Abstract
Genetic somatic alterations are fundamental hallmarks of cancer. In addition to point and other small mutations targeting cancer genes, solid tumors often exhibit aneuploidy as well as multiple chromosomal rearrangements of large fragments of the genome. Whether somatic chromosomal alterations and aneuploidy are a driving force or a mere consequence of tumorigenesis remains controversial. Recently it became apparent that not only genetic but also epigenetic alterations play a major role in carcinogenesis. Epigenetic regulation mechanisms underlie the maintenance of cell identity crucial for development and differentiation. These epigenetic regulatory mechanisms have been found substantially altered during cancer development and progression. In this review, we discuss approaches designed to analyze genetic and epigenetic alterations in colorectal cancer, especially DNA fingerprinting approaches to detect changes in DNA copy number and methylation. DNA fingerprinting techniques, despite their modest throughput, played a pivotal role in significant discoveries in the molecular basis of colorectal cancer. The aim of this review is to revisit the fingerprinting technologies employed and the oncogenic processes that they unveiled.
1. Introduction. Genetic and epigenetic aberrations in human cancers
The isolation of the first human oncogenes and tumor suppressors [1–4] led to the prevailing hypothesis during the last decades postulating that the origin of cancer resides in the accumulation of somatic mutations in cancer genes, i.e. proto-oncogenes and tumor suppressors [5,6]. In addition to these oncogeneic mutations, the vast majority of solid tumors exhibit aneuploidy and chromosomal rearrangements. Whether somatic chromosomal alterations and aneuploidy are a driving force or a consequence of tumorigenesis remains controversial [7–9]. With the exception of the DNA mismatch repair mutator genes that underlie microsatellite instability, diagnostic of a strong mutator phenotype [10–12] the search for somatic mutations in genes involved in the preservation of genome integrity has been disappointing [13–17]. With the information at hand, it appears that somatic genetic alterations leading to such an active chromosomal instability do not have a great significance in cancer. The chromosomal alterations universally found in solid tumors may be in a greater extent due to clonal selection and evolution of errors of chromosomal segregation that, in some cases, can be originated by age-related epigenetic alterations.
A shift has recently occurred in cancer research with the realization that not only genetic but also epigenetic alterations play a major role in carcinogenesis. To date, three major types of epigenetic mechanisms have been identified in humans: DNA methylation, histone modifications, and, more recently, non-coding RNAs. These mechanisms, responsible for the cell identity and differentiation, have been found substantially altered during cancer development. Consequently, the study of cancer epigenetics has attracted considerable attention.
In vertebrates, DNA methylation occurs almost exclusively at the position 5 of cytosine residues within the dinucleotide CpG. These sites are found concentrated in some genomic regions denominated CpG islands (CGIs), generally associated to gene promoters. While most of the CpG sites outside CGIs are constitutively methylated, the CpG sites inside CGIs are frequently devoid of methylation. The methylation status of the CGIs located in promoters and other gene regulatory regions can exert a drastic effect on the transcriptional levels of downstream genes, providing an epigenetic mechanism to control gene expression. Up to 72% of the human gene promoters are specially rich in CpG sites [18], consistent with the notion that many genes are susceptible of epigenetic regulation through histone modifications and DNA-methylation.
Global DNA hypomethylation of cancer cells was discovered more than twenty-five years ago [19,20]. Soon after, the first example of somatic hypomethylation of cellular oncogenes in human cancer was reported [21]. Years later, it was found that some sporadic retinoblastoma tumors exhibited hypermethylation of the promoter of the tumor suppressor gene RB1, leading to transcriptional repression [22]. Since that seminal discovery, numerous genes have been found to undergo promoter hypermethylation in a large variety of cancers [23]. The role of gene promoter hypermethylation in carcinogenesis has been extensively studied, yielding cancer detection markers and chemotherapy predictors for cancer patients, as well as fostering the development of epigenetic drugs approved for the treatment of hematological malignancies [24].
Technologies to detect mutations, chromosomal copy number alterations and DNA methylation alterations have improved exponentially, fostered by the generalization of the microarray platforms in the mid nineties [25] and, more recently, the development of massively parallel sequencing platforms [26–29]. These technologies have reached an impressive throughput: today it is possible to analyze over one million genomic locations in just a single microarray chip and the massively parallel sequencing platforms are capable of delivering more than one hundred million sequences in a single experiment. Very recently, the analysis of colorectal tumors using a comprehensive high-throughput array-based relative methylation (CHARM) method [30], not biased for CpG island or promoter sequences, yielded a surprising discovery: the authors found that methylation alterations in colon cancer occur not only in promoters and CpG islands, but also in sequences up to 2 kb distant, which they termed 'CpG island shores'. CpG island shore methylation showed an inverse relationship with gene expression [31].
However, before these remarkable technological advances were achieved, a handful of ingenious techniques based on the generation of DNA fingerprints of matched normal and tumor tissues were successfully employed to detect genetic and epigenetic alterations and, despite their modest throughput, they were germane to significant discoveries that provided insights in the molecular basis of colorectal cancer. Some of these technologies still have a place in everyday lab research, as they usually require less complex and inexpensive equipment. We will revisit in this review some of the fingerprint technologies employed in colorectal cancer research and their role in the discovery of fundamental oncogenic processes.
2. Fingerprinting techniques for the detection of chromosomal copy number changes
Few years after the development of the polymerase chain reaction (PCR) technique [32], two PCR-based fingerprinting techniques were almost simultaneously published, Arbitrarily Primed PCR (AP-PCR) [33] and Random Amplification of Polymorphic DNA (RAPD) [34]. RAPD fingerprints are obtained using a pair of very short primers, usually no longer than 10-mer, favoring unspecific annealing in the first several cycles of PCR amplification. The basis of the AP-PCR is the amplification of random genomic DNA sequences using a single primer of arbitrary sequence [33]. The first cycles of amplification are performed at a low annealing temperature, typically 40–50ºC, favoring the primer annealing to many non-specific sequences in the genomic DNA. The annealing temperature is raised in subsequent cycles. A complex process of competition between the many initial amplification products results in only a subset of the initial amplicons to be further amplified in a very reproducible manner. Each arbitrary primer can amplify more than 100 DNA fragments ranging in size from less than 100bp to more than 2000bp, although the fragment size is typically less than 1kb [35]. AP-PCR amplicons are radioactively labeled, either adding [α-32P]-labeled dNTPs to the PCR reaction or labeling the 5’ end of the arbitrary primer with [γ-32P]-ATP prior to PCR amplification, and detected by autoradiography after electrophoretic separation in a denaturing polyacrylamide gel. In some protocols, 32P is replaced with the lower-emission isotopes 33P or 35S, in order to obtain sharper autoradiography bands at the expense of longer exposure times [36,37]. Non-radioactive AP-PCR fingerprints have been also developed [38,39].
RAPD fingerprints have been employed in phylogenetic studies to distinguish between different cell strains or organisms, and have been also used to analyze chromosomal copy number changes in cancer cells [40–42]. Applications and achievements of RAPD technology have been recently reviewed by F.A. Atienzar and A.N. Jha [43]. Similarly, AP-PCR was originally designed to identify differences in the genomes of different prokaryotic and eukaryotic organisms and, consequently, it has been often used in genotyping and phylogenetic studies [43]. However, soon after its invention, the potential of this technique to detect DNA copy number alterations in tumorigenesis was established [44].
2.a. AP-PCR and the detection of chromosomal copy number changes
Comparison of AP-PCR fingerprints of genomic DNA from tumor cells and non-tumor tissue from the same individual allows the identification of both deletions and amplifications in randomly amplified loci. Losses are visualized as a reduction in the intensity of a band in AP-PCR fingerprint of the tumor genome compared to the normal, while an increase in the intensity reflect amplification in the tumor genome of the target sequence and hence their adjacent chromosomal sequences. A decrease in band intensity could, in theory, result from mutations in the primer annealing site preventing amplification [35], but this possibility is extremely infrequent. Theoretically, homozygous deletions could also be detected [45], but their frequency in cancer is very low. Thus, the vast majority of losses detected in the AP-PCR fingerprints represent heterozygous deletions often accompanying the loss of heterozygosity (LOH) unmasking tumor suppressor genes. Amplifications are observed as an increase in the intensity of the AP-PCR bands in the tumor genomic DNA fingerprint. The lower limit of detection is at the level of trisomies - tetrasomies [46].
AP-PCR offered several advantages over the previously available methods: the ability to analyze many arbitrary locations in a single experiment while being an inexpensive method that does not require complex equipment. AP-PCR amplicons exhibiting alterations can be easily eluted from the dried gel, re-amplified and subsequently sequenced to determine their genomic location. Once the AP-PCR fragments are identified, this technique can be used to generate a molecular karyotype, or amplotype, reflecting the chromosomal gains and losses in the tumor sample. Amplotyping confirmed many of the previously identified chromosomal alterations in colorectal cancer (ie. loss of 5q, and 17p), and discovered several alterations that were subsequently confirmed by other techniques, especially Comparative Genomic Hybridization (CGH). These include loss of chromosomes 4 and 14 in colorectal cancer and of 2q and 6p in breast cancers [47] as well as the very frequent gains of chromosomes 8q, 13q and 20 and the gains of chromosome 6 associated to metastatic colon cancers [44,46]. Of particular interest is the observation that gains of the region at 8q24 encompassing c-Myc were detected in around and above 50% of all of the tumors analyzed, including colorectal, gastric, liver, pancreas, lung, and breast cancers that made up over 50% of all human cancer worldwide, thus implicating c-Myc as perhaps the most common altered gene in human cancer ([48,46,47,49] and unpublished observations).
In addition, the fraction of bands exhibiting alterations in the AP-PCR fingerprints of tumors – or Genomic Damage Fraction (GDF) - provides an estimation of chromosomal copy number alterations. The distribution of the tumor specimens according to the extent of their genomic damage was found to be gradual but remarkably GDF provided an independent prognosis indicator in both gastric and colon cancers. Patients with low-GDF tumors had significantly better prognosis than patients with high-GDF tumors, independently of the cut-off point to stratify the tumors in either the low-GDF or high-GDF groups [36,37,49,50].
2.b. AP-PCR and the discovery of microsatellite instability and the mutator phenotype
In addition to copy number changes, AP-PCR unexpectedly detected small insertions and deletions in the template DNA, visualized as mobility shifts of one or a few fingerprint bands. This observation led to the discovery in 1993 of microsatellite instability (MSI) or the microsatellite mutator phenotype (MMP), a novel pathway for carcinogenesis [11]. About 12% of unselected colorectal cancers studied by AP-PCR exhibited mobility shifts of several of the AP-PCR fingerprint bands. When these bands were isolated and sequenced, it was found that all of them contained repeated Alu sequences, which had undergone somatic deletions of a few nucleotides in their poly(A) tails. Further experiments confirmed that microdeletions also occurred in other mononucleotide repeats and insertions and deletions in dinucleotide and trinucleotide microsatellite repeats.
The unbiased nature of the AP-PCR fingerprints allowed to extrapolate that these mutations originated by errors of replication due to slippage by strand misalignment surpassed the hundreds of thousands in the subset of tumors with MSI. Among these many irrelevant non-coding microsatellite sequence mutations some mutations will also hit cancer genes (oncogenes and tumor suppressors), ultimately leading to cancer [51,52]. Soon after the initial observation it was found that mutations in these microsatellite sequences accumulated because of mutations occurring in the DNA mismatch repair machinery genes [53,54]. MMR deficiency underlies the genome-wide microsatellite instability (MSI) phenotype in most of the hereditary non-polyposis colorectal cancers (HNPCC) and some sporadic gastrointestinal tumors [52,55].
