qRT-PCR | miR-200 (a–c), let-7 (a– f), miR-146a |
ZEB1, EGFR | Pancreas | 25 µM | 48h | (225; 226) | |
| Curcumin | Turmeric | Microarray; qRT-PCR | miR-22, miR-199a*, miR-21, miR-200 |
SP1, ESR1, PTEN |
Pancreas | 1–10 µM | 72h | (222; 223) | |
|
Epigalocatechin-3- gallate |
Green tea | Microarray; qRT-PCR | miR-16 | BCLl2 | Liver | 100 µM | 24h | (224) | |
|
Genistein (with diadzein, glycetein) |
Soy | Microarray; qRT-PCR | miR-200 (a–c), let-7 (a– f), mir-27b, miR-146a |
ZEB1, ZBTB10, EGFR |
Pancreas; Uveal; Melanoma; Ovarian |
25 µM | 48h | (225–228) | |
| Isothiocyanates | Broccoli, wasabi | Microarray | Rat: lung cancer | 500 mg/kg | 28d | (229) |
Figure 4. Effect of dietary polyphenols on microRNA (miRNA) expression.
miRNA are transcribed in the nucleus into primary miRNA (pri-miRNA) which is further cleaved by Drosha into precursor miRNA (pre-miRNA). Pre-miRNA is exported from nucleus to the cytoplasm and further processed by Dicer into miRNA duplex. Single strand of miRNA duplex (also called mature miRNA) leads this complex to mRNA cleavage or translation repression, which is dependent on miRNA:mRNA complementarity. Dependent on various factors, miRNA can have either an oncogenic role (called onco-miRNAs) if the target mRNA is a tumor suppressor gene, or a tumor suppressive role (tumor-suppressor miRNAs) if the target molecule is an oncogene. Dietary polyphenols can impact expression level of miRNAs and participate in gene expression regulation.
7.1 Curcumin
The antitumor activity of curcumin has been recently linked to changes in miRNA expression. Sun et al. treated BxPC-3 pancreatic cancer cell lines with curcumin and performed microRNA expression profile, showing significant changes in the level of expression of 29 miRNAs (222). Upregulation of miR-22 was one of the most significant curcumin-induced changes. Functional studies showed that upregulation of miRNA-22 expression by curcumin or by transfection with miRNA-22 mimetics in the PxBC-3 pancreatic cancer cell line suppressed expression of two of its known target genes, SP1 and ESR1. In other recently published study, Ali et al. have further analyzed potential of curcumin in treatment of pancreatic cancer in combination with Gemcitabine, a standard agent for the pancreatic cancer chemotherapy (223). Using BxPC-3 and MIAPaCa cell lines, which show difference in gemcitabine-resistance, they demonstrated that treatment with either curcumin or curcumin-analogues potentiated apoptotic effects of gemcitabine, which resulted in induction of miR-200b/c and inhibition of miR-21 expression. In addition, the authors showed increased PTEN expression as a result of miR-21 expression (223).
7.2 EGCG
EGCG has been recently found to modulate the miRNA expression in human hepatocellular carcinoma HepG2 cells. Tsang et al. performed microarray analysis in this cell line after EGCG treatment and found that this compound modified the expressions of 61 miRNAs (224). miR-16, one of the miRNAs up-regulated by EGCG, is known to regulate the anti-apoptotic protein Bcl-2, and interestingly EGCG treatment induced apoptosis and down-regulated Bcl-2 in HepG2 cells. Transfection with anti-miR-16 inhibitor suppressed miR-16 expression and counteracted the EGCG effects on Bcl-2 down-regulation and also induction of apoptosis in cells..
7.3 Isoflavones: genistein and 3,3′-diindolylmethane (DIM)
Soybean isoflavones and DIM have been found to regulate the miRNA expression in pancreatic cancer cells. Li et al. recently compared the expression of miRNAs between gemcitabine-sensitive and gemcitabine-resistant pancreatic cancer cells and investigated whether the treatment of cells with these two dietary compounds could affect the expression of miRNAs (225). The expression of the miR-200 and let-7 family was significantly down-regulated in gemcitabine-resistant cells, which showed epithelial-mesenchymal transition (EMT) characteristics. Interestingly, reexpression of miR-200 or treatment of gemcitabine-resistant cells with DIM or isoflavones (a combination of genistein, diadzin, and glycitein) resulted in the reversal of the EMT features, leading to epithelial morphology. These results indicate that natural compounds could function as miRNA regulators leading to the reversal of EMT phenotype, which is likely to be important for designing novel therapies for pancreatic cancer. The same group has further shown that treatment with the same compounds up-regulates miR-146a in pancreatic cancer cell, and this was associated with reduction in cell invasion and metastasis (226). This effect was associated with downregulation of EGFR and NF-κB regulatory kinase interleukin 1 receptor associated kinase. Reexpression of miR-146a inhibited the invasive capacity of pancreatic cancer cells with concomitant down-regulation of EGFR and interleukin 1 receptor–associated kinase 1 (IRAK-1) further pointing the significance of polyphenol mediated anticancer effect (226).
Another study has shown that genistein inhibits cell growth and modulate the expression of miR-27a and one of its targets (gene zinc finger and BTB domain containing 10 or ZBTB10) in human uveal melanoma cell lines (227). Further, in an observational study, Parker et al. observed that genistein is able to induce changes in miRNA expression in ovarian cancer cell lines (228).
7.4 Indole-3-carbinol and phenethyl isothiocyanate
Izzotti et al. recently published a study focused on the potential of natural compounds as chemopreventive agents after environmental cigarette smoke (ECS) exposure in animals (229). In this study, microarray miRNA expression analysis was performed in the lungs of either ECS-free or ECS-exposed rats treated with the orally administered chemopreventive agents including indole-3-carbinol and phenethyl isothiocyanate (both found in cruciferous vegetables). Interestingly, none of the above chemopreventive agents appreciably affected the baseline microRNA expression in non-exposed lungs, indicating potential safety. However, all of them attenuated ECS-induced alterations to a variable extent and with different patterns, indicating potential preventive efficacy.
8. SUMMARY AND CONCLUSIONS
There is traditional and widespread use of dietary polyphenols all around the world. While the anecdotal epidemiological evidence has historically supported the idea of different diet and good health, experimental evidence accumulated in the recent years from various pre-clinical and clinical studies clearly support the idea that dietary polyphenols have potentially beneficial effects on multitude of health conditions, including cancer. This review article provides a novel perspective on the potential chemoprevention by diet and dietary agents, as the extensive data summarized here suggest that beneficial effects of different dietary polyphenols may in part be attributable to their epigenetic properties, including changes in the DNA methylation pattern, regulation of histone modifications and changes in the expression of some miRNAs. Although the health effects of dietary polyphenols in humans are generally considered promising, there are definite challenges and limitations of the current data in better understanding the molecular mechanisms responsible for this effect, together with the possible interactions between different polyphenols and other dietary constituents. While in vitro models have enormously contributed to the understanding of polyphenols mediated regulation of the epigenetic network, there is still a paucity of in vivo data for the majority of these dietary compounds. Therefore, until sufficient preclinical and clinical data has been gathered on the epigenetic changes induced by some of the dietary polyphenols, one should be cautious while interpreting and extrapolating the significance of current in vitro evidence. Once such evidence is established, the next and more important step would be to determine the most effective doses of these ‘dietary nutraceuticals’ in order to obtain various beneficial effects in human subjects. Additional clinical work is required to examine the safety profile of various doses of dietary polyphenols, and more basic science studies are needed to improve our understanding of the molecular mechanisms underlying the chemopreventive effect of various dietary polyphenols. It is really exciting to witness that we have at least begun to explore the molecular mechanisms underpinning the “goodness” of certain diets and diet-related factors that has been in existence for centuries. The mere fact that currently hundreds of dietary polyphenols are being characterized from an “epigenomic” perspective clearly reflects our enthusiasm and trust we pose in the concept of safe and natural agents for cancer chemoprevention. Of course, the current evidence is thin and it is a long and treacherous road ahead of us; nonetheless, given the promise and potential of these polyphenols it is realistic to fathom that some of these compounds may become integral for the cancer chemoprevention in future.
Acknowledgement
The present work was supported in part by grant R01 CA129286 from the National Cancer Institute, National Institutes of Health.
Abbreviations
- DNMT
DNA methyltransferase
- HMT
histone methyltranferase
- HDAC
histone deacetylase
- HAT
histone acetyltransferase
- DIM
3,3’-diindolylmethane
- SGR
sanguinarine
- SAM
S-adenosyl methionine
- SAH
S-adenosyl-L-homocysteine
- miRNA
microRNA
- EGCG
epigallocatechin-3-gallate
- PsA
psammaplin A
- SFN
sulforaphane
- AM
allyl mercaptan
- DADS
diallyl disulfide
- COPD
chronic obstructive pulmonary disease
- PEITC
phenethyl isothiocyanate
- EMT
epithelial-mesenchymal transition
- LINE
Long Interspersed Nuclear Elements
- 5-aza-CdR
5-aza-2-deoxycytidine
- TSA
Trichostatin A
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None of the authors have any potential conflicts to disclose.
REFERENCES
- 1.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007 Nov;7(11):834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ducasse M, Brown MA. Epigenetic aberrations and cancer. Mol Cancer. 2006;5:60. doi: 10.1186/1476-4598-5-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ellis L, Atadja PW, Johnstone RW. Epigenetics in cancer: targeting chromatin modifications. Mol Cancer Ther. 2009 Jun;8(6):1409–1420. doi: 10.1158/1535-7163.MCT-08-0860. [DOI] [PubMed] [Google Scholar]
- 4.Esteller M. Epigenetics in cancer. N Engl J Med. 2008 Mar 13;358(11):1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 5.Dolinoy DC, Weidman JR, Jirtle RL. Epigenetic gene regulation: linking early developmental environment to adult disease. Reprod Toxicol. 2007 Apr;23(3):297–307. doi: 10.1016/j.reprotox.2006.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Herceg Z. Epigenetics and cancer: towards an evaluation of the impact of environmental and dietary factors. Mutagenesis. 2007 Mar;22(2):91–103. doi: 10.1093/mutage/gel068. [DOI] [PubMed] [Google Scholar]
- 7.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006 Apr;114(4):567–572. doi: 10.1289/ehp.8700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dolinoy DC, Huang D, Jirtle RL. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proc Natl Acad Sci U S A. 2007 Aug 7;104(32):13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin HJ, Zuo T, Chao JR, Peng Z, Asamoto LK, Yamashita SS, et al. Seed in soil, with an epigenetic view. Biochim Biophys Acta. 2009 Sep;1790(9):920–924. doi: 10.1016/j.bbagen.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003 Mar;33 Suppl:245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 11.Dehan P, Kustermans G, Guenin S, Horion J, Boniver J, Delvenne P. DNA methylation and cancer diagnosis: new methods and applications. Expert Rev Mol Diagn. 2009 Oct;9(7):651–657. doi: 10.1586/erm.09.53. [DOI] [PubMed] [Google Scholar]
- 12.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009 Jun 15;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002 Jan 1;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y, Leung FC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004 May 1;20(7):1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008 Jun;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 16.Prendergast GC, Ziff EB. Methylation-sensitive sequence-specific DNA binding by the c-Myc basic region. Science. 1991 Jan 11;251(4990):186–189. doi: 10.1126/science.1987636. [DOI] [PubMed] [Google Scholar]
- 17.Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998 Jun;19(2):187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 18.Plass C. Cancer epigenomics. Hum Mol Genet. 2002 Oct 1;11(20):2479–2488. doi: 10.1093/hmg/11.20.2479. [DOI] [PubMed] [Google Scholar]
- 19.Plass C, Soloway PD. DNA methylation, imprinting and cancer. Eur J Hum Genet. 2002 Jan;10(1):6–16. doi: 10.1038/sj.ejhg.5200768. [DOI] [PubMed] [Google Scholar]
- 20.Berletch JB, Phipps SM, Walthall SL, Andrews LG, Tollefsbol TO. A method to study the expression of DNA methyltransferases in aging systems in vitro. Methods Mol Biol. 2007;371:81–87. doi: 10.1007/978-1-59745-361-5_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berletch JB, Andrews LG, Tollefsbol TO. A method to detect DNA methyltransferase I gene transcription in vitro in aging systems. Methods Mol Biol. 2007;371:73–80. doi: 10.1007/978-1-59745-361-5_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005 Apr 15;14(Spec No 1):R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 23.Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003 Nov 20;349(21):2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- 24.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009 Jun 15;15(12):3927–3937. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Razin A, Kantor B. DNA methylation in epigenetic control of gene expression. Prog Mol Subcell Biol. 2005;38:151–167. doi: 10.1007/3-540-27310-7_6. [DOI] [PubMed] [Google Scholar]
- 26.Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007 Jan;1775(1):138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 27.Ushijima T, Asada K. Aberrant DNA methylation in contrast with mutations. Cancer Sci. 2010 Feb;101(2):300–305. doi: 10.1111/j.1349-7006.2009.01434.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999 Aug 1;13(15):1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lund AH, van LM. Epigenetics and cancer. Genes Dev. 2004 Oct 1;18(19):2315–2335. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 30.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997 Sep 18;389(6648):251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 31.Kouzarides T. Chromatin modifications and their function. Cell. 2007 Feb 23;128(4):693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Liang G, Lin JC, Wei V, Yoo C, Cheng JC, Nguyen CT, et al. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004 May 11;101(19):7357–7362. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iacobuzio-Donahue CA. Epigenetic changes in cancer. Annu Rev Pathol. 2009;4:229–249. doi: 10.1146/annurev.pathol.3.121806.151442. [DOI] [PubMed] [Google Scholar]
- 34.Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet. 2009 Jan;10(1):32–42. doi: 10.1038/nrg2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y. Histone lysine demethylases: emerging roles in development, physiology and disease. Nat Rev Genet. 2007 Nov;8(11):829–833. doi: 10.1038/nrg2218. [DOI] [PubMed] [Google Scholar]
- 36.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008 Jul;40(7):897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rouhi A, Mager DL, Humphries RK, Kuchenbauer F. MiRNAs, epigenetics, and cancer. Mamm Genome. 2008 Aug;19(7–8):517–525. doi: 10.1007/s00335-008-9133-x. [DOI] [PubMed] [Google Scholar]
- 38.Chuang JC, Jones PA. Epigenetics and microRNAs. Pediatr Res. 2007 May;61(5 Pt 2):24R–29R. doi: 10.1203/pdr.0b013e3180457684. [DOI] [PubMed] [Google Scholar]
- 39.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004 Jul;5(7):522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 40.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004 Mar 2;101(9):2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs--the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009 Apr;9(4):293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 42.Peter ME. Let-7 and miR-200 microRNAs: guardians against pluripotency and cancer progression. Cell Cycle. 2009 Mar 15;8(6):843–852. doi: 10.4161/cc.8.6.7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saito Y, Jones PA. Epigenetic activation of tumor suppressor microRNAs in human cancer cells. Cell Cycle. 2006 Oct;5(19):2220–2222. doi: 10.4161/cc.5.19.3340. [DOI] [PubMed] [Google Scholar]
- 44.Paranjape T, Slack FJ, Weidhaas JB. MicroRNAs: tools for cancer diagnostics. Gut. 2009 Nov;58(11):1546–1554. doi: 10.1136/gut.2009.179531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat Rev Mol Cell Biol. 2010 Apr;11(4):252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 46.Kai ZS, Pasquinelli AE. MicroRNA assassins: factors that regulate the disappearance of miRNAs. Nat Struct Mol Biol. 2010 Jan;17(1):5–10. doi: 10.1038/nsmb.1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teodoridis JM, Strathdee G, Brown R. Epigenetic silencing mediated by CpG island methylation: potential as a therapeutic target and as a biomarker. Drug Resist Updat. 2004 Aug;7(4–5):267–278. doi: 10.1016/j.drup.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 48.Yoo CB, Jones PA. Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov. 2006 Jan;5(1):37–50. doi: 10.1038/nrd1930. [DOI] [PubMed] [Google Scholar]
- 49.Ross SA. Diet and DNA methylation interactions in cancer prevention. Ann N Y Acad Sci. 2003 Mar;983:197–207. doi: 10.1111/j.1749-6632.2003.tb05974.x. [DOI] [PubMed] [Google Scholar]
- 50.Hede K. Histone deacetylase inhibitors sit at crossroads of diet, aging, cancer. J Natl Cancer Inst. 2006 Mar 15;98(6):377–379. doi: 10.1093/jnci/djj120. [DOI] [PubMed] [Google Scholar]