The sequence of events in the MMP pathway can be summarized as follows: inactivation of MMR (mutator) genes causes a mutator phenotype, which causes oncogenic mutations, which cause cancer [56]. Therefore, cancer driven by mutator genes represents a “remote control” mechanism for carcinogenesis, as mutator gene inactivation does not immediately lead to altered cell growth or survival [57,52,58]. MSI represents a distinctive pathway to cancer, with a particular spectrum of mutated cancer genes in MSI positive tumors [59–62]. MSI tumors display a low incidence in the APC and P53 tumor suppressor genes and K-ras oncogene, prototypical of colon cancer [11,63,57,64–66]. Instead, MSI positive colon tumors carry a plethora of different mutated genes, such as TGFβRII and Bax, which are rarely found in microsatellite stable (MSS) tumors [59,60,67,68]. This is because in a MMR deficiency background, mutations occur preferentially in genes with simple repeats in their coding or regulatory sequences [69,70].
2.c. DNA microarray platforms for detection of copy number alterations
Simultaneous hybridization of AP-PCR products (SHARP) was developed to determine the chromosomal location of the amplified products, without having to isolate, clone and sequence the individual AP-PCR bands [71]. In this technique, AP-PCR fingerprints are performed on genomic DNA from human/rodent monochromosome cell hybrids. Only the bands of human origin are visualized after hybridization, allowing the immediate determination of the chromosomal location of the different bands.
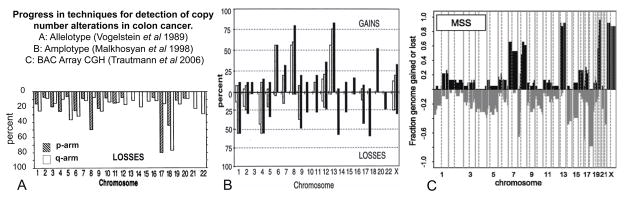
To increase the number of loci that could be analyzed in a single experiment, AP-PCR was transferred to a microarray format. The resulting technology was termed Chromosomal Hybridization of AP-PCR Amplicons (CHAPA) [72]. CHAPA employed a modified AP-PCR primer favoring the amplification of CA repeats, which are very common in the human genome, to maximize the number of different amplified PCR fragments. After AP-PCR amplification, the products were cloned into E. coli and the inserts of thousands of clones were individually identified by sequencing and printed onto microarray slides. DNA from normal and tumor samples was subsequently amplified using the same primer, labeled with different fluorescent dyes and hybridized onto the microarray. Differences in the ratio of the dyes accurately reflected copy number differences in the samples. Albeit CHAPA was proven to be suitable to detect copy number changes, its further development was hampered by the contemporary commercialization of CGH platforms facilitated by the information available form the Human Genome Project [73]. The higher resolution of the microarray platforms confirmed and extended the previous results obtained by allelotyping and AP-PCR amplotyping. Figure 2 shows the concordance in interpretation of results obtained with the original allelotype approach [74] that only detected losses (loss of heterozygosity or LOH) but not gains using polymorphic minisatellite sequences, AP-PCR amplotyping [46] and BAC CGH arrays [75].
Figure 2.
Comparison of the copy number alterations in colorectal cancers detected by: (A) Allelotyping, (B) AP-PCR Amplotyping and (C) Array CGH. Allelotyping detects loss of heterozygosis by individually analyzing polymorphic minisatellite sequences in every chromosome arm. The use of AP-PCR made possible the detection of gains and losses in multiple genomic regions in a single PCR reaction. The results obtained with these two techniques were later confirmed by the higher resolution CGH arrays.
In addition to AP-PCR, several technologies were developed in the early nineties to detect copy number alterations. One was Comparative Genomic Hybridization (CGH) [76–78]. Another approach, Representation Differential Analysis (RDA) [79,80] utilized PCR-derived genomic representations instead of the total genomic DNA to reduce complexity and increase hybridization signal intensity [81]. More recently, these technologies have been transformed to DNA microarray format. Pollack et al. utilized arrays of cDNA fragments for CGH analysis [82]. Pinkel et al. used arrays of BAC clones to increase the resolution up to 1 Mbp [83–85] and later tiling arrays were generated [86]. The next advance was the development of high density genomic arrays using oligoprobes derived from in silico analysis [87]. These platforms offer a coverage of the human genome without requiring cloning and sequencing individual probes before printing them onto microarray slides.
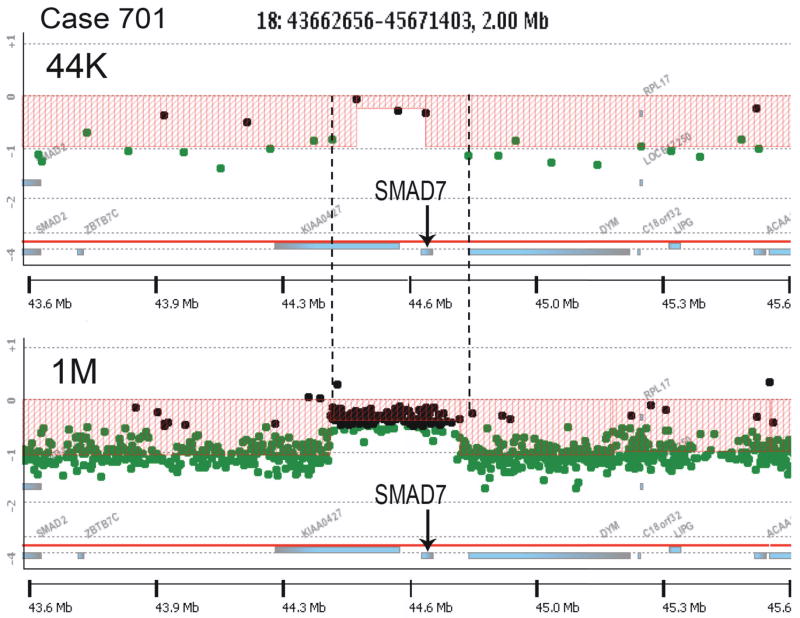
The analysis of colon cancer with Agilent 44K CGH arrays for copy number changes further validated the AP-PCR and CHAPA results. The larger number of loci analyzed by the CGH platform facilitated the detection of small alterations that might be missed by the AP-PCR amplotyping analysis. This CGH study revealed an interesting novel type of small genomic alteration: within large deleted regions there are smaller sequences that do not undergo the deletion. They appear to have escaped from the loss. In some cases, the size of these alterations was just at the limit of the 44K arrays resolution, with just 3 or 4 probes within the escaped region, preventing a conclusive identification. Re-analysis of some of these alterations on the highest resolution arrays currently available from Agilent, the SurePrint G3 Human CGH Microarray Kit 1x1M, which have more than 974,000 coding and non-coding human sequences represented, confirmed the existence of some genes that escape from a deletion that affects the surrounding region (Figure 3). The implication is that these genes are under strong selection pressure against their loss during colorectal cancer development or progression. Some of the escaped genes participate in essential cellular functions, such as RNPOLII. Other genes, however, might be involved in the carcinogenesis process in a more direct way, like SMAD7, which has been found to be a factor in the risk to develop colorectal cancer [88].
Figure 3.
Array CGH analysis with the Agilent 44K (top) and 1M (bottom) arrays of a 2Mb region of chromosome 18q21 from a colon cancer. Filled circles represent probes with no changes (black) and probes lost (green). 44K array analysis revealed a region close to SMAD7 that ‘escaped’ from loss (indent in the red area). The vertical dashed lines indicate the estimated maximum size of the region (~337Kb), determined by the closest neighbor probes that are lost (in green). The 1M array analysis confirmed that this ~300Kb region, containing the complete gene SMAD7, had escaped from the loss that affected the surrounding area.
3. Fingerprinting technologies for the analysis of epigenetic alterations
Most current DNA methylation analysis technologies rely on the DNA treatment with sodium bisulfite after chemical denaturation. This treatment deaminates all the unmethylated cytosine residues, converting them to uracil. Methylated cytosines, however, are resistant to deamination and remain unaffected. The bisulfite treated DNA is subsequently analyzed by PCR amplification, where the converted uracile residues in the sequence are substituted by thymine in the amplification product and therefore easily identified. Several techniques have been designed to study the methylation of regions of interest. Among them, the most popular technologies are bisulfite genomic sequencing [89], methylation-specific PCR (MSP) [90], MethyLight [91], combined bisulfite and restriction analysis (COBRA) [92], and more recently methylation microarrays [93]. Most of these techniques have been thoroughly reviewed [94,95] (see also Jorda and Peinado in this issue). However, before the widespread use of bisulfite-based techniques, several fingerprint approaches were developed based on methylation-sensitive restriction enzymes, whose activity is blocked by methylation of their target sequences.
3.a. Restriction Landmark Genomic Scanning for Methylation (RLGS-M)
RLGS is a high-resolution two-dimensional gel electrophoresis method based on the use of rare-cutter restriction enzymes to produce a discrete number of fragments [96]. The original method, based on a methylation-insensitive rare-cutter PacI, was capable of detecting polymorphisms and copy number changes but not methylation differences. RLGS-M [97] is a variation of RLGS in which methylation-sensitive rare-cutters are employed, like NotI (GCGGCCGC) and AscI (GGCGCGCC) that only cut when both internal CpG sites (in bold type) are unmethylated. RLGS-M is suitable to assess copy number changes as well as differences in methylation. After digestion, the 5’-protruding ends are labeled by filling the overhanging strand with radioactive nucleotides. The DNA is then subjected to a second digestion to produce smaller DNA fragments, most commonly with the restriction enzyme EcoRV, and separated in a first dimension agarose gel. After this electrophoresis, the sample is subjected to an in situ digestion with a third restriction enzyme, usually HinfI, allowing adequate separation of the resultant smaller fragments in a second-dimension polyacrylamide gel electrophoresis. After autoradiography, the end result is a reproducible RLGS profile that displays hundreds of spots reflecting the combination of copy number and methylation status of individual loci [98]. RLGS-M profiles from different tissues can be compared to assess for tissue-specific methylation, or from tumor and adjacent normal tissue to investigate cancer-associated methylation aberrations. Deletions or hypermethylation are detected by the loss or reduction of signal intensity of a RGLS-M spot, while amplification or hypomethylation result in increased signal intensity or new spots. Using the surrounding single copy spots as internal controls, the loss or gain of intensity of a locus can be visually estimated or quantified by densitometry. To facilitate the identification task, it is now possible to conduct automated RLGS fragment prediction and to download corresponding sequences for mouse [99] and human studies [100].
RLGS-M fingerprinting is one of the first methods capable of identifying many landmark fragments in a single run and the simultaneous identification of both gene copy number and methylation status [101]. However, this method has some drawbacks: it is rather labor-intensive, and only one sample per gel can be analyzed. Also, it requires at least a few micrograms of high-molecular-weight DNA to ensure the integrity of large pieces of DNA and to prevent nonspecific labeling of degraded fragments [98], so sample availability might be a limitation. Finally, the cloning step is critical and difficult as no PCR amplification is possible. [102]. Despite these limitations, RLGS-M has been successfully employed to analyze aberrant DNA methylation in breast, ovarian, colon, gastric, lung and hepatocellular cancers [103–110]. One RLGS-M study on colorectal cancer found that methylation of some CpG islands located in non-promoter regions of genes was associated with gene expression upregulation, indicating that alterations in the methylation status within CpG islands in colon tumors may have complex consequences on gene expression and tumorigenesis. [110].