- 51.Pogribny IP, Ross SA, Tryndyak VP, Pogribna M, Poirier LA, Karpinets TV. Histone H3 lysine 9 and H4 lysine 20 trimethylation and the expression of Suv4-20h2 and Suv-39h1 histone methyltransferases in hepatocarcinogenesis induced by methyl deficiency in rats. Carcinogenesis. 2006 Jun;27(6):1180–1186. doi: 10.1093/carcin/bgi364. [DOI] [PubMed] [Google Scholar]
- 52.Davis CD, Ross SA. Dietary components impact histone modifications and cancer risk. Nutr Rev. 2007 Feb;65(2):88–94. doi: 10.1111/j.1753-4887.2007.tb00285.x. [DOI] [PubMed] [Google Scholar]
- 53.Davis CD, Ross SA. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr Rev. 2008 Aug;66(8):477–482. doi: 10.1111/j.1753-4887.2008.00080.x. [DOI] [PubMed] [Google Scholar]
- 54.Poirier LA. Methyl group deficiency in hepatocarcinogenesis. Drug Metab Rev. 1994;26(1–2):185–199. doi: 10.3109/03602539409029790. [DOI] [PubMed] [Google Scholar]
- 55.Pogribny IP, Ross SA, Wise C, Pogribna M, Jones EA, Tryndyak VP, et al. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006 Jan 29;593(1–2):80–87. doi: 10.1016/j.mrfmmm.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 56.Davis CD, Uthus EO, Finley JW. Dietary selenium and arsenic affect DNA methylation in vitro in Caco-2 cells and in vivo in rat liver and colon. J Nutr. 2000 Dec;130(12):2903–2909. doi: 10.1093/jn/130.12.2903. [DOI] [PubMed] [Google Scholar]
- 57.Yen TT, Gill AM, Frigeri LG, Barsh GS, Wolff GL. Obesity, diabetes, and neoplasia in yellow A(vy)/- mice: ectopic expression of the agouti gene. FASEB J. 1994 May;8(8):479–488. doi: 10.1096/fasebj.8.8.8181666. [DOI] [PubMed] [Google Scholar]
- 58.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998 Aug;12(11):949–957. [PubMed] [Google Scholar]
- 59.Waterland RA, Jirtle RL. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol. 2003 Aug;23(15):5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waterland RA. Do maternal methyl supplements in mice affect DNA methylation of offspring? J Nutr. 2003 Jan;133(1):238. doi: 10.1093/jn/133.1.238. [DOI] [PubMed] [Google Scholar]
- 61.Blewitt ME, Vickaryous NK, Paldi A, Koseki H, Whitelaw E. Dynamic reprogramming of DNA methylation at an epigenetically sensitive allele in mice. PLoS Genet. 2006 Apr;2(4):e49. doi: 10.1371/journal.pgen.0020049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004 Aug;7(8):847–854. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 63.Weaver IC, Diorio J, Seckl JR, Szyf M, Meaney MJ. Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Ann N Y Acad Sci. 2004 Jun;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- 64.Weaver IC, Meaney MJ, Szyf M. Maternal care effects on the hippocampal transcriptome and anxiety-mediated behaviors in the offspring that are reversible in adulthood. Proc Natl Acad Sci U S A. 2006 Feb 28;103(9):3480–3485. doi: 10.1073/pnas.0507526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheldon CC, Finnegan EJ, Rouse DT, Tadege M, Bagnall DJ, Helliwell CA, et al. The control of flowering by vernalization. Curr Opin Plant Biol. 2000 Oct;3(5):418–422. doi: 10.1016/s1369-5266(00)00106-0. [DOI] [PubMed] [Google Scholar]
- 66.Sheldon CC, Rouse DT, Finnegan EJ, Peacock WJ, Dennis ES. The molecular basis of vernalization: the central role of FLOWERING LOCUS C (FLC) Proc Natl Acad Sci U S A. 2000 Mar 28;97(7):3753–3758. doi: 10.1073/pnas.060023597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Aggarwal BB, Shishodia S, Sandur SK, Pandey MK, Sethi G. Inflammation and cancer: how hot is the link? Biochem Pharmacol. 2006 Nov 30;72(11):1605–1621. doi: 10.1016/j.bcp.2006.06.029. [DOI] [PubMed] [Google Scholar]
- 68.Gunter MJ, Leitzmann MF. Obesity and colorectal cancer: epidemiology, mechanisms and candidate genes. J Nutr Biochem. 2006 Mar;17(3):145–156. doi: 10.1016/j.jnutbio.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 69.Hodge DR, Xiao W, Clausen PA, Heidecker G, Szyf M, Farrar WL. Interleukin-6 regulation of the human DNA methyltransferase (HDNMT) gene in human erythroleukemia cells. J Biol Chem. 2001 Oct 26;276(43):39508–39511. doi: 10.1074/jbc.C100343200. [DOI] [PubMed] [Google Scholar]
- 70.Hodge DR, Peng B, Cherry JC, Hurt EM, Fox SD, Kelley JA, et al. Interleukin 6 supports the maintenance of p53 tumor suppressor gene promoter methylation. Cancer Res. 2005 Jun 1;65(11):4673–4682. doi: 10.1158/0008-5472.CAN-04-3589. [DOI] [PubMed] [Google Scholar]
- 71.Issa JP. Epigenetic variation and human disease. J Nutr. 2002 Aug;132(8 Suppl):2388S–2392S. doi: 10.1093/jn/132.8.2388S. [DOI] [PubMed] [Google Scholar]
- 72.Miyamoto K, Ushijima T. Diagnostic and therapeutic applications of epigenetics. Jpn J Clin Oncol. 2005 Jun;35(6):293–301. doi: 10.1093/jjco/hyi088. [DOI] [PubMed] [Google Scholar]
- 73.Gilbert J, Gore SD, Herman JG, Carducci MA. The clinical application of targeting cancer through histone acetylation and hypomethylation. Clin Cancer Res. 2004 Jul 15;10(14):4589–4596. doi: 10.1158/1078-0432.CCR-03-0297. [DOI] [PubMed] [Google Scholar]
- 74.Horrobin DF. Are large clinical trials in rapidly lethal diseases usually unethical? Lancet. 2003 Feb 22;361(9358):695–697. doi: 10.1016/S0140-6736(03)12571-8. [DOI] [PubMed] [Google Scholar]
- 75.Komashko VM, Farnham PJ. 5-azacytidine treatment reorganizes genomic histone modification patterns. Epigenetics. 2010 Apr 4;5(3) doi: 10.4161/epi.5.3.11409. [DOI] [PubMed] [Google Scholar]
- 76.Lee BH, Yegnasubramanian S, Lin X, Nelson WG. Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem. 2005 Dec 9;280(49):40749–40756. doi: 10.1074/jbc.M505593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tada M, Imazeki F, Fukai K, Sakamoto A, Arai M, Mikata R, et al. Procaine inhibits the proliferation and DNA methylation in human hepatoma cells. Hepatol Int. 2007 Sep;1(3):355–364. doi: 10.1007/s12072-007-9014-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gao Z, Xu Z, Hung MS, Lin YC, Wang T, Gong M, et al. Promoter demethylation of WIF-1 by epigallocatechin-3-gallate in lung cancer cells. Anticancer Res. 2009 Jun;29(6):2025–2030. [PubMed] [Google Scholar]
- 79.Huang HC, Way TD, Lin CL, Lin JK. EGCG stabilizes p27kip1 in E2-stimulated MCF-7 cells through down-regulation of the Skp2 protein. Endocrinology. 2008 Dec;149(12):5972–5983. doi: 10.1210/en.2008-0408. [DOI] [PubMed] [Google Scholar]
- 80.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008 Feb 1;103(2):509–519. doi: 10.1002/jcb.21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gu B, Ding Q, Xia G, Fang Z. EGCG inhibits growth and induces apoptosis in renal cell carcinoma through TFPI-2 overexpression. Oncol Rep. 2009 Mar;21(3):635–640. [PubMed] [Google Scholar]
- 82.Kim SH, Kang HJ, Na H, Lee MO. Trichostatin A enhances acetylation as well as protein stability of ER alpha through induction of p300 protein. Breast Cancer Res. 2010 Apr 13;12(2):R22. doi: 10.1186/bcr2562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Choo QY, Ho PC, Tanaka Y, Lin HS. Histone deacetylase inhibitors MS-275 and SAHA induced growth arrest and suppressed lipopolysaccharide-stimulated NF-{kappa}B p65 nuclear accumulation in human rheumatoid arthritis synovial fibroblastic E11 cells. Rheumatology (Oxford) 2010 Apr 25; doi: 10.1093/rheumatology/keq108. [DOI] [PubMed] [Google Scholar]
- 84.Jones K, Nourse J, Corbett G, Gandhi MK. Sodium valproate in combination with ganciclovir induces lysis of EBV-infected lymphoma cells without impairing EBV-specific T-cell immunity. Int J Lab Hematol. 2010 Feb;32(1 Pt 1):e169–e174. doi: 10.1111/j.1751-553X.2008.01130.x. [DOI] [PubMed] [Google Scholar]
- 85.Hurtubise A, Bernstein ML, Momparler RL. Preclinical evaluation of the antineoplastic action of 5-aza-2'-deoxycytidine and different histone deacetylase inhibitors on human Ewing's sarcoma cells. Cancer Cell Int. 2008;8:16. doi: 10.1186/1475-2867-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zelent A, Waxman S, Carducci M, Wright J, Zweibel J, Gore SD. State of the translational science: summary of Baltimore workshop on gene re-expression as a therapeutic target in cancer January 2003. Clin Cancer Res. 2004 Jul 15;10(14):4622–4629. doi: 10.1158/1078-0432.CCR-1219-03. [DOI] [PubMed] [Google Scholar]