3.b. Methylation-Sensitive Arbitrarily Primed PCR (MS-AP-PCR) and Restriction Fingerprinting (MSRF)
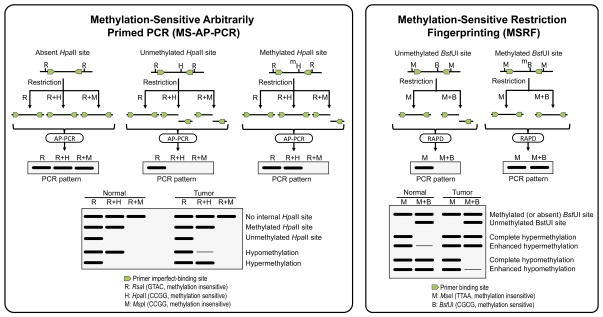
In 1997, two similar methods to generate methylation-sensitive fingerprints were published almost simultaneously, Methylation-Sensitive Arbitrarily Primed PCR method (MS-AP-PCR) [111] and Methylation-sensitive Restriction Fingerprinting method (MSRF) [112] (Figure 4). Both methods rely on the treatment of the genomic DNA with a methylation-insensitive frequent cutter yielding a methylation-independent DNA fragments library and, in a separate aliquot, with the same frequent-cutter and a methylation-sensitive restriction enzyme yielding a methylation-dependent DNA fragments library. In MSRF, the methylation-independent library is generated with the frequent cutter MseI (TTAA) and the methylation-dependent library with a mixture of MseI and the methylation-sensitive BstUI (CGCG), which cuts only if both CpG sites within its recognition sequence are unmethylated. In MS-AP-PCR, two different methylation independent libraries are generated: one with RsaI (GATC), and another one with a mixture of RsaI and MspI, which digests in CCGG sequences regardless the methylation status of the internal CG dinucleotide. The methylation-dependent library is generated with a mixture RsaI and the methylation-sensitive HpaII, which recognizes the same sequence than MspI (CCGG) but only digests if the internal CG dinucleotide is demethylated. After enzymatic restriction, these DNA fragments libraries serve as template for PCR amplification in AP-PCR or RAPD conditions.
Figure 4.
Methylation-Sensitive Arbitrarily-Primed PCR (left) and Methylation-Sensitive Restriction Fingerprinting (right) rely on the amplification of DNA libraries generated by methylation-sensitive and methylation-insensitive restriction enzymes. MS-AP-PCR libraries are generated by digestion with RsaI (GTAC) alone or in combination with HpaII (CCGG, methylation-sensitive) or MspI (CCGG, methylation-insensitive). After digestion, PCR amplification of the DNA fragments libraries is performed with 20-mer primers following the principle of AP-PCR. MSRF libraries are generated by treatment of the genomic DNA with MseI (TTAA, methylation-insensitive) alone or in combination with BstUI (CGCG, methylation-sensitive), and the amplification is performed with a pair of shorter primers, following the RAPD protocol. In both techniques, changes in the intensity of the fingerprinting bands reflect changes in methylation of the methylation-sensitive enzyme recognition sites.
To maximize the likelihood of amplifying regions susceptible of being differentially methylated, primers containing at least one CpG dinucleotide in their 3’ end are recommended. The rationale in both methods is that the methylated sequences, which are protected from restriction, will amplify in both libraries, whereas sequences containing unmethylated HpaII (in MS-AP-PCR method) or BstUI (in MSFR) sites are cleaved and will not yield any product in the methylation-dependent library. MS-AP-PCR includes an additional methylation-independent library generated with a mix of RsaI and MspI to discern whether the fragments amplified in both the RsaI methylation-independent and the RsaI-HpaII methylation-dependent libraries contain internal HpaII sites. The radioactive MS-AP-PCR and MSFR PCR products are resolved on high-resolution polyacrylamide gels under denaturing (MS-AP-PCR) or non-denaturing (MSRF) conditions. Comparison of methylation-dependent fingerprints from two samples, for instance a tumor and the surrounding normal tissue, reveals methylation differences. In both methods, hypermethylation is detected by increased intensity, whereas hypomethylation result in decreased intensity of the fingerprint bands. In principle, changes in intensity of the fingerprints bands from the methylation-dependent libraries can arise also from copy number alterations. The nature of the alteration can be ascertained by comparing the fingerprints from the methylation-independent libraries. Since these libraries are generated with methylation-insensitive restriction enzymes, changes in band intensity in their fingerprints exclusively result from copy number alterations.
MSFR and MS-AP-PCR offered several advantages over previously published methods. Since the original sample is amplified by PCR, only a small amount of genomic DNA is required (100ng-1μg). Also, they provide a direct way to determine whether the change in intensity of a particular band reflects copy number and/or methylation changes. However, the number of loci that can be interrogated with these technologies is limited, especially with MSFR, and even when 4 to 8 sets of arbitrary primers are run on each gel, only a small number of candidates (30–40) can be identified in one autoradiography [102]. MS-AP-PCR, and to a lesser extent MSFR, have been widely used to identify aberrantly methylated genes in human cancers [111–127]. Specifically, since its implementation, MSFR has been instrumental in the detection of hypermethylation in the 5’ CpG island of genes, such as EDNRB [117], PMP24 [119], ZHX2 [122], as well as hypomethylation of a repetitive element SATR-1 [120] in several human cancers.
Peter A Jones and colleagues developed MS-AP-PCR by screening genomic DNA from colon primary tumors, cell lines and normal tissues for methylation alterations [111]. They detected hypermethylation of a CpG island in the 5’ end of the gene TPEF (transmembrane protein containing epidermal growth factor and follistatin domains) in both colon tumors and cell lines, which was shown to be associated with a reduction in expression [114]. In another study, a CpG-rich region in exon 5 of PAX6, which is a highly conserved transcription regulatory factor involved in embryogenesis, was found to be hypermethylated in colon and bladder cancer [115].
3.c. Amplification of Inter Methylated Sites (AIMS)
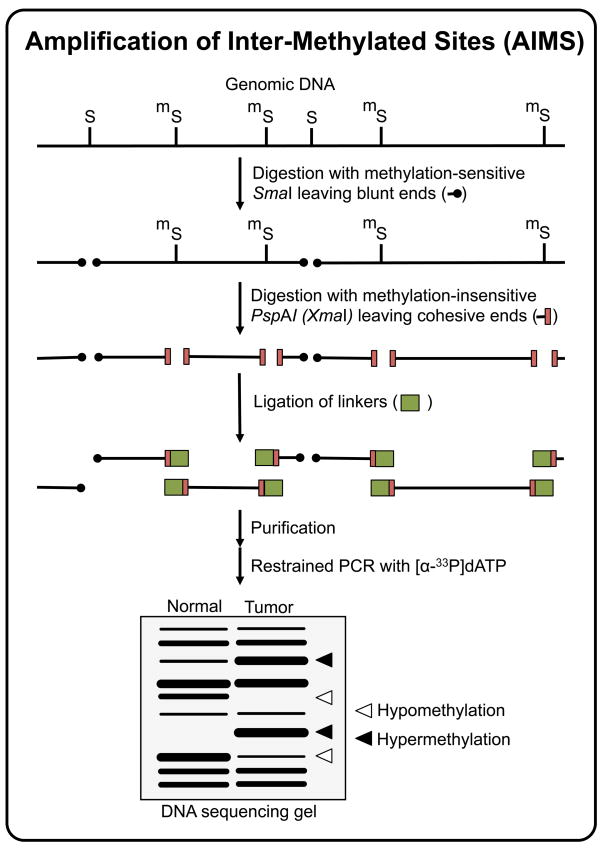
In 2002, Frigola and colleagues published an epigenetic fingerprinting method that they named Amplification of Inter Methylated Sites (AIMS, Figure 5) [128]. AIMS is a modified version of the methylated CpG island amplification (MCA) method [129], based on the differential cleavage of isoschizomers with distinct methylation sensitivity SmaI and XmaI. Both enzymes recognize the CCCGGG octamer, but whereas SmaI activity is blocked by methylation of the central CpG site (in bold type), XmaI is methylation-insensitive. These isoschizomers also differ in the type of DNA ends generated after digestion. While SmaI generates blunt ends, XmaI generates 4-nt 5’ protruding ends. In AIMS, the genomic DNA is first treated with SmaI, which cleaves all the unmethylated CpG sites leaving blunt ended DNA molecules. Then, the DNA is treated with XmaI, which cuts all the remaining methylated sites producing cohesive ends. Specific adaptors are ligated to the cohesive ends of the digested genomic DNA. The ligated sequences amplified by PCR using adaptor-specific primers extended at the 3’ end with two to four arbitrarily chosen nucleotidic residues to reduce the complexity of the product. This method amplifies DNA sequences that have two close methylated SmaI sites and show homology to the nucleotides extended at the 3’ end of the primer. Lack of methylation at either site will allow the digestion by SmaI, leaving blunt ends not compatible to the adaptor and therefore preventing amplification. Different fingerprints can be generated from the same sample by modifying the arbitrarily chosen 3’ end of the amplification primer. After PCR, the amplicons are resolved in denaturing polyacrylamide-sequencing gels generating fingerprints that consist of multiple anonymous bands, ranging in size from about 200 to 1200 bp, representing DNA sequences flanked by two methylated sites. Individual bands can be excised from the gel, reamplified using the same fingerprinting primer and characterized by sequencing [128,130,131].
Figure 5.
Amplification of Intermethylated Sites (AIMS) technique. Non-methylated SmaI recognition sites (CCCGGG) are first digested using the methylation-sensitive SmaI restriction endonuclease, leaving blunt ends. A second digestion is then performed using the methylation-insensitive isoschizomer XmaI, leaving 5’ protruding ends to which compatible adaptors are ligated. The DNA fragments in the range of ~200 to ~2000 bp and flanked by two ligated adaptors are subsequently amplified using primers that anneal to the adaptor sequence plus the restriction sequence and, to limit the number of amplified fragments, 1 or more arbitrarily chosen additional nucleotides. The increase of intensity of a particular AIMS fingerprint band reflects the hypermethylation of the flanking SmaI sites in the original sequence, while a decrease of intensity indicates hypomethylation.
AIMS is suitable to compare large numbers of samples and the simultaneous identification of hypomethylation and hypermethylation events. Hypomethylation is visualized as the loss of the fingerprint band in the tumor sample compared to its normal counterpart and hypermethylation is detected as the appearance of a new band in the tumor specimen. The method is especially powerful in detecting hypermethylation. Dilution experiments indicate that the technique is able to detect hypermethylated sequences even if they are present in <1% of the cells [128]. It detects hypomethylation as well, however the contamination of normal cells in the tumor samples limits its sensitivity.
Applications of AIMS to investigate epigenetic changes in cancer include the identification of recurrent hypermethylation associated with gene silencing [132,133], screening for both hypomethylation and hypermethylation in cancer cell lines with altered DNA methylation function [130,131]; and the genome-wide estimation of abnormal DNA methylation in cancers [132,133]. It has also been applied to identify epigenetic differences arisen during the lifetime of monozygotic twins [134]. In 2005, Frigola et al. employed AIMS to compare the levels of hypermethylation and hypomethylation alterations in colorectal carcinomas and adenomas. They found that the premalignant lesions already attained high levels of hypomethylation, but not hypermethylation, which suggested that this factor might play a key role in conferring the malignant potential since early stages. Also, they found that hypomethylation, but not hypermethylation, associated to poor prognosis [132]. Later, and based on their findings with AIMS, they described a novel mechanism in carcinogenesis involving the common repression of the entire 4-Mb band of chromosome 2q.14.2, by a coordinate long-range epigenetic gene silencing involving DNA hypermethylation and global methylation of histone H3K9 [133]. Remarkably, they found the transcriptional silencing of not only the genes undergoing promoter hypermethylation, but also the unmethylated neighboring genes, which also exhibited H3K9 methylation. Their work changed the view about DNA hypermethylation in cancer, which was previously envisaged as a local event silencing discrete genes, demonstrating that epigenetic silencing can span large chromosomal regions, affecting DNA-methylated and neighboring unmethylated genes that result coordinately suppressed by global changes in their histone methylation profiles.