- 87.Sato N, Maitra A, Fukushima N, van Heek NT, Matsubayashi H, Iacobuzio-Donahue CA, et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res. 2003 Jul 15;63(14):4158–4166. [PubMed] [Google Scholar]
- 88.Costello JF, Plass C. Methylation matters. J Med Genet. 2001 May;38(5):285–303. doi: 10.1136/jmg.38.5.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sporn MB, Suh N. Chemoprevention: an essential approach to controlling cancer. Nat Rev Cancer. 2002 Jul;2(7):537–543. doi: 10.1038/nrc844. [DOI] [PubMed] [Google Scholar]
- 90.Yang CS, Landau JM, Huang MT, Newmark HL. Inhibition of carcinogenesis by dietary polyphenolic compounds. Annu Rev Nutr. 2001;21:381–406. doi: 10.1146/annurev.nutr.21.1.381. [DOI] [PubMed] [Google Scholar]
- 91.Singh UP, Singh N, Singh B, Hofseth LJ, Price BL, Nagarkatti M, et al. Resveratrol (trans-3, 5, 4'-trihydroxystilbene) induces SIRT1 and down-regulates NF-{kappa}B activation to abrogate DSS-induced colitis. J Pharmacol Exp Ther. 2009 Nov 30; doi: 10.1124/jpet.109.160838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cui X, Jin Y, Hofseth AB, Pena E, Habiger J, Chumanevich A, et al. Resveratrol suppresses colitis and colon cancer associated with colitis. Cancer Prev Res (Phila Pa) 2010 Apr;3(4):549–559. doi: 10.1158/1940-6207.CAPR-09-0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Singh UP, Singh NP, Singh B, Hofseth LJ, Price RL, Nagarkatti M, et al. Resveratrol (trans-3,5,4'-trihydroxystilbene) induces silent mating type information regulation-1 and down-regulates nuclear transcription factor-kappaB activation to abrogate dextran sulfate sodium-induced colitis. J Pharmacol Exp Ther. 2010 Mar;332(3):829–839. doi: 10.1124/jpet.109.160838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bravo L. Polyphenols: chemistry, dietary sources, metabolism, and nutritional significance. Nutr Rev. 1998 Nov;56(11):317–333. doi: 10.1111/j.1753-4887.1998.tb01670.x. [DOI] [PubMed] [Google Scholar]
- 95.Manson MM. Cancer prevention -- the potential for diet to modulate molecular signalling. Trends Mol Med. 2003 Jan;9(1):11–18. doi: 10.1016/s1471-4914(02)00002-3. [DOI] [PubMed] [Google Scholar]
- 96.Surh YJ. Cancer chemoprevention with dietary phytochemicals. Nat Rev Cancer. 2003 Oct;3(10):768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 97.Aggarwal BB, Shishodia S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem Pharmacol. 2006 May 14;71(10):1397–1421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 98.Shishodia S, Chaturvedi MM, Aggarwal BB. Role of curcumin in cancer therapy. Curr Probl Cancer. 2007 Jul;31(4):243–305. doi: 10.1016/j.currproblcancer.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 99.Russo GL. Ins and outs of dietary phytochemicals in cancer chemoprevention. Biochem Pharmacol. 2007 Aug 15;74(4):533–544. doi: 10.1016/j.bcp.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 100.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003 Nov 15;63(22):7563–7570. [PubMed] [Google Scholar]
- 101.Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005 Oct;68(4):1018–1030. doi: 10.1124/mol.104.008367. [DOI] [PubMed] [Google Scholar]
- 102.Navarro-Peran E, Cabezas-Herrera J, Campo LS, Rodriguez-Lopez JN. Effects of folate cycle disruption by the green tea polyphenol epigallocatechin-3-gallate. Int J Biochem Cell Biol. 2007;39(12):2215–2225. doi: 10.1016/j.biocel.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 103.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007 Jan;137(1 Suppl):223S–228S. doi: 10.1093/jn/137.1.223S. [DOI] [PubMed] [Google Scholar]
- 104.Kato K, Long NK, Makita H, Toida M, Yamashita T, Hatakeyama D, et al. Effects of green tea polyphenol on methylation status of RECK gene and cancer cell invasion in oral squamous cell carcinoma cells. Br J Cancer. 2008 Aug 19;99(4):647–654. doi: 10.1038/sj.bjc.6604521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Pandey M, Shukla S, Gupta S. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int J Cancer. 2009 Oct 23; doi: 10.1002/ijc.24988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Guilleret I, Benhattar J. Unusual distribution of DNA methylation within the hTERT CpG island in tissues and cell lines. Biochem Biophys Res Commun. 2004 Dec 17;325(3):1037–1043. doi: 10.1016/j.bbrc.2004.10.137. [DOI] [PubMed] [Google Scholar]
- 107.Quante M, Heeg S, von WA, Goessel G, Fulda C, Doebele M, et al. Differential transcriptional regulation of human telomerase in a cellular model representing important genetic alterations in esophageal squamous carcinogenesis. Carcinogenesis. 2005 Nov;26(11):1879–1889. doi: 10.1093/carcin/bgi153. [DOI] [PubMed] [Google Scholar]
- 108.Chuang JC, Yoo CB, Kwan JM, Li TW, Liang G, Yang AS, et al. Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2'-deoxycytidine. Mol Cancer Ther. 2005 Oct;4(10):1515–1520. doi: 10.1158/1535-7163.MCT-05-0172. [DOI] [PubMed] [Google Scholar]
- 109.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006 Mar 1;66(5):2794–2800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 110.Mittal A, Piyathilake C, Hara Y, Katiyar SK. Exceptionally high protection of photocarcinogenesis by topical application of (--)-epigallocatechin-3-gallate in hydrophilic cream in SKH-1 hairless mouse model: relationship to inhibition of UVB-induced global DNA hypomethylation. Neoplasia. 2003 Nov;5(6):555–565. doi: 10.1016/s1476-5586(03)80039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Morey Kinney SR, Zhang W, Pascual M, Greally JM, Gillard BM, Karasik E, et al. Lack of evidence for green tea polyphenols as DNA methylation inhibitors in murine prostate. Cancer Prev Res (Phila Pa) 2009 Dec;2(12):1065–1075. doi: 10.1158/1940-6207.CAPR-09-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yuasa Y, Nagasaki H, Akiyama Y, Hashimoto Y, Takizawa T, Kojima K, et al. DNA methylation status is inversely correlated with green tea intake and physical activity in gastric cancer patients. Int J Cancer. 2009 Jun 1;124(11):2677–2682. doi: 10.1002/ijc.24231. [DOI] [PubMed] [Google Scholar]
- 113.Yuasa Y, Nagasaki H, Akiyama Y, Sakai H, Nakajima T, Ohkura Y, et al. Relationship between CDX2 gene methylation and dietary factors in gastric cancer patients. Carcinogenesis. 2005 Jan;26(1):193–200. doi: 10.1093/carcin/bgh304. [DOI] [PubMed] [Google Scholar]
- 114.Tsao AS, Liu D, Martin J, Tang XM, Lee JJ, El-Naggar AK, et al. Phase II randomized, placebo-controlled trial of green tea extract in patients with high-risk oral premalignant lesions. Cancer Prev Res (Phila Pa) 2009 Nov;2(11):931–941. doi: 10.1158/1940-6207.CAPR-09-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, Yang CS. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005 Oct 1;11(19 Pt 1):7033–7041. doi: 10.1158/1078-0432.CCR-05-0406. [DOI] [PubMed] [Google Scholar]
- 116.Li Y, Liu L, Andrews LG, Tollefsbol TO. Genistein depletes telomerase activity through cross-talk between genetic and epigenetic mechanisms. Int J Cancer. 2009 Jul 15;125(2):286–296. doi: 10.1002/ijc.24398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008 Jan;49(1):36–45. doi: 10.1002/em.20363. [DOI] [PubMed] [Google Scholar]
- 118.Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009 Apr;30(4):662–670. doi: 10.1093/carcin/bgp042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Majid S, Dar AA, Shahryari V, Hirata H, Ahmad A, Saini S, et al. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 2009 Nov 2; doi: 10.1002/cncr.24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Day JK, Bauer AM, desBordes C, Zhuang Y, Kim BE, Newton LG, et al. Genistein alters methylation patterns in mice. J Nutr. 2002 Aug;132(8 Suppl):2419S–2423S. doi: 10.1093/jn/132.8.2419S. [DOI] [PubMed] [Google Scholar]