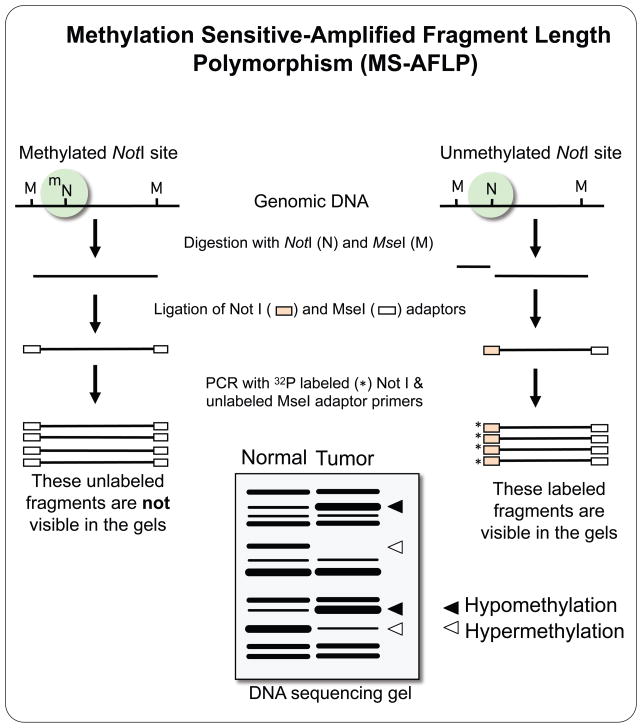
3.d. Methylation Sensitive-Amplified Fragment Length Polymorphism (MS-AFLP)
The NotI-MseI methylation sensitive-amplified fragment length polymorphism method (MS-AFLP) is also a DNA fingerprinting technology that allows for the simultaneous genome-wide detection of DNA hypermethylation and hypomethylation in tumor samples compared with normal tissue samples [135]. MS-AFLP is based on a previously published method, AFLP [136], but the restriction enzyme EcoRI was replaced with the methylation-sensitive restriction endonuclease NotI. This enzyme recognizes the GCGGCCGC sequence, which is frequent inside or in the proximity of CpG islands. The DNA methylation statuses of the two CpG sites in the octamer sequence are analyzed at hundreds of NotI sites in the genome by using this gel-electrophoresis based DNA fingerprinting technique. Epigenetic alterations, both hypo- and hypermethylation, in tumor tissue DNA are detected by comparing band intensities between normal and tumor tissue DNA fingerprints (Figure 6).
Figure 6.
Methylation-Sensitive Amplification Fragment Length Polymorphism (MS-AFLP). Genomic DNA is treated with a mix of MseI (TTAA, methylation-insensitive) and NotI (GCGGCCGC, methylation-sensitive) restriction endonucleases. MseI cuts all TTAA sites leaving 5’ protruding ends. NotI digests the unmethylated but not the methylated GCGGCCGC, leaving 5’ protruding ends. Compatible adaptors are ligated to the NotI and MseI cohesive ends, and the resulting DNA fragment library is amplified using primers that anneal on these adaptors. To limit the number of fragments amplified during the PCR, one or two arbitrarily chosen nucleotides are added to the sequence of the MseI primer. Three different types of fragments are amplified, methylation-independent MseI-MseI fragments, and methylation-dependent MseI-NotI and NotI-NotI fragments. The primer that anneals on the NotI adapter is labeled, so only the methylation-dependent fragments are visualized after autoradiography of the fingerprinting gels. Differences in MS-AFLP fingerprints band intensity between normal tissue and tumor reflect changes in DNA methylation.
In short, genomic DNA is digested with the methylation-sensitive rare-cutter NotI and the methylation-insensitive frequent-cutter MseI. All the MseI sites are cleaved, and the MseI ends are ligated to specific MseI adaptors. The NotI sites are cleaved only when both of the cytosines of the two CpG dinucleotides in the octamer NotI recognition sequence are unmethylated. The NotI ends are ligated to specific NotI adaptors. When either one or both of the two cytosines are methylated, the NotI site is protected from cleavage and, therefore, no adaptors are ligated. The fragment library is subsequently amplified by PCR with a 32P-labeled primer complementary to the NotI adaptors, and a non-labeled primer complementary to the MseI adaptors. Different combinations of primers differing in their 3’ ends can be employed to increase the number of NotI sites analyzed. The PCR products are then resolved in standard sequencing gels and autoradiographied. Three different types of fragments are amplified: MseI-MseI, MseI-NotI and NotI-NotI. The MseI-MseI fragments do not provide any information about DNA methylation, but they are not detected in the autoradiography since only the NotI-primer is labeled. The amplified products are DNA fragments in the range of 50–1,000bp, representing random anonymous genomic sequences.
MS-AFLP is highly reproducible and a relatively large number of bands (~100) are amplified with a high ratio of band signal/background noise [56]. Additionally, MS-AFLP requires little amount of template DNA. As little as five nanograms are sufficient for one MS-AFLP experiment with one pair of primers. In contrast with MSFR and MS-AP-PCR, bands in MS-AFLP fingerprints originate from the unmethylated (cleaved) NotI sites and therefore hypermethylation results in decreased intensity while hypomethylations results in increased intensity of the bands. DNA fragments exhibiting alterations can be directly cloned from fingerprint bands by amplification of gel-eluted DNA with the same pair of primers used for the fingerprint [135]. Several fingerprints generated with different primer combinations can be analyzed in parallel to maximize de number of loci per gel. As some MS-AFLP bands are common to several primer combinations, this provides an internal control for the alterations. Identification of consistent changes in a particular type of cancer can be facilitated by the analysis of multiple samples in the same gels.
Fluorescent-MS-AFLP (FL-MS-AFLP) is a non-radioactive adaptation of this technique in which fluorescently labeled NotI primers are employed. In FL-MS-AFLP the differences in methylation levels between normal and tumor samples are visualized by the difference in signal intensities in the electropherogram. MS-AFLP products were labeled with a fluorescent primer, TAMRA, and analyzed the changes in DNA methylation in cancer using an ABI Prism 377 automatic DNA Sequencer [135]. In a recent paper Kageyama and colleagues further developed the FL-MS-AFLP method to analyze the methylation levels of blood DNA from gastric cancer patients, as well as methylation differences between gastric tumor samples and adjacent normal tissue samples [137]. They labeled the NotI primer with the fluorescent dye FITC, and measured fluorescence intensity using the DSQ-2000 automatic DNA sequencer. They quantitatively evaluated the methylation status of more than 350 NotI sites in the human genome per run by FL-MS-AFLP. The use of fluorescence for MS-AFLP has not only increased the number of bands that can be analyzed, but also made the analysis of longer sequences (up to 1000 bp) possible. FL-MS-AFLP is safer than MS-AFLP because it does not require the use of radioactivity, which may be an important factor in clinical settings. In addition, multiple dyes with different absorption and emission range can be utilized [135]. Furthermore, as opposed to the short half-life of 32P (14.3 days), fluorescently labeled primers have a longer shelf life if they are kept in the dark. However, using fluorescent tags for NotI-MseI MS-AFLP has also disadvantages. The critical one is that the method does not allow direct cloning of DNA fragments after the identification of alterations.
MS-AFLP has also been applied to a DNA microarray hybridization format (DNA Microarray MS-AFLP) [138]. In short, in the microarray hybridization method PCR products are labeled with CY5 and CY3 (using the Klenow fragment and CY5-dCTP or CY3-dCTP), the two probes are mixed, and unincorporated dNTPs are removed. The probes are then used to hybridize DNA microarrays on glass slides in the CGH manner [77,83,139]. Fluorescent signal associated with each target spot is measured using a fluorescence scanner. The fluorescence intensity correlates with the number of unmethylated NotI sites cleaved by the enzyme.
In our study describing the method, we performed a total of 30 (15x2 reciprocal labeling) DNA Microarray MS-AFLP hybridization experiments using genomic DNA from two breast and three prostate cancer cell lines in all the pairwise combinations. We also performed hundreds of Southern hybridization experiments to calculate the rates of false positives and negatives, and obtained the values of 2.6% and 3.6%, respectively. We also determined the sensitivity (78.5%) and specificity (97.1%) of the DNA Microarray MS-AFLP method. In the original gel electrophoresis-based MS-AFLP it is often difficult to quantify the signal due to noise from the bands in the background. An advantage with the DNA Microarray MS-AFLP method is that it is more quantitative, and even differences in copy number could also be detected, in addition to DNA methylation alterations. Furthermore, as in the FL-MS-AFLP, no radioisotopes are used. On the other hand, in the microarray format only two samples can be compared at a time. Another drawback is its limited use as a gene discovery tool because only the sequences on the microarrays can be analyzed. Nevertheless, gene identification is straightforward as each dot in the microarray is represented by a single sequence. This feature makes DNA Microarray MS-AFLP a valid method for DNA methylation analysis of larger number of NotI sites in a larger scale of analysis [138].
3.e. MS-AFLP and the absence of CpG island methylator phenotype in colorectal cancer
As previously mentioned, NotI sites are uncommon in the genome. Just 9853 NotI sites are found in the latest release (GRCh37) of the human genome sequence from the Genome Reference Consortium. Most of them, however, are located within CpG islands [140] and frequently associated with the 5’ region of genes, but not in the highly repetitive elements Alu and LINEs. Hence, analysis of the methylation of NotI sites by MS-AFLP fingerprinting provides an unbiased estimation of the methylation status of CpG islands throughout the genome. This unbiased approach was key to demonstrate that CpG island hypermethylation in gastrointestinal cancers follows a gradual distribution [56,50], challenging the hypothesis of the existence of a cancer-specific CpG island methylator phenotype (CIMP). This hypothesis postulates the existence of two distinctive types of tumors: with and without a methylator phenotype that causes the concordant hypermethylation of some CpG islands. Which CpG islands and how many of them should be found methylated to unambiguously classify a particular tumor into CIMP+ or CIMP- have been a moving target in constant redefinition since the inception of the concept [141,129].
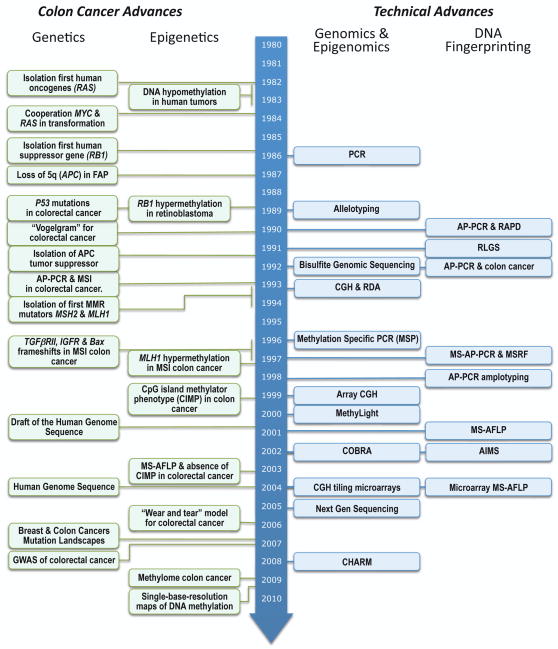
A recent study aimed to characterize loci markers for CIMP demonstrated that the original “classic” panel indeed exhibited a unimodal distribution and a completely redefined panel of CIMP markers was proposed [142]. It is still unexplained, however, what distinguishes these CpG islands that are targets and markers for CIMP from the many hundreds and thousands of CpG islands found also methylated in each one of the CIMP-negative tumors. Unquestionably, some CpG islands are more frequently methylated than others in tumors. It remains to be established whether the preferential hypermethylation of a subgroup of CGIs reflects an underlying mechanism targeting a particular subset of CGIs or is a mere consequence of the selective pressure over a stochastic widespread hypermethylation phenomenon, if the concomitant silencing of the affected genes favors the clonal expansion of the malignant cells. It has been over a decade now since the proposal of the CIMP hypothesis, and despite numerous CIMP publications, including editorials reassuringly confirmatory of the hypothesis reiterated every couple of years [143,144], we are still waiting to know what the methylator genes are [145]. This is in contrast with the identification and isolation of the first MMR mutator genes soon after the initial discovery of MSI (See Figure 7). The CIMP concept has been a remarkable example of the chase of the wild geese and a red herring that for over a decade has confused and continues to confuse an important aspect of cancer epigenetics.