- 121.Guerrero-Bosagna CM, Sabat P, Valdovinos FS, Valladares LE, Clark SJ. Epigenetic and phenotypic changes result from a continuous pre and post natal dietary exposure to phytoestrogens in an experimental population of mice. BMC Physiol. 2008;8:17. doi: 10.1186/1472-6793-8-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tang WY, Newbold R, Mardilovich K, Jefferson W, Cheng RY, Medvedovic M, et al. Persistent hypomethylation in the promoter of nucleosomal binding protein 1 (Nsbp1) correlates with overexpression of Nsbp1 in mouse uteri neonatally exposed to diethylstilbestrol or genistein. Endocrinology. 2008 Dec;149(12):5922–5931. doi: 10.1210/en.2008-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Qin W, Zhu W, Shi H, Hewett JE, Ruhlen RL, MacDonald RS, et al. Soy isoflavones have an antiestrogenic effect and alter mammary promoter hypermethylation in healthy premenopausal women. Nutr Cancer. 2009;61(2):238–244. doi: 10.1080/01635580802404196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chalabi N, Satih S, Delort L, Bignon YJ, Bernard-Gallon DJ. Expression profiling by whole-genome microarray hybridization reveals differential gene expression in breast cancer cell lines after lycopene exposure. Biochim Biophys Acta. 2007 Feb;1769(2):124–130. doi: 10.1016/j.bbaexp.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 125.Chalabi N, Delort L, Le CL, Satih S, Bignon YJ, Bernard-Gallon D. Gene signature of breast cancer cell lines treated with lycopene. Pharmacogenomics. 2006 Jul;7(5):663–672. doi: 10.2217/14622416.7.5.663. [DOI] [PubMed] [Google Scholar]
- 126.Lee WJ, Zhu BT. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006 Feb;27(2):269–277. doi: 10.1093/carcin/bgi206. [DOI] [PubMed] [Google Scholar]
- 127.Traka M, Gasper AV, Smith JA, Hawkey CJ, Bao Y, Mithen RF. Transcriptome analysis of human colon Caco-2 cells exposed to sulforaphane. J Nutr. 2005 Aug;135(8):1865–1872. doi: 10.1093/jn/135.8.1865. [DOI] [PubMed] [Google Scholar]
- 128.Wang LG, Beklemisheva A, Liu XM, Ferrari AC, Feng J, Chiao JW. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol Carcinog. 2007 Jan;46(1):24–31. doi: 10.1002/mc.20258. [DOI] [PubMed] [Google Scholar]
- 129.Wilkinson JT, Morse MA, Kresty LA, Stoner GD. Effect of alkyl chain length on inhibition of N-nitrosomethylbenzylamine-induced esophageal tumorigenesis and DNA methylation by isothiocyanates. Carcinogenesis. 1995 May;16(5):1011–1015. doi: 10.1093/carcin/16.5.1011. [DOI] [PubMed] [Google Scholar]
- 130.Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009 Feb 1;19(3):706–709. doi: 10.1016/j.bmcl.2008.12.041. [DOI] [PubMed] [Google Scholar]
- 131.Kuck D, Singh N, Lyko F, Medina-Franco JL. Novel and selective DNA methyltransferase inhibitors: Docking-based virtual screening and experimental evaluation. Bioorg Med Chem. 2009 Nov 27; doi: 10.1016/j.bmc.2009.11.050. [DOI] [PubMed] [Google Scholar]
- 132.Paluszczak J, Krajka-Kuzniak V, Baer-Dubowska W. The effect of dietary polyphenols on the epigenetic regulation of gene expression in MCF7 breast cancer cells. Toxicol Lett. 2009 Oct 17; doi: 10.1016/j.toxlet.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 133.Stefanska B, Rudnicka K, Bednarek A, Fabianowska-Majewska K. Hypomethylation and induction of retinoic acid receptor beta 2 by concurrent action of adenosine analogues and natural compounds in breast cancer cells. Eur J Pharmacol. 2010 Jul 25;638(1–3):47–53. doi: 10.1016/j.ejphar.2010.04.032. [DOI] [PubMed] [Google Scholar]
- 134.Singh N, Misra K. Computational screening of molecular targets in Plasmodium for novel non resistant anti-malarial drugs. Bioinformation. 2009;3(6):255–262. doi: 10.6026/97320630003255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, et al. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006 Mar;2(2):169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]
- 136.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, et al. Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J Biol Chem. 2004 Dec 3;279(49):51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 137.Kang J, Chen J, Shi Y, Jia J, Zhang Y. Curcumin-induced histone hypoacetylation: the role of reactive oxygen species. Biochem Pharmacol. 2005 Apr 15;69(8):1205–1213. doi: 10.1016/j.bcp.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 138.Sbardella G, Castellano S, Vicidomini C, Rotili D, Nebbioso A, Miceli M, et al. Identification of long chain alkylidenemalonates as novel small molecule modulators of histone acetyltransferases. Bioorg Med Chem Lett. 2008 May 1;18(9):2788–2792. doi: 10.1016/j.bmcl.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 139.Mai A, Rotili D, Tarantino D, Ornaghi P, Tosi F, Vicidomini C, et al. Small-molecule inhibitors of histone acetyltransferase activity: identification and biological properties. J Med Chem. 2006 Nov 16;49(23):6897–6907. doi: 10.1021/jm060601m. [DOI] [PubMed] [Google Scholar]
- 140.Kutluay SB, Doroghazi J, Roemer ME, Triezenberg SJ. Curcumin inhibits herpes simplex virus immediate-early gene expression by a mechanism independent of p300/CBP histone acetyltransferase activity. Virology. 2008 Apr 10;373(2):239–247. doi: 10.1016/j.virol.2007.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Araki Y, Fann M, Wersto R, Weng NP. Histone acetylation facilitates rapid and robust memory CD8 T cell response through differential expression of effector molecules (eomesodermin and its targets: perforin and granzyme B) J Immunol. 2008 Jun 15;180(12):8102–8108. doi: 10.4049/jimmunol.180.12.8102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kang SK, Cha SH, Jeon HG. Curcumin-induced histone hypoacetylation enhances caspase-3-dependent glioma cell death and neurogenesis of neural progenitor cells. Stem Cells Dev. 2006 Apr;15(2):165–174. doi: 10.1089/scd.2006.15.165. [DOI] [PubMed] [Google Scholar]
- 143.Cui L, Miao J, Cui L. Cytotoxic effect of curcumin on malaria parasite Plasmodium falciparum: inhibition of histone acetylation and generation of reactive oxygen species. Antimicrob Agents Chemother. 2007 Feb;51(2):488–494. doi: 10.1128/AAC.01238-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Cui L, Miao J, Furuya T, Li X, Su XZ, Cui L. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot Cell. 2007 Jul;6(7):1219–1227. doi: 10.1128/EC.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sng JC, Taniura H, Yoneda Y. Histone modifications in kainate-induced status epilepticus. Eur J Neurosci. 2006 Mar;23(5):1269–1282. doi: 10.1111/j.1460-9568.2006.04641.x. [DOI] [PubMed] [Google Scholar]
- 146.Chiu J, Khan ZA, Farhangkhoee H, Chakrabarti S. Curcumin prevents diabetes-associated abnormalities in the kidneys by inhibiting p300 and nuclear factor-kappaB. Nutrition. 2009 Sep;25(9):964–972. doi: 10.1016/j.nut.2008.12.007. [DOI] [PubMed] [Google Scholar]
- 147.Tikoo K, Meena RL, Kabra DG, Gaikwad AB. Change in post-translational modifications of histone H3, heat-shock protein-27 and MAP kinase p38 expression by curcumin in streptozotocin-induced type I diabetic nephropathy. Br J Pharmacol. 2008 Mar;153(6):1225–1231. doi: 10.1038/sj.bjp.0707666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Li HL, Liu C, de CG, Ouzounian M, Sun M, Wang AB, et al. Curcumin prevents and reverses murine cardiac hypertrophy. J Clin Invest. 2008 Mar;118(3):879–893. doi: 10.1172/JCI32865. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 149.Morimoto T, Sunagawa Y, Kawamura T, Takaya T, Wada H, Nagasawa A, et al. The dietary compound curcumin inhibits p300 histone acetyltransferase activity and prevents heart failure in rats. J Clin Invest. 2008 Mar;118(3):868–878. doi: 10.1172/JCI33160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Liu HL, Chen Y, Cui GH, Zhou JF. Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol Sin. 2005 May;26(5):603–609. doi: 10.1111/j.1745-7254.2005.00081.x. [DOI] [PubMed] [Google Scholar]
- 151.Chen Y, Shu W, Chen W, Wu Q, Liu H, Cui G. Curcumin, both histone deacetylase and p300/CBP-specific inhibitor, represses the activity of nuclear factor kappa B and Notch 1 in Raji cells. Basic Clin Pharmacol Toxicol. 2007 Dec;101(6):427–433. doi: 10.1111/j.1742-7843.2007.00142.x. [DOI] [PubMed] [Google Scholar]