Figure 7.
Chronological diagram of some of the relevant advances in colorectal cancer research and the technical advances that made some of them possible.
In conclusion, we are still essentially in the dark as the cause of the aberrant hypermethylation common in cancer. Recent growing evidence suggests a role of the Trithorax (Trx) and Polycomb repressor complexes (PRCs), the two main cell memory systems involved in the maintenance of a stem cell state [146–148]. It has been found that PRC-target genes are up to 12-fold more likely to have cancer-specific promoter DNA hypermethylation than non-targets [148] and that methylation on Lys27 of histone H3, a modification mediated by the Polycomb repressor complex 2 (PRC2), pre-marks genes for de novo methylation in cancer [147]. These findings support a stem cell origin of cancer in which reversible gene repression mediated by PRCs is replaced by permanent silencing (DNA methylation), locking the cell into a perpetual state of self-renewal and thereby predisposing to subsequent malignant transformation [148]. According to this hypothesis, the abnormal crosstalk between the Polycomb proteins and the DNA methylation machinery would be mechanistically underlying the aberrant promoter methylation of some genes, although only a minority of all the genes frequently hypermethylated in cancers seem to be PRCs targets. A notable exception is hMLH1, which is hypermethylated in the majority of sporadic MSI cases but has not been categorized as a PRCs target [149–151].
3.f. MS-AFLP and the association between DNA hypomethylation, aging and genomic damage
The results obtained by MS-AFLP on gastrointestinal cancers showed that not only hyper- but also hypomethylation accumulates gradually and in an age-associated fashion [50]. In addition, DNA hypomethylation detected by MS-AFLP strongly associated with the degree of aneuploidy estimated by AP-PCR. These findings led to the proposal of a novel “wear and tear” model linking aging and cancer through DNA hypomethylation. This pathway would explain the accumulation of genetic alterations in a certain fraction of gastrointestinal cancers not caused by an underlying microsatellite mismatch repair defect and subsequent mutator phenotype, or “spontaneous” mutations in cancer genes such as APC and p53. In this model, genome-wide DNA demethylation of CpG sites occurs as a result of a gradual and accumulative age-associated failure to preserve methylation replication fidelity, without the involvement of any particular genetic or epigenetic defect. When certain regions of the genome are hit, such as pericentromeric repetitive sequences, normally heavily methylated at birth, the probability of mitotic errors increases, leading to the accumulation of genomic damage and eventually to cancer [50].
4. Concluding remarks
In this review we have presented several fingerprinting techniques that have been applied to the study of genetic and epigenetic alterations in human colorectal cancers, and some of the discoveries that they made possible (Figure 7). These methods have now been surpassed in sensitivity and throughput by modern technologies, especially after the widespread commercialization of microarrays for both genetic and epigenetic alterations and, more recently, the development of massively parallel sequencing platforms [26–28]. As a discovery tool, the use of most of the fingerprinting methods presented in this review will be soon relegated by their more powerful successors. Some of them, however, can have a place in the day-to-day laboratory since they still offer certain advantages over the more sophisticated but expensive new methods. Its simplicity and affordability make them a reasonable option for the initial screenings of a large set of samples that, in a second phase, can be classified and analyzed in more detail using more powerful technologies.
AP-PCR was developed contemporarily with CGH and RDA. All of these approaches, designed to detect and estimate moderate changes in genome copy number, underwent a process of convergent evolution that after taking advantage of the information obtained from the human genome project, ended in a winner array CGH format with long synthetic oligos that are commonly used not only for the analysis of somatic alterations associated to the development and progression of sporadic cancer, but also for the study of germline copy number variations (CNV) and their impact in genome-wide association studies (GWAS) for cancer susceptibility.
The analysis of DNA methylation alterations by MS-AFLP in the NotI restriction site yields information of the methylation alterations occurring not only in CpG islands but also in CpG islands shores, which have been proven to be an important target of DNA methylation alterations [31]. This unforeseen lucky feature allowed to detect the association between aging and both hypermethylation and hypomethylation in gastrointestinal cancer and revitalized the interest on DNA hypomethylation as a relevant driving force in oncogenesis [50], which was overshadowed by the predominant focus on gene promoter hypermethylation.
It is worthy to reflect on how the use of relatively simple DNA fingerprinting techniques permitted original discoveries because of the panoramic views that they provided of the somatic alterations occurring in the colon cancer cell genome. For instance, due to the ability to study many samples in a few experiments, AP-PCR made possible a first insightful estimation of the high prevalence of altered 8q24 region pointing to a major role of c-Myc in human cancer. Also AP-PCR, due to its unbiased nature, allowed the extrapolation from the mobility shifts of a few fingerprint bands to the huge number of somatic clonal microsatellite mutations diagnostic of a genetic defect leading to a profound genomic instability. On the other hand, MS-AFLP fingerprinting also provided a panoramic view of the gradual distribution of hypermethyaltion alterations in colon cancers thus making impossible the distinction of tumors with and without a methylator phenotype. Thus, two DNA fingerprinting approaches, AP-PCR and MS-AFLP, directed to the analysis of somatic genetic and epigenetic alterations respectively, were instrumental for the discovery of the existence of a mutator phenotype and the non-existence of a methylator phenotype.
Figure 1.
The first cycles of amplification at low temperature allow the annealing of an arbitrary primer (in red) to multiple sites along the genome, and the amplification of sequences within two close annealing sites. The originally imperfect-match sequences will be transformed to perfect matches in the second round of amplification (in blue), where the DNA synthesized in the first round serves as template. DNA regions flanked by two primer-annealing sites are further amplified in the subsequent high-annealing temperature cycles, generating a reproducible fingerprinting pattern. Copy number alterations, both gains and losses, can be detected by comparing AP-PCR fingerprints from normal and tumor samples.
Table 1.
Comparative features of DNA fingerprinting technologies
| Technique (year) | Typea | Basisb | Resolutionc | Detectiond | Amounte | Throughputf | Costg |
|---|---|---|---|---|---|---|---|
| RAPD (1990) | CNA | Arbitrary primer PCR amplification | Native electrophoresis | Ethidium bromide staining | >10ng | L: ~20–50 S: ~20 |
$1–$2 |
| AP-PCR (1990) | CNA | Arbitrary primer PCR amplification | Denaturing electrophoresis | Autoradiography, silver staining or fluorescence | >10ng | L: ~50–100 S: ~40 |
$2–$3 |
| CHAPA (2001) | CNA | Arbitrary primer PCR amplification | Microarray slides | Fluorescence | >100ng | L: >1,000 S: 2 |
~$100–$200 |
| RLGS-M (1993) | MET | Digestion with NotI or AscI | Bidimensional electrophoresis | Autoradiography | >1μg | L: ~1,000 S: 1 |
~$20 |
| MS-AP-PCR (1997) | MET | Arbitrary primer PCR amplification after digestion with HpaII and RsaI | Denaturing electrophoresis | Autoradiography | >100ng* | L: ~50 S: ~40 |
$3–$4 |
| MSRF (1997) | MET | Arbitrary primer PCR amplification after digestion with BstUI and MseI | Native electrophoresis | Autoradiography | >100ng* | L: ~50 S: ~40 |
$3–$4 |
| AIMS (2002) | MET | Adaptor-mediated PCR amplification after digestion with SmaI and XmaI | Denaturing electrophoresis | Autoradiography | >100ng* | L: ~50–100 S: ~20 |
$3–$5 |
| MS-AFLP (2001) | MET | Adaptor-mediated PCR amplification after digestion with NotI and MseI | Denaturing electrophoresis | Autoradiography or fluorescence | >100ng* | L: ~50–100 S: ~20 |
$3–$5 |
| Array-MS-AFLP (2004) | MET CNA |
Adaptor-mediated PCR amplification after digestion with NotI and MseI | Microarray slides | Fluorescence | >1μg | L: ~10,000h S: 2 |
~$100–$200 |
| OTHER NON-DNA FINGERPRINTING TECHNIQUESi | |||||||
| Agilent CGH arrays | CNA | Hybridization of labeled genomic DNA fragments | Microarray slides | Fluorescence | >500ng | L: 104–106 S: 1–8 |
~$200–$500 |
| Illumina Goldengate | CNA MET |
Hybridization or single nucleotide extension of labeled DNA fragments | Microarray slides | Fluorescence | 200ng-1μg | L: 104–106 S: 1–12 |
~$200–$500 |
| Affimetrix arrays | CNA | Hybridization of labeled genomic DNA fragments | Microarray slides | Fluorescence | >500ng | L: 104–106 S: 1–96 |
~$200–$500 |
| CHARM | MET | McrBC-fractionated DNA libraries | Microarray slides | Fluorescence | >3μg | L: 44,000 S: 2 |
~$200 |
CNA: copy number alterations. MET: DNA methylation alterations.
Key feature that underlies the method is listed.
Method for resolving DNA sequences. Except RAPD that may use agarose gel electrophpresis, the rest of the fingerprinting techniques use polyacrylamide or derivatives (i.e., sequagel) gel electrophoresis.
Method of detection of the DNA sequences.
Values indicate amount of genomic DNA for the initial digestions. Amounts of sample loaded in the electrophesis for the fingerprinting techniques (*) are less (5–10ng).
L: loci per sample; S: number of samples per gel. The values given for the fingerprinting techniques
The cost in US dollars per DNA sample is given as an approximation in a wide range (i.e., depending on whether the analysis is by autoradiography or fluorescence). Costs for fingerprinting techniques include the costs of electrophoresis but not the costs of sample DNA preparation. Costs can be calculated per Mb of genome covered by the techniques, by increasing the number of experiments with different primers. For instance, a map by AP-PCR of random genome sites at a 10Mb resolution for copy number alterations that could provide panoramic views of the degree of disruption of the cancer genome would cost around 6 (primers) X $2–3 = ~$15 per sample, and a map by MS-AFLP of NotI sites at ~10Mb resolution for DNA methylation alterations would cost around 6 (primers) X $3–$5 = ~$25. To reach 1Mb resolution would cost 10 times more, without counting the labor. More than 1Mb resolution would be impractical for DNA fingerprinting technologies.
The current limitation is imposed by the number of NotI sites in the genome, not by the technique itself.
The list is not exhaustive, and is only intended to contrast some differences in throughput and cost for comparative purposes. Massive sequencing approaches (i.e., Illumina Solexa Technology, Roche 454 or Applied Biosystems SOLiD) are not included because it is difficult to calculate the cost per sample depending on the extent of the analysis.
List of abbreviations
- AIMS
Amplification of Intermethylated Sites
- AP-PCR
Arbitrarily Primed PCR
- CGH
Comparative Genomic Hybridization
- CGI
CpG Island
- CHAPA
Chromosomal Hybridization of AP-PCR Amplicons
- CHARM
Comprehensive High-throughput Array-based Relative Methylation
- CIMP
CpG Island Methylator Phenotype
- CNV
Copy Number Variation
- COBRA
Combined Bisulfite and Restriction Analysis
- FL-MS-AFLP
Fluorescence MS-AFLP
- GDF
Genomic Damage Fraction
- GWAS
Genome-Wide Association Study
- HNPCC
Human Nonpolyposis Colorectal Cancer
- LOH
Loss of Heterozygosity
- MCA
Methylated CpG-Island Amplification
- MMP
Microsatellite Mutator Phenotype
- MS-AFLP
Methylation Sensitive Amplified Fragment Length Polymorphism
- MS-AP-PCR
Methylation-Sensitive AP-PCR
- MSI
Microsatellite Instability/Instable
- MSP
Methylation-Sensitive PCR
- MSRF
Methylation-Sensitive Restriction Fiongerprinting
- MSS
Microsatellite Stability/Stable
- PCR
Polymerase Chain Reaction
- PRC
Polycomb Repressor Complex
- RAPD
Random Amplification of Polymorphic DNA
- RDA
Representation Differential Analysis
- RLGS
Restriction Landmark Genomic Scanning
- RLGS-M
Restriction Landmark Genomic Scanning for Methylation
- SHARP
Simultaneous Hybridization of AP-PCR Products
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Goldfarb M, Shimizu K, Perucho M, Wigler M. Isolation and preliminary characterization of a human transforming gene from T24 bladder carcinoma cells. Nature. 1982;296:404–409. doi: 10.1038/296404a0. [DOI] [PubMed] [Google Scholar]
- 2.Pulciani S, Santos E, Lauver AV, Long LK, Robbins KC, Barbacid M. Oncogenes in human tumor cell lines: molecular cloning of a transforming gene from human bladder carcinoma cells. Proc Natl Acad Sci U S A. 1982;79:2845–2849. doi: 10.1073/pnas.79.9.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shih C, Weinberg RA. Isolation of a transforming sequence from a human bladder carcinoma cell line. Cell. 1982;29:161–169. doi: 10.1016/0092-8674(82)90100-3. [DOI] [PubMed] [Google Scholar]
- 4.Dryja TP, Friend S, Weinberg RA. Genetic sequences that predispose to retinoblastoma and osteosarcoma. Symp Fundam Cancer Res. 1986;39:115–119. [PubMed] [Google Scholar]
- 5.Alberts B. Molecular biology of the cell. Garland Science; New York: 2002. [Google Scholar]
- 6.Lewin B. Genes IX. Jones and Bartlett Publishers; Sudbury, Mass: 2008. [Google Scholar]
- 7.Tomlinson IP, Novelli MR, Bodmer WF. The mutation rate and cancer. Proc Natl Acad Sci U S A. 1996;93:14800–14803. doi: 10.1073/pnas.93.25.14800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li R, Sonik A, Stindl R, Rasnick D, Duesberg P. Aneuploidy vs. gene mutation hypothesis of cancer: recent study claims mutation but is found to support aneuploidy. Proc Natl Acad Sci U S A. 2000;97:3236–3241. doi: 10.1073/pnas.040529797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pellman D. Cell biology: aneuploidy and cancer. Nature. 2007;446:38–39. doi: 10.1038/446038a. [DOI] [PubMed] [Google Scholar]
- 10.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 11.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 12.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 13.Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 14.Forrest WF, Cavet G. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138179. author reply 1500. [DOI] [PubMed] [Google Scholar]
- 15.Getz G, Hofling H, Mesirov JP, Golub TR, Meyerson M, Tibshirani R, Lander ES. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138764. [DOI] [PubMed] [Google Scholar]
- 16.Rubin AF, Green P. Comment on “The consensus coding sequences of human breast and colorectal cancers”. Science. 2007;317:1500. doi: 10.1126/science.1138956. [DOI] [PubMed] [Google Scholar]
- 17.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, Silliman N, Szabo S, Dezso Z, Ustyanksky V, Nikolskaya T, Nikolsky Y, Karchin R, Wilson PA, Kaminker JS, Zhang Z, Croshaw R, Willis J, Dawson D, Shipitsin M, Willson JK, Sukumar S, Polyak K, Park BH, Pethiyagoda CL, Pant PV, Ballinger DG, Sparks AB, Hartigan J, Smith DR, Suh E, Papadopoulos N, Buckhaults P, Markowitz SD, Parmigiani G, Kinzler KW, Velculescu VE, Vogelstein B. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 18.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diala ES, Hoffman RM. Hypomethylation of HeLa cell DNA and the absence of 5-methylcytosine in SV40 and adenovirus (type 2) DNA: analysis by HPLC. Biochem Biophys Res Commun. 1982;107:19–26. doi: 10.1016/0006-291x(82)91663-1. [DOI] [PubMed] [Google Scholar]
- 20.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 21.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 22.Greger V, Passarge E, Hopping W, Messmer E, Horsthemke B. Epigenetic changes may contribute to the formation and spontaneous regression of retinoblastoma. Hum Genet. 1989;83:155–158. doi: 10.1007/BF00286709. [DOI] [PubMed] [Google Scholar]
- 23.Esteller M. Epigenetic gene silencing in cancer: the DNA hypermethylome. Hum Mol Genet. 2007;16(Spec No 1):R50–59. doi: 10.1093/hmg/ddm018. [DOI] [PubMed] [Google Scholar]
- 24.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–1506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 25.Southern EM. DNA microarrays. History and overview. Methods Mol Biol. 2001;170:1–15. doi: 10.1385/1-59259-234-1:1. [DOI] [PubMed] [Google Scholar]
- 26.Bennett S. Solexa Ltd. Pharmacogenomics. 2004;5:433–438. doi: 10.1517/14622416.5.4.433. [DOI] [PubMed] [Google Scholar]
- 27.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Godwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer ML, Jarvie TP, Jirage KB, Kim JB, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li J, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate multiplex polony sequencing of an evolved bacterial genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 29.Mardis ER. Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]
- 30.Irizarry RA, Ladd-Acosta C, Carvalho B, Wu H, Brandenburg SA, Jeddeloh JA, Wen B, Feinberg AP. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Irizarry RA, Ladd-Acosta C, Wen B, Wu Z, Montano C, Onyango P, Cui H, Gabo K, Rongione M, Webster M, Ji H, Potash JB, Sabunciyan S, Feinberg AP. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–186. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific enzymatic amplification of DNA in vitro: the polymerase chain reaction. Cold Spring Harb Symp Quant Biol 51 Pt. 1986;1:263–273. doi: 10.1101/sqb.1986.051.01.032. [DOI] [PubMed] [Google Scholar]
- 33.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18:6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navarro JM, Jorcano JL. The use of arbitrarily primed polymerase chain reaction in cancer research. Electrophoresis. 1999;20:283–290. doi: 10.1002/(SICI)1522-2683(19990201)20:2<283::AID-ELPS283>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 36.Arribas R, Capella G, Tortola S, Masramon L, Grizzle WE, Perucho M, Peinado MA. Assessment of genomic damage in colorectal cancer by DNA fingerprinting: prognostic applications. J Clin Oncol. 1997;15:3230–3240. doi: 10.1200/JCO.1997.15.10.3230. [DOI] [PubMed] [Google Scholar]
- 37.Risques RA, Moreno V, Ribas M, Marcuello E, Capella G, Peinado MA. Genetic pathways and genome-wide determinants of clinical outcome in colorectal cancer. Cancer Res. 2003;63:7206–7214. [PubMed] [Google Scholar]
- 38.Vogt T, Stolz W, Landthaler M, Ruschoff J, Schlegel J. Nonradioactive arbitrarily primed polymerase chain reaction: a novel technique for detecting genetic defects in skin tumors. J Invest Dermatol. 1996;106:194–197. doi: 10.1111/1523-1747.ep12329949. [DOI] [PubMed] [Google Scholar]
- 39.Yasuda J, Kashiwabara H, Kawakami K, Uematsu K, Sugano K, Perucho M, Sekiya T. Detection of microsatellite instability in cancers by arbitrarily primed-PCR fingerprinting using a fluorescently labeled primer (FAP-PCR) Biol Chem. 1996;377:563–570. doi: 10.1515/bchm3.1996.377.9.563. [DOI] [PubMed] [Google Scholar]
- 40.Dil A, Misra A, Sulaiman IM, Sinha S, Sarkar C, Mahapatra AK, Hasnain SE. Genetic alterations in brain tumors identified by RAPD analysis. Gene. 1998;206:45–48. doi: 10.1016/s0378-1119(97)00579-9. [DOI] [PubMed] [Google Scholar]
- 41.Ong TM, Song B, Qian HW, Wu ZL, Whong WZ. Detection of genomic instability in lung cancer tissues by random amplified polymorphic DNA analysis. Carcinogenesis. 1998;19:233–235. doi: 10.1093/carcin/19.1.233. [DOI] [PubMed] [Google Scholar]
- 42.Wang J, Wang Q, Ye F. Genetic instability in cancer tissues analyzed by random amplified polymorphic DNA PCR. Chin Med J (Engl) 2002;115:430–432. [PubMed] [Google Scholar]
- 43.Atienzar FA, Jha AN. The random amplified polymorphic DNA (RAPD) assay and related techniques applied to genotoxicity and carcinogenesis studies: a critical review. Mutat Res. 2006;613:76–102. doi: 10.1016/j.mrrev.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 44.Peinado MA, Malkhosyan S, Velazquez A, Perucho M. Isolation and characterization of allelic losses and gains in colorectal tumors by arbitrarily primed polymerase chain reaction. Proc Natl Acad Sci U S A. 1992;89:10065–10069. doi: 10.1073/pnas.89.21.10065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohno T, Morishita K, Takano H, Shapiro DN, Yokota J. Homozygous deletion at chromosome 2q33 in human small-cell lung carcinoma identified by arbitrarily primed PCR genomic fingerprinting. Oncogene. 1994;9:103–108. [PubMed] [Google Scholar]
- 46.Malkhosyan S, Yasuda J, Soto JL, Sekiya T, Yokota J, Perucho M. Molecular karyotype (amplotype) of metastatic colorectal cancer by unbiased arbitrarily primed PCR DNA fingerprinting. Proc Natl Acad Sci U S A. 1998;95:10170–10175. doi: 10.1073/pnas.95.17.10170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piao Z, Lee KS, Kim H, Perucho M, Malkhosyan S. Identification of novel deletion regions on chromosome arms 2q and 6p in breast carcinomas by amplotype analysis. Genes Chromosomes Cancer. 2001;30:113–122. [PubMed] [Google Scholar]
- 48.Achille A, Biasi MO, Zamboni G, Bogina G, Magalini AR, Pederzoli P, Perucho M, Scarpa A. Chromosome 7q allelic losses in pancreatic carcinoma. Cancer Res. 1996;56:3808–3813. [PubMed] [Google Scholar]
- 49.Suzuki K, Ohnami S, Tanabe C, Sasaki H, Yasuda J, Katai H, Yoshimura K, Terada M, Perucho M, Yoshida T. The genomic damage estimated by arbitrarily primed PCR DNA fingerprinting is useful for the prognosis of gastric cancer. Gastroenterology. 2003;125:1330–1340. doi: 10.1016/j.gastro.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 50.Suzuki K, Suzuki I, Leodolter A, Alonso S, Horiuchi S, Yamashita K, Perucho M. Global DNA demethylation in gastrointestinal cancer is age dependent and precedes genomic damage. Cancer Cell. 2006;9:199–207. doi: 10.1016/j.ccr.2006.02.016. [DOI] [PubMed] [Google Scholar]