- 152.Bora-Tatar G, Dayangac-Erden D, Demir AS, Dalkara S, Yelekci K, Erdem-Yurter H. Molecular modifications on carboxylic acid derivatives as potent histone deacetylase inhibitors: Activity and docking studies. Bioorg Med Chem. 2009 Jul 15;17(14):5219–5228. doi: 10.1016/j.bmc.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 153.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. Small molecule modulators of histone acetyltransferase p300. J Biol Chem. 2003 May 23;278(21):19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 154.Sun Y, Jiang X, Chen S, Price BD. Inhibition of histone acetyltransferase activity by anacardic acid sensitizes tumor cells to ionizing radiation. FEBS Lett. 2006 Aug 7;580(18):4353–4356. doi: 10.1016/j.febslet.2006.06.092. [DOI] [PubMed] [Google Scholar]
- 155.Eliseeva ED, Valkov V, Jung M, Jung MO. Characterization of novel inhibitors of histone acetyltransferases. Mol Cancer Ther. 2007 Sep;6(9):2391–2398. doi: 10.1158/1535-7163.MCT-07-0159. [DOI] [PubMed] [Google Scholar]
- 156.Chandregowda V, Kush A, Reddy GC. Synthesis of benzamide derivatives of anacardic acid and their cytotoxic activity. Eur J Med Chem. 2009 Jun;44(6):2711–2719. doi: 10.1016/j.ejmech.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 157.Souto JA, Conte M, Alvarez R, Nebbioso A, Carafa V, Altucci L, et al. Synthesis of benzamides related to anacardic acid and their histone acetyltransferase (HAT) inhibitory activities. ChemMedChem. 2008 Sep;3(9):1435–1442. doi: 10.1002/cmdc.200800096. [DOI] [PubMed] [Google Scholar]
- 158.Sung B, Pandey MK, Ahn KS, Yi T, Chaturvedi MM, Liu M, et al. Anacardic acid (6-nonadecyl salicylic acid), an inhibitor of histone acetyltransferase, suppresses expression of nuclear factor-kappaB-regulated gene products involved in cell survival, proliferation, invasion, and inflammation through inhibition of the inhibitory subunit of nuclear factor-kappaBalpha kinase, leading to potentiation of apoptosis. Blood. 2008 May 15;111(10):4880–4891. doi: 10.1182/blood-2007-10-117994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kim MK, Shin JM, Eun HC, Chung JH. The role of p300 histone acetyltransferase in UV-induced histone modifications and MMP-1 gene transcription. PLoS One. 2009;4(3):e4864. doi: 10.1371/journal.pone.0004864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cui L, Miao J, Furuya T, Fan Q, Li X, Rathod PK, et al. Histone acetyltransferase inhibitor anacardic acid causes changes in global gene expression during in vitro Plasmodium falciparum development. Eukaryot Cell. 2008 Jul;7(7):1200–1210. doi: 10.1128/EC.00063-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Balasubramanyam K, Altaf M, Varier RA, Swaminathan V, Ravindran A, Sadhale PP, et al. Polyisoprenylated benzophenone, garcinol, a natural histone acetyltransferase inhibitor, represses chromatin transcription and alters global gene expression. J Biol Chem. 2004 Aug 6;279(32):33716–33726. doi: 10.1074/jbc.M402839200. [DOI] [PubMed] [Google Scholar]
- 162.Arif M, Pradhan SK, Thanuja GR, Vedamurthy BM, Agrawal S, Dasgupta D, et al. Mechanism of p300 specific histone acetyltransferase inhibition by small molecules. J Med Chem. 2009 Jan 22;52(2):267–277. doi: 10.1021/jm800657z. [DOI] [PubMed] [Google Scholar]
- 163.Arif M, Kumar GV, Narayana C, Kundu TK. Autoacetylation induced specific structural changes in histone acetyltransferase domain of p300: probed by surface enhanced Raman spectroscopy. J Phys Chem B. 2007 Oct 18;111(41):11877–11879. doi: 10.1021/jp0762931. [DOI] [PubMed] [Google Scholar]
- 164.Mantelingu K, Reddy BA, Swaminathan V, Kishore AH, Siddappa NB, Kumar GV, et al. Specific inhibition of p300-HAT alters global gene expression and represses HIV replication. Chem Biol. 2007 Jun;14(6):645–657. doi: 10.1016/j.chembiol.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 165.Choi KC, Jung MG, Lee YH, Yoon JC, Kwon SH, Kang HB, et al. Epigallocatechin-3-gallate, a histone acetyltransferase inhibitor, inhibits EBV-induced B lymphocyte transformation via suppression of RelA acetylation. Cancer Res. 2009 Jan 15;69(2):583–592. doi: 10.1158/0008-5472.CAN-08-2442. [DOI] [PubMed] [Google Scholar]
- 166.Nair S, Hebbar V, Shen G, Gopalakrishnan A, Khor TO, Yu S, et al. Synergistic effects of a combination of dietary factors sulforaphane and (-) epigallocatechin-3-gallate in HT-29 AP-1 human colon carcinoma cells. Pharm Res. 2008 Feb;25(2):387–399. doi: 10.1007/s11095-007-9364-7. [DOI] [PubMed] [Google Scholar]
- 167.Balasubramanian S, Adhikary G, Eckert RL. The Bmi-1 Polycomb Protein Antagonizes the (-)- Epigallocatechin-3-Gallate Dependent Suppression of Skin Cancer Cell Survival. Carcinogenesis. 2009 Dec 16; doi: 10.1093/carcin/bgp314. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 168.Murugan RS, Vinothini G, Hara Y, Nagini S. Black tea polyphenols target matrix metalloproteinases, RECK, proangiogenic molecules and histone deacetylase in a rat hepatocarcinogenesis model. Anticancer Res. 2009 Jun;29(6):2301–2305. [PubMed] [Google Scholar]
- 169.Cosio BG, Mann B, Ito K, Jazrawi E, Barnes PJ, Chung KF, et al. Histone acetylase and deacetylase activity in alveolar macrophages and blood mononocytes in asthma. Am J Respir Crit Care Med. 2004 Jul 15;170(2):141–147. doi: 10.1164/rccm.200305-659OC. [DOI] [PubMed] [Google Scholar]
- 170.Cosio BG, Iglesias A, Rios A, Noguera A, Sala E, Ito K, et al. Low-dose theophylline enhances the anti-inflammatory effects of steroids during exacerbations of COPD. Thorax. 2009 May;64(5):424–429. doi: 10.1136/thx.2008.103432. [DOI] [PubMed] [Google Scholar]
- 171.Cosio BG, Tsaprouni L, Ito K, Jazrawi E, Adcock IM, Barnes PJ. Theophylline restores histone deacetylase activity and steroid responses in COPD macrophages. J Exp Med. 2004 Sep 6;200(5):689–695. doi: 10.1084/jem.20040416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ito K, Lim S, Caramori G, Cosio B, Chung KF, Adcock IM, et al. A molecular mechanism of action of theophylline: Induction of histone deacetylase activity to decrease inflammatory gene expression. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8921–8926. doi: 10.1073/pnas.132556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lea MA, Randolph VM, Patel M. Increased acetylation of histones induced by diallyl disulfide and structurally related molecules. Int J Oncol. 1999 Aug;15(2):347–352. doi: 10.3892/ijo.15.2.347. [DOI] [PubMed] [Google Scholar]
- 174.Lea MA, Randolph VM, Lee JE, desBordes C. Induction of histone acetylation in mouse erythroleukemia cells by some organosulfur compounds including allyl isothiocyanate. Int J Cancer. 2001 Jun 15;92(6):784–789. doi: 10.1002/ijc.1277. [DOI] [PubMed] [Google Scholar]
- 175.Lea MA, Randolph VM. Induction of histone acetylation in rat liver and hepatoma by organosulfur compounds including diallyl disulfide. Anticancer Res. 2001 Jul;21(4A):2841–2845. [PubMed] [Google Scholar]
- 176.Lea MA, Rasheed M, Randolph VM, Khan F, Shareef A, desBordes C. Induction of histone acetylation and inhibition of growth of mouse erythroleukemia cells by S-allylmercaptocysteine. Nutr Cancer. 2002;43(1):90–102. doi: 10.1207/S15327914NC431_11. [DOI] [PubMed] [Google Scholar]
- 177.Druesne N, Pagniez A, Mayeur C, Thomas M, Cherbuy C, Duee PH, et al. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004 Jul;25(7):1227–1236. doi: 10.1093/carcin/bgh123. [DOI] [PubMed] [Google Scholar]
- 178.Nian H, Delage B, Pinto JT, Dashwood RH. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis. 2008 Sep;29(9):1816–1824. doi: 10.1093/carcin/bgn165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Druesne-Pecollo N, Pagniez A, Thomas M, Cherbuy C, Duee PH, Martel P, et al. Diallyl disulfide increases CDKN1A promoter-associated histone acetylation in human colon tumor cell lines. J Agric Food Chem. 2006 Oct 4;54(20):7503–7507. doi: 10.1021/jf061369w. [DOI] [PubMed] [Google Scholar]