- 51.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 52.Perucho M. Cancer of the microsatellite mutator phenotype. Biol Chem. 1996;377:675–684. [PubMed] [Google Scholar]
- 53.Leach FS, Nicolaides NC, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen LA, Nystrom-Lahti M, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 54.Papadopoulos N, Nicolaides NC, Wei YF, Ruben SM, Carter KC, Rosen CA, Haseltine WA, Fleischmann RD, Fraser CM, Adams MD, et al. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 55.Kolodner RD, Marsischky GT. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 56.Yamashita K, Dai T, Dai Y, Yamamoto F, Perucho M. Genetics supersedes epigenetics in colon cancer phenotype. Cancer Cell. 2003;4:121–131. doi: 10.1016/s1535-6108(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 57.Perucho M, Peinado MA, Ionov Y, Casares S, Malkhosyan S, Stanbridge E. Defects in replication fidelity of simple repeated sequences reveal a new mutator mechanism for oncogenesis. Cold Spring Harb Symp Quant Biol. 1994;59:339–348. doi: 10.1101/sqb.1994.059.01.038. [DOI] [PubMed] [Google Scholar]
- 58.Cahill DP, Kinzler KW, Vogelstein B, Lengauer C. Genetic instability and darwinian selection in tumours. Trends Cell Biol. 1999;9:M57–60. [PubMed] [Google Scholar]
- 59.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 60.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 61.Yamamoto H, Sawai H, Perucho M. Frameshift somatic mutations in gastrointestinal cancer of the microsatellite mutator phenotype. Cancer Res. 1997;57:4420–4426. [PubMed] [Google Scholar]
- 62.Lindblom A. Different mechanisms in the tumorigenesis of proximal and distal colon cancers. Curr Opin Oncol. 2001;13:63–69. doi: 10.1097/00001622-200101000-00013. [DOI] [PubMed] [Google Scholar]
- 63.Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- 64.Konishi M, Kikuchi-Yanoshita R, Tanaka K, Muraoka M, Onda A, Okumura Y, Kishi N, Iwama T, Mori T, Koike M, Ushio K, Chiba M, Nomizu S, Konishi F, Utsunomiya J, Miyaki M. Molecular nature of colon tumors in hereditary nonpolyposis colon cancer, familial polyposis, and sporadic colon cancer. Gastroenterology. 1996;111:307–317. doi: 10.1053/gast.1996.v111.pm8690195. [DOI] [PubMed] [Google Scholar]
- 65.Breivik J, Lothe RA, Meling GI, Rognum TO, Borresen-Dale AL, Gaudernack G. Different genetic pathways to proximal and distal colorectal cancer influenced by sex-related factors. Int J Cancer. 1997;74:664–669. doi: 10.1002/(sici)1097-0215(19971219)74:6<664::aid-ijc18>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 66.Olschwang S, Hamelin R, Laurent-Puig P, Thuille B, De Rycke Y, Li YJ, Muzeau F, Girodet J, Salmon RJ, Thomas G. Alternative genetic pathways in colorectal carcinogenesis. Proc Natl Acad Sci U S A. 1997;94:12122–12127. doi: 10.1073/pnas.94.22.12122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Woerner SM, Benner A, Sutter C, Schiller M, Yuan YP, Keller G, Bork P, Doeberitz MK, Gebert JF. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative Real Common Target genes. Oncogene. 2003;22:2226–2235. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- 68.Royrvik EC, Ahlquist T, Rognes T, Lothe RA. Slip slidin' away: a duodecennial review of targeted genes in mismatch repair deficient colorectal cancer. Crit Rev Oncog. 2007;13:229–257. doi: 10.1615/critrevoncog.v13.i3.20. [DOI] [PubMed] [Google Scholar]
- 69.Perucho M. Microsatellite instability: the mutator that mutates the other mutator. Nat Med. 1996;2:630–631. doi: 10.1038/nm0696-630. [DOI] [PubMed] [Google Scholar]
- 70.Suzuki K, Dai T, Suzuki I, Dai Y, Yamashita K, Perucho M. Low mutation incidence in polymorphic noncoding short mononucleotide repeats in gastrointestinal cancer of the microsatellite mutator phenotype pathway. Cancer Res. 2002;62:1961–1965. [PubMed] [Google Scholar]
- 71.Yasuda J, Navarro JM, Malkhosyan S, Velazquez A, Arribas R, Sekiya T, Perucho M. Chromosomal assignment of human DNA fingerprint sequences by simultaneous hybridization to arbitrarily primed PCR products from human/rodent monochromosome cell hybrids. Genomics. 1996;34:1–8. doi: 10.1006/geno.1996.0235. [DOI] [PubMed] [Google Scholar]
- 72.Malkhosyan SR, Baranovskaya S. Comparative Hybridization of AP-PCR Arrays, New Method for Analysis of Single Copy Chromosomal Losses and Gains in Cancer Cell Genome Innovative Molecular Analysis Technologies. 2nd Principal Investigators Meeting, Riitz-Carlton; Washington, DC. 2001. [Google Scholar]
- 73.Barrett MT, Scheffer A, Ben-Dor A, Sampas N, Lipson D, Kincaid R, Tsang P, Curry B, Baird K, Meltzer PS, Yakhini Z, Bruhn L, Laderman S. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc Natl Acad Sci U S A. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vogelstein B, Fearon ER, Kern SE, Hamilton SR, Preisinger AC, Nakamura Y, White R. Allelotype of colorectal carcinomas. Science. 1989;244:207–211. doi: 10.1126/science.2565047. [DOI] [PubMed] [Google Scholar]
- 75.Trautmann K, Terdiman JP, French AJ, Roydasgupta R, Sein N, Kakar S, Fridlyand J, Snijders AM, Albertson DG, Thibodeau SN, Waldman FM. Chromosomal instability in microsatellite-unstable and stable colon cancer. Clin Cancer Res. 2006;12:6379–6385. doi: 10.1158/1078-0432.CCR-06-1248. [DOI] [PubMed] [Google Scholar]
- 76.du Manoir S, Speicher MR, Joos S, Schrock E, Popp S, Dohner H, Kovacs G, Robert-Nicoud M, Lichter P, Cremer T. Detection of complete and partial chromosome gains and losses by comparative genomic in situ hybridization. Hum Genet. 1993;90:590–610. doi: 10.1007/BF00202476. [DOI] [PubMed] [Google Scholar]
- 77.Kallioniemi OP, Kallioniemi A, Sudar D, Rutovitz D, Gray JW, Waldman F, Pinkel D. Comparative genomic hybridization: a rapid new method for detecting and mapping DNA amplification in tumors. Semin Cancer Biol. 1993;4:41–46. [PubMed] [Google Scholar]
- 78.Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D. Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer. 1994;10:231–243. doi: 10.1002/gcc.2870100403. [DOI] [PubMed] [Google Scholar]
- 79.Lisitsyn N, Wigler M. Cloning the differences between two complex genomes. Science. 1993;259:946–951. doi: 10.1126/science.8438152. [DOI] [PubMed] [Google Scholar]
- 80.Lisitsyn NA, Lisitsina NM, Dalbagni G, Barker P, Sanchez CA, Gnarra J, Linehan WM, Reid BJ, Wigler MH. Comparative genomic analysis of tumors: detection of DNA losses and amplification. Proc Natl Acad Sci U S A. 1995;92:151–155. doi: 10.1073/pnas.92.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lucito R, West J, Reiner A, Alexander J, Esposito D, Mishra B, Powers S, Norton L, Wigler M. Detecting gene copy number fluctuations in tumor cells by microarray analysis of genomic representations. Genome Res. 2000;10:1726–1736. doi: 10.1101/gr.138300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, Jeffrey SS, Botstein D, Brown PO. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 83.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 84.Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, Hamilton G, Hindle AK, Huey B, Kimura K, Law S, Myambo K, Palmer J, Ylstra B, Yue JP, Gray JW, Jain AN, Pinkel D, Albertson DG. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29:263–264. doi: 10.1038/ng754. [DOI] [PubMed] [Google Scholar]
- 85.Pinkel D, Albertson DG. Comparative genomic hybridization. Annu Rev Genomics Hum Genet. 2005;6:331–354. doi: 10.1146/annurev.genom.6.080604.162140. [DOI] [PubMed] [Google Scholar]
- 86.Ishkanian AS, Malloff CA, Watson SK, DeLeeuw RJ, Chi B, Coe BP, Snijders A, Albertson DG, Pinkel D, Marra MA, Ling V, MacAulay C, Lam WL. A tiling resolution DNA microarray with complete coverage of the human genome. Nat Genet. 2004;36:299–303. doi: 10.1038/ng1307. [DOI] [PubMed] [Google Scholar]
- 87.Kallioniemi A. CGH microarrays and cancer. Curr Opin Biotechnol. 2008;19:36–40. doi: 10.1016/j.copbio.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 88.Thompson CL, Plummer SJ, Acheson LS, Tucker TC, Casey G, Li L. Association of common genetic variants in SMAD7 and risk of colon cancer. Carcinogenesis. 2009;30:982–986. doi: 10.1093/carcin/bgp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. doi: 10.1093/nar/28.8.e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Eads CA, Laird PW. Combined bisulfite restriction analysis (COBRA) Methods Mol Biol. 2002;200:71–85. doi: 10.1385/1-59259-182-5:071. [DOI] [PubMed] [Google Scholar]
- 93.Bibikova M, Lin Z, Zhou L, Chudin E, Garcia EW, Wu B, Doucet D, Thomas NJ, Wang Y, Vollmer E, Goldmann T, Seifart C, Jiang W, Barker DL, Chee MS, Floros J, Fan JB. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–393. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fraga MF, Esteller M. DNA methylation: a profile of methods and applications. Biotechniques. 2002;33:632, 634, 636–649. doi: 10.2144/02333rv01. [DOI] [PubMed] [Google Scholar]
- 95.Zuo T, Tycko B, Liu TM, Lin HJ, Huang TH. Methods in DNA methylation profiling. Epigenomics. 2009;1:331–345. doi: 10.2217/epi.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hatada I, Hayashizaki Y, Hirotsune S, Komatsubara H, Mukai T. A genomic scanning method for higher organisms using restriction sites as landmarks. Proc Natl Acad Sci U S A. 1991;88:9523–9527. doi: 10.1073/pnas.88.21.9523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hayashizaki Y, Hirotsune S, Okazaki Y, Hatada I, Shibata H, Kawai J, Hirose K, Watanabe S, Fushiki S, Wada S, et al. Restriction landmark genomic scanning method and its various applications. Electrophoresis. 1993;14:251–258. doi: 10.1002/elps.1150140145. [DOI] [PubMed] [Google Scholar]
- 98.Rush LJ, Plass C. Restriction landmark genomic scanning for DNA methylation in cancer: past, present, and future applications. Anal Biochem. 2002;307:191–201. doi: 10.1016/s0003-2697(02)00033-7. [DOI] [PubMed] [Google Scholar]