- 180.Druesne-Pecollo N, Chaumontet C, Pagniez A, Vaugelade P, Bruneau A, Thomas M, et al. In vivo treatment by diallyl disulfide increases histone acetylation in rat colonocytes. Biochem Biophys Res Commun. 2007 Mar 2;354(1):140–147. doi: 10.1016/j.bbrc.2006.12.158. [DOI] [PubMed] [Google Scholar]
- 181.Zhao J, Huang WG, He J, Tan H, Liao QJ, Su Q. Diallyl disulfide suppresses growth of HL-60 cell through increasing histone acetylation and p21WAF1 expression in vivo and in vitro. Acta Pharmacol Sin. 2006 Nov;27(11):1459–1466. doi: 10.1111/j.1745-7254.2006.00433.x. [DOI] [PubMed] [Google Scholar]
- 182.Arunkumar A, Vijayababu MR, Gunadharini N, Krishnamoorthy G, Arunakaran J. Induction of apoptosis and histone hyperacetylation by diallyl disulfide in prostate cancer cell line PC-3. Cancer Lett. 2007 Jun 18;251(1):59–67. doi: 10.1016/j.canlet.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 183.Lee JH, Kim KA, Kwon KB, Kim EK, Lee YR, Song MY, et al. Diallyl disulfide accelerates adipogenesis in 3T3-L1 cells. Int J Mol Med. 2007 Jul;20(1):59–64. [PubMed] [Google Scholar]
- 184.Chu Q, Ling MT, Feng H, Cheung HW, Tsao SW, Wang X, et al. A novel anticancer effect of garlic derivatives: inhibition of cancer cell invasion through restoration of E-cadherin expression. Carcinogenesis. 2006 Nov;27(11):2180–2189. doi: 10.1093/carcin/bgl054. [DOI] [PubMed] [Google Scholar]
- 185.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004 Aug 15;64(16):5767–5774. doi: 10.1158/0008-5472.CAN-04-1326. [DOI] [PubMed] [Google Scholar]
- 186.Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006 Apr;27(4):811–819. doi: 10.1093/carcin/bgi265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Myzak MC, Tong P, Dashwood WM, Dashwood RH, Ho E. Sulforaphane retards the growth of human PC-3 xenografts and inhibits HDAC activity in human subjects. Exp Biol Med (Maywood ) 2007 Feb;232(2):227–234. [PMC free article] [PubMed] [Google Scholar]
- 188.Pledgie-Tracy A, Sobolewski MD, Davidson NE. Sulforaphane induces cell type-specific apoptosis in human breast cancer cell lines. Mol Cancer Ther. 2007 Mar;6(3):1013–1021. doi: 10.1158/1535-7163.MCT-06-0494. [DOI] [PubMed] [Google Scholar]
- 189.Myzak MC, Dashwood WM, Orner GA, Ho E, Dashwood RH. Sulforaphane inhibits histone deacetylase in vivo and suppresses tumorigenesis in Apc-minus mice. FASEB J. 2006 Mar;20(3):506–508. doi: 10.1096/fj.05-4785fje. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hu R, Khor TO, Shen G, Jeong WS, Hebbar V, Chen C, et al. Cancer chemoprevention of intestinal polyposis in ApcMin/+ mice by sulforaphane, a natural product derived from cruciferous vegetable. Carcinogenesis. 2006 Oct;27(10):2038–2046. doi: 10.1093/carcin/bgl049. [DOI] [PubMed] [Google Scholar]
- 191.Schwab M, Reynders V, Loitsch S, Steinhilber D, Schroder O, Stein J. The dietary histone deacetylase inhibitor sulforaphane induces human beta-defensin-2 in intestinal epithelial cells. Immunology. 2008 Oct;125(2):241–251. doi: 10.1111/j.1365-2567.2008.02834.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Fang MZ, Jin Z, Wang Y, Liao J, Yang GY, Wang LD, et al. Promoter hypermethylation and inactivation of O(6)-methylguanine-DNA methyltransferase in esophageal squamous cell carcinomas and its reactivation in cell lines. Int J Oncol. 2005 Mar;26(3):615–622. [PubMed] [Google Scholar]
- 193.Ma X, Fang Y, Beklemisheva A, Dai W, Feng J, Ahmed T, et al. Phenylhexyl isothiocyanate inhibits histone deacetylases and remodels chromatins to induce growth arrest in human leukemia cells. Int J Oncol. 2006 May;28(5):1287–1293. [PubMed] [Google Scholar]
- 194.Beklemisheva AA, Fang Y, Feng J, Ma X, Dai W, Chiao JW. Epigenetic mechanism of growth inhibition induced by phenylhexyl isothiocyanate in prostate cancer cells. Anticancer Res. 2006 Mar;26(2A):1225–1230. [PubMed] [Google Scholar]
- 195.Wang LG, Liu XM, Fang Y, Dai W, Chiao FB, Puccio GM, et al. De-repression of the p21 promoter in prostate cancer cells by an isothiocyanate via inhibition of HDACs and c-Myc. Int J Oncol. 2008 Aug;33(2):375–380. [PubMed] [Google Scholar]
- 196.Bhatnagar N, Li X, Chen Y, Zhou X, Garrett SH, Guo B. 3,3'-diindolylmethane enhances the efficacy of butyrate in colon cancer prevention through down-regulation of survivin. Cancer Prev Res (Phila Pa) 2009 Jun;2(6):581–589. doi: 10.1158/1940-6207.CAPR-08-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Li Y, Li X, Guo B. Chemopreventive agent 3,3'-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer Res. 2010 Jan 15;70(2):646–654. doi: 10.1158/0008-5472.CAN-09-1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Degner SC, Papoutsis AJ, Selmin O, Romagnolo DF. Targeting of aryl hydrocarbon receptor-mediated activation of cyclooxygenase-2 expression by the indole-3-carbinol metabolite 3,3'-diindolylmethane in breast cancer cells. J Nutr. 2009 Jan;139(1):26–32. doi: 10.3945/jn.108.099259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hong T, Nakagawa T, Pan W, Kim MY, Kraus WL, Ikehara T, et al. Isoflavones stimulate estrogen receptor-mediated core histone acetylation. Biochem Biophys Res Commun. 2004 Apr 23;317(1):259–264. doi: 10.1016/j.bbrc.2004.03.041. [DOI] [PubMed] [Google Scholar]
- 200.Majid S, Kikuno N, Nelles J, Noonan E, Tanaka Y, Kawamoto K, et al. Genistein induces the p21WAF1/CIP1 and p16INK4a tumor suppressor genes in prostate cancer cells by epigenetic mechanisms involving active chromatin modification. Cancer Res. 2008 Apr 15;68(8):2736–2744. doi: 10.1158/0008-5472.CAN-07-2290. [DOI] [PubMed] [Google Scholar]
- 201.Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, et al. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008 Aug 1;123(3):552–560. doi: 10.1002/ijc.23590. [DOI] [PubMed] [Google Scholar]
- 202.Artaud-Wild SM, Connor SL, Sexton G, Connor WE. Differences in coronary mortality can be explained by differences in cholesterol and saturated fat intakes in 40 countries but not in France and Finland. A paradox. Circulation. 1993 Dec;88(6):2771–2779. doi: 10.1161/01.cir.88.6.2771. [DOI] [PubMed] [Google Scholar]
- 203.Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003 Sep 11;425(6954):191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 204.Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004 Aug 5;430(7000):686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 205.Wang RH, Sengupta K, Li C, Kim HS, Cao L, Xiao C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008 Oct 7;14(4):312–323. doi: 10.1016/j.ccr.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gracia-Sancho J, Villarreal G, Jr, Zhang Y, Garcia-Cardena G. Activation of SIRT1 by resveratrol induces KLF2 expression conferring an endothelial vasoprotective phenotype. Cardiovasc Res. 2010 Feb 1;85(3):514–519. doi: 10.1093/cvr/cvp337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sulaiman M, Matta MJ, Sundaresan NR, Gupta MP, Periasamy M, Gupta M. Resveratrol, an activator of SIRT1 up-regulates sarcoplasmic calcium ATPase and improves cardiac function in diabetic cardiomyopathy. Am J Physiol Heart Circ Physiol. 2009 Dec 11; doi: 10.1152/ajpheart.00418.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Beher D, Wu J, Cumine S, Kim KW, Lu SC, Atangan L, et al. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem Biol Drug Des. 2009 Dec;74(6):619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 209.Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005 Apr 29;280(17):17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 210.Kaeberlein M, McDonagh T, Heltweg B, Hixon J, Westman EA, Caldwell SD, et al. Substrate-specific activation of sirtuins by resveratrol. J Biol Chem. 2005 Apr 29;280(17):17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 211.Malik R, Kashyap A, Bansal K, Sharma P, Rayasam GV, Davis JA, et al. Comparative deacetylase activity of wild type and mutants of SIRT1. Biochem Biophys Res Commun. 2009 Nov 26; doi: 10.1016/j.bbrc.2009.11.130. [DOI] [PubMed] [Google Scholar]
- 212.Boily G, He XH, Pearce B, Jardine K, McBurney MW. SirT1-null mice develop tumors at normal rates but are poorly protected by resveratrol. Oncogene. 2009 Aug 13;28(32):2882–2893. doi: 10.1038/onc.2009.147. [DOI] [PubMed] [Google Scholar]
- 213.Wang RH, Zheng Y, Kim HS, Xu X, Cao L, Luhasen T, et al. Interplay among BRCA1, SIRT1, and Survivin during BRCA1-associated tumorigenesis. Mol Cell. 2008 Oct 10;32(1):11–20. doi: 10.1016/j.molcel.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Kai L, Samuel SK, Levenson AS. Resveratrol enhances p53 acetylation and apoptosis in prostate cancer by inhibiting MTA1/NuRD complex. Int J Cancer. 2010 Apr 1;126(7):1538–1548. doi: 10.1002/ijc.24928. [DOI] [PubMed] [Google Scholar]
- 215.Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006 Dec 15;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 216.Allard JS, Perez E, Zou S, de CR. Dietary activators of Sirt1. Mol Cell Endocrinol. 2009 Feb 5;299(1):58–63. doi: 10.1016/j.mce.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Knutson MD, Leeuwenburgh C. Resveratrol and novel potent activators of SIRT1: effects on aging and age-related diseases. Nutr Rev. 2008 Oct;66(10):591–596. doi: 10.1111/j.1753-4887.2008.00109.x. [DOI] [PubMed] [Google Scholar]
- 218.Chaudhary N, Pfluger PT. Metabolic benefits from Sirt1 and Sirt1 activators. Curr Opin Clin Nutr Metab Care. 2009 Jul;12(4):431–437. doi: 10.1097/MCO.0b013e32832cdaae. [DOI] [PubMed] [Google Scholar]
- 219.Ruiz PA, Braune A, Holzlwimmer G, Quintanilla-Fend L, Haller D. Quercetin inhibits TNF-induced NF-kappaB transcription factor recruitment to proinflammatory gene promoters in murine intestinal epithelial cells. J Nutr. 2007 May;137(5):1208–1215. doi: 10.1093/jn/137.5.1208. [DOI] [PubMed] [Google Scholar]
- 220.Olaharski AJ, Rine J, Marshall BL, Babiarz J, Zhang L, Verdin E, et al. The flavoring agent dihydrocoumarin reverses epigenetic silencing and inhibits sirtuin deacetylases. PLoS Genet. 2005 Dec;1(6):e77. doi: 10.1371/journal.pgen.0010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Selvi BR, Pradhan SK, Shandilya J, Das C, Sailaja BS, Shankar GN, et al. Sanguinarine interacts with chromatin, modulates epigenetic modifications, and transcription in the context of chromatin. Chem Biol. 2009 Feb 27;16(2):203–216. doi: 10.1016/j.chembiol.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 222.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008 Mar;7(3):464–473. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 223.Ali S, Ahmad A, Banerjee S, Padhye S, Dominiak K, Schaffert JM, et al. Gemcitabine Sensitivity Can Be Induced in Pancreatic Cancer Cells through Modulation of miR-200 and miR-21 Expression by Curcumin or Its Analogue CDF. Cancer Res. 2010 Apr 13; doi: 10.1158/0008-5472.CAN-09-4598. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 224.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2009 Mar 5; doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 225.Li Y, Vandenboom TG, Kong D, Wang Z, Ali S, Philip PA, et al. Up-regulation of miR-200 and let-7 by Natural Agents Leads to the Reversal of Epithelial-to-Mesenchymal Transition in Gemcitabine-Resistant Pancreatic Cancer Cells. Cancer Res. 2009 Aug 4; doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Li Y, Vandenboom TG, Wang Z, Kong D, Ali S, Philip PA, et al. miR-146a Suppresses Invasion of Pancreatic Cancer Cells. Cancer Res. 2010 Feb 2; doi: 10.1158/0008-5472.CAN-09-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sun Q, Cong R, Yan H, Gu H, Zeng Y, Liu N, et al. Genistein inhibits growth of human uveal melanoma cells and affects microRNA-27a and target gene expression. Oncol Rep. 2009 Sep;22(3):563–567. doi: 10.3892/or_00000472. [DOI] [PubMed] [Google Scholar]
- 228.Parker LP, Taylor DD, Kesterson J, Metzinger DS, Gercel-Taylor C. Modulation of microRNA associated with ovarian cancer cells by genistein. Eur J Gynaecol Oncol. 2009;30(6):616–621. [PubMed] [Google Scholar]
- 229.Izzotti A, Calin GA, Steele VE, Cartiglia C, Longobardi M, Croce CM, et al. Chemoprevention of cigarette smoke-induced alterations of MicroRNA expression in rat lungs. Cancer Prev Res (Phila Pa) 2010 Jan;3(1):62–72. doi: 10.1158/1940-6207.CAPR-09-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Vandegehuchte MB, Lemiere F, Vanhaecke L, Vanden Berghe W, Janssen CR. Direct and transgenerational impact on Daphnia magna of chemicals with a known effect on DNA methylation. Comp Biochem Physiol C Toxicol Pharmacol. 2009 Dec 2; doi: 10.1016/j.cbpc.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 231.Volate SR, Muga SJ, Issa AY, Nitcheva D, Smith T, Wargovich MJ. Epigenetic modulation of the retinoid X receptor alpha by green tea in the azoxymethane-Apc(Min/+) mouse model of intestinal cancer. Mol Carcinog. 2009 Apr 17; doi: 10.1002/mc.20542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Spurling CC, Suhl JA, Boucher N, Nelson CE, Rosenberg DW, Giardina C. The short chain fatty acid butyrate induces promoter demethylation and reactivation of RARbeta2 in colon cancer cells. Nutr Cancer. 2008;60(5):692–702. doi: 10.1080/01635580802008278. [DOI] [PubMed] [Google Scholar]
- 233.Tan S, Wang C, Lu C, Zhao B, Cui Y, Shi X, et al. Quercetin is able to demethylate the p16INK4a gene promoter. Chemotherapy. 2009;55(1):6–10. doi: 10.1159/000166383. [DOI] [PubMed] [Google Scholar]
- 234.Chung IM, Kim MY, Park WH, Moon HI. Histone deacetylase inhibitors from the rhizomes of Zingiber zerumbet. Pharmazie. 2008 Oct;63(10):774–776. [PubMed] [Google Scholar]
- 235.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008 Sep;19(9):587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 236.Druesne N, Pagniez A, Mayeur C, Thomas M, Cherbuy C, Duee PH, et al. Repetitive treatments of colon HT-29 cells with diallyl disulfide induce a prolonged hyperacetylation of histone H3 K14. Ann N Y Acad Sci. 2004 Dec;1030:612–621. doi: 10.1196/annals.1329.071. [DOI] [PubMed] [Google Scholar]
- 237.Bontempo P, Mita L, Miceli M, Doto A, Nebbioso A, De BF, et al. Feijoa sellowiana derived natural Flavone exerts anti-cancer action displaying HDAC inhibitory activities. Int J Biochem Cell Biol. 2007;39(10):1902–1914. doi: 10.1016/j.biocel.2007.05.010. [DOI] [PubMed] [Google Scholar]
- 238.Mantelingu K, Kishore AH, Balasubramanyam K, Kumar GV, Altaf M, Swamy SN, et al. Activation of p300 histone acetyltransferase by small molecules altering enzyme structure: probed by surface-enhanced Raman spectroscopy. J Phys Chem B. 2007 May 3;111(17):4527–4534. doi: 10.1021/jp067655s. [DOI] [PubMed] [Google Scholar]
- 239.Yang SR, Wright J, Bauter M, Seweryniak K, Kode A, Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF-kappaB in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging. Am J Physiol Lung Cell Mol Physiol. 2007 Feb;292(2):L567–L576. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 240.Hou X, Xu S, Maitland-Toolan KA, Sato K, Jiang B, Ido Y, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008 Jul 18;283(29):20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Binda O, Nassif C, Branton PE. SIRT1 negatively regulates HDAC1-dependent transcriptional repression by the RBP1 family of proteins. Oncogene. 2008 May 29;27(24):3384–3392. doi: 10.1038/sj.onc.1211014. [DOI] [PubMed] [Google Scholar]
- 242.Yang J, Kong X, Martins-Santos ME, Aleman G, Chaco E, Liu GE, et al. The activation of SIRT1 by resveratrol represses transcription of the gene for the cytosolic form of phosphoenolpyruvate carboxykinase (GTP) by deacetylating HNF4alpha. J Biol Chem. 2009 Aug 3; doi: 10.1074/jbc.M109.047340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Lah JJ, Cui W, Hu KQ. Effects and mechanisms of silibinin on human hepatoma cell lines. World J Gastroenterol. 2007 Oct 28;13(40):5299–5305. doi: 10.3748/wjg.v13.i40.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Chen IH, Lu MC, Du YC, Yen MH, Wu CC, Chen YH, et al. Cytotoxic Triterpenoids from the Stems of Microtropis japonica. J Nat Prod. 2009 Jun 17; doi: 10.1021/np800694b. [DOI] [PubMed] [Google Scholar]