- 99.Matsuyama T, Kimura MT, Koike K, Abe T, Nakano T, Asami T, Ebisuzaki T, Held WA, Yoshida S, Nagase H. Global methylation screening in the Arabidopsis thaliana and Mus musculus genome: applications of virtual image restriction landmark genomic scanning (Vi-RLGS) Nucleic Acids Res. 2003;31:4490–4496. doi: 10.1093/nar/gkg488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rouillard JM, Erson AE, Kuick R, Asakawa J, Wimmer K, Muleris M, Petty EM, Hanash S. Virtual genome scan: a tool for restriction landmark-based scanning of the human genome. Genome Res. 2001;11:1453–1459. doi: 10.1101/gr.181601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zardo G, Tiirikainen MI, Hong C, Misra A, Feuerstein BG, Volik S, Collins CC, Lamborn KR, Bollen A, Pinkel D, Albertson DG, Costello JF. Integrated genomic and epigenomic analyses pinpoint biallelic gene inactivation in tumors. Nat Genet. 2002;32:453–458. doi: 10.1038/ng1007. [DOI] [PubMed] [Google Scholar]
- 102.Ho SM, Tang WY. Techniques used in studies of epigenome dysregulation due to aberrant DNA methylation: an emphasis on fetal-based adult diseases. Reprod Toxicol. 2007;23:267–282. doi: 10.1016/j.reprotox.2007.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim YS, Yoo HS, Lee KT, Goh SH, Jung JS, Oh SW, Baba M, Yasuda T, Matsubara K, Nagai H. Detection of genetic alterations in the human gastric cancer cell lines by two-dimensional analysis of genomic DNA. Int J Oncol. 2000;17:297–308. [PubMed] [Google Scholar]
- 104.Dai Z, Lakshmanan RR, Zhu WG, Smiraglia DJ, Rush LJ, Fruhwald MC, Brena RM, Li B, Wright FA, Ross P, Otterson GA, Plass C. Global methylation profiling of lung cancer identifies novel methylated genes. Neoplasia. 2001;3:314–323. doi: 10.1038/sj.neo.7900162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dai Z, Zhu WG, Morrison CD, Brena RM, Smiraglia DJ, Raval A, Wu YZ, Rush LJ, Ross P, Molina JR, Otterson GA, Plass C. A comprehensive search for DNA amplification in lung cancer identifies inhibitors of apoptosis cIAP1 and cIAP2 as candidate oncogenes. Hum Mol Genet. 2003;12:791–801. doi: 10.1093/hmg/ddg083. [DOI] [PubMed] [Google Scholar]
- 106.Motiwala T, Ghoshal K, Das A, Majumder S, Weichenhan D, Wu YZ, Holman K, James SJ, Jacob ST, Plass C. Suppression of the protein tyrosine phosphatase receptor type O gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene. 2003;22:6319–6331. doi: 10.1038/sj.onc.1206750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu R, Lin L, Beer DG, Ellenson LH, Lamb BJ, Rouillard JM, Kuick R, Hanash S, Schwartz DR, Fearon ER, Cho KR. Amplification and overexpression of the L-MYC proto-oncogene in ovarian carcinomas. Am J Pathol. 2003;162:1603–1610. doi: 10.1016/S0002-9440(10)64294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Park J, Brena RM, Gruidl M, Zhou J, Huang T, Plass C, Tockman MS. CpG island hypermethylation profiling of lung cancer using restriction landmark genomic scanning (RLGS) analysis. Cancer Biomark. 2005;1:193–200. doi: 10.3233/cbm-2005-12-307. [DOI] [PubMed] [Google Scholar]
- 109.Asaga S, Ueda M, Jinno H, Kikuchi K, Itano O, Ikeda T, Kitajima M. Identification of a new breast cancer-related gene by restriction landmark genomic scanning. Anticancer Res. 2006;26:35–42. [PubMed] [Google Scholar]
- 110.Smith JF, Mahmood S, Song F, Morrow A, Smiraglia D, Zhang X, Rajput A, Higgins MJ, Krumm A, Petrelli NJ, Costello JF, Nagase H, Plass C, Held WA. Identification of DNA methylation in 3' genomic regions that are associated with upregulation of gene expression in colorectal cancer. Epigenetics. 2007;2:161–172. doi: 10.4161/epi.2.3.4805. [DOI] [PubMed] [Google Scholar]
- 111.Gonzalgo ML, Liang G, Spruck CH, 3rd, Zingg JM, Rideout WM, 3rd, Jones PA. Identification and characterization of differentially methylated regions of genomic DNA by methylation-sensitive arbitrarily primed PCR. Cancer Res. 1997;57:594–599. [PubMed] [Google Scholar]
- 112.Huang TH, Laux DE, Hamlin BC, Tran P, Tran H, Lubahn DB. Identification of DNA methylation markers for human breast carcinomas using the methylation-sensitive restriction fingerprinting technique. Cancer Res. 1997;57:1030–1034. [PubMed] [Google Scholar]
- 113.Kohno T, Kawanishi M, Inazawa J, Yokota J. Identification of CpG islands hypermethylated in human lung cancer by the arbitrarily primed-PCR method. Hum Genet. 1998;102:258–264. doi: 10.1007/s004390050689. [DOI] [PubMed] [Google Scholar]
- 114.Liang G, Robertson KD, Talmadge C, Sumegi J, Jones PA. The gene for a novel transmembrane protein containing epidermal growth factor and follistatin domains is frequently hypermethylated in human tumor cells. Cancer Res. 2000;60:4907–4912. [PubMed] [Google Scholar]
- 115.Salem CE, Markl ID, Bender CM, Gonzales FA, Jones PA, Liang G. PAX6 methylation and ectopic expression in human tumor cells. Int J Cancer. 2000;87:179–185. doi: 10.1002/1097-0215(20000715)87:2<179::aid-ijc4>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 116.Markl ID, Cheng J, Liang G, Shibata D, Laird PW, Jones PA. Global and gene-specific epigenetic patterns in human bladder cancer genomes are relatively stable in vivo and in vitro over time. Cancer Res. 2001;61:5875–5884. [PubMed] [Google Scholar]
- 117.Lo KW, Tsang YS, Kwong J, To KF, Teo PM, Huang DP. Promoter hypermethylation of the EDNRB gene in nasopharyngeal carcinoma. Int J Cancer. 2002;98:651–655. doi: 10.1002/ijc.10271. [DOI] [PubMed] [Google Scholar]
- 118.Bahar A, Simpson DJ, Cutty SJ, Bicknell JE, Hoban PR, Holley S, Mourtada-Maarabouni M, Williams GT, Clayton RN, Farrell WE. Isolation and characterization of a novel pituitary tumor apoptosis gene. Mol Endocrinol. 2004;18:1827–1839. doi: 10.1210/me.2004-0087. [DOI] [PubMed] [Google Scholar]
- 119.Wu M, Ho SM. PMP24, a gene identified by MSRF, undergoes DNA hypermethylation-associated gene silencing during cancer progression in an LNCaP model. Oncogene. 2004;23:250–259. doi: 10.1038/sj.onc.1207076. [DOI] [PubMed] [Google Scholar]
- 120.Costa FF, Paixao VA, Cavalher FP, Ribeiro KB, Cunha IW, Rinck JA, Jr, O'Hare M, Mackay A, Soares FA, Brentani RR, Camargo AA. SATR-1 hypomethylation is a common and early event in breast cancer. Cancer Genet Cytogenet. 2006;165:135–143. doi: 10.1016/j.cancergencyto.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 121.Estecio MR, Youssef EM, Rahal P, Fukuyama EE, Gois-Filho JF, Maniglia JV, Goloni-Bertollo EM, Issa JP, Tajara EH. LHX6 is a sensitive methylation marker in head and neck carcinomas. Oncogene. 2006;25:5018–5026. doi: 10.1038/sj.onc.1209509. [DOI] [PubMed] [Google Scholar]
- 122.Lv Z, Zhang M, Bi J, Xu F, Hu S, Wen J. Promoter hypermethylation of a novel gene, ZHX2, in hepatocellular carcinoma. Am J Clin Pathol. 2006;125:740–746. doi: 10.1309/09B4-52V7-R76K-7D6K. [DOI] [PubMed] [Google Scholar]
- 123.Tryndyak V, Kovalchuk O, Pogribny IP. Identification of differentially methylated sites within unmethylated DNA domains in normal and cancer cells. Anal Biochem. 2006;356:202–207. doi: 10.1016/j.ab.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 124.Kuznetsova EB, Kekeeva TV, Larin SS, Zemlyakova VV, Khomyakova AV, Babenko OV, Nemtsova MV, Zaletayev DV, Strelnikov VV. Methylation of the BIN1 gene promoter CpG island associated with breast and prostate cancer. J Carcinog. 2007;6:9. doi: 10.1186/1477-3163-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhao BJ, Tan SN, Cui Y, Sun DG, Ma X. Aberrant promoter methylation of the TPEF gene in esophageal squamous cell carcinoma. Dis Esophagus. 2008;21:582–588. doi: 10.1111/j.1442-2050.2007.00808.x. [DOI] [PubMed] [Google Scholar]
- 126.Liu WB, Liu JY, Ao L, Zhou ZY, Zhou YH, Cui ZH, Yang H, Cao J. Dynamic changes in DNA methylation during multistep rat lung carcinogenesis induced by 3-methylcholanthrene and diethylnitrosamine. Toxicol Lett. 2009;189:5–13. doi: 10.1016/j.toxlet.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 127.Zhao BJ, Sun DG, Zhang M, Tan SN, Ma X. Identification of aberrant promoter methylation of EDNRB gene in esophageal squamous cell carcinoma. Dis Esophagus. 2009;22:55–61. doi: 10.1111/j.1442-2050.2008.00848.x. [DOI] [PubMed] [Google Scholar]
- 128.Frigola J, Ribas M, Risques RA, Peinado MA. Methylome profiling of cancer cells by amplification of inter-methylated sites (AIMS) Nucleic Acids Res. 2002;30:e28. doi: 10.1093/nar/30.7.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Toyota M, Ho C, Ahuja N, Jair KW, Li Q, Ohe-Toyota M, Baylin SB, Issa JP. Identification of differentially methylated sequences in colorectal cancer by methylated CpG island amplification. Cancer Res. 1999;59:2307–2312. [PubMed] [Google Scholar]
- 130.Paz MF, Wei S, Cigudosa JC, Rodriguez-Perales S, Peinado MA, Huang TH, Esteller M. Genetic unmasking of epigenetically silenced tumor suppressor genes in colon cancer cells deficient in DNA methyltransferases. Hum Mol Genet. 2003;12:2209–2219. doi: 10.1093/hmg/ddg226. [DOI] [PubMed] [Google Scholar]
- 131.Sadikovic B, Haines TR, Butcher DT, Rodenhiser DI. Chemically induced DNA hypomethylation in breast carcinoma cells detected by the amplification of intermethylated sites. Breast Cancer Res. 2004;6:R329–337. doi: 10.1186/bcr799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Frigola J, Sole X, Paz MF, Moreno V, Esteller M, Capella G, Peinado MA. Differential DNA hypermethylation and hypomethylation signatures in colorectal cancer. Hum Mol Genet. 2005;14:319–326. doi: 10.1093/hmg/ddi028. [DOI] [PubMed] [Google Scholar]
- 133.Frigola J, Song J, Stirzaker C, Hinshelwood RA, Peinado MA, Clark SJ. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–549. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 134.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, Boix-Chornet M, Sanchez-Aguilera A, Ling C, Carlsson E, Poulsen P, Vaag A, Stephan Z, Spector TD, Wu YZ, Plass C, Esteller M. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Yamamoto F, Yamamoto M, Soto JL, Kojima E, Wang EN, Perucho M, Sekiya T, Yamanaka H. Notl-Msell methylation-sensitive amplied fragment length polymorhism for DNA methylation analysis of human cancers. Electrophoresis. 2001;22:1946–1956. doi: 10.1002/1522-2683(200106)22:10<1946::AID-ELPS1946>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 136.Vos P, Hogers R, Bleeker M, Reijans M, van de Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kageyama S, Shinmura K, Yamamoto H, Goto M, Suzuki K, Tanioka F, Tsuneyoshi T, Sugimura H. Fluorescence-labeled methylation-sensitive amplified fragment length polymorphism (FL-MS-AFLP) analysis for quantitative determination of DNA methylation and demethylation status. Jpn J Clin Oncol. 2008;38:317–322. doi: 10.1093/jjco/hyn021. [DOI] [PubMed] [Google Scholar]
- 138.Yamamoto F, Yamamoto M. A DNA microarray-based methylation-sensitive (MS)-AFLP hybridization method for genetic and epigenetic analyses. Mol Genet Genomics. 2004;271:678–686. doi: 10.1007/s00438-004-1017-5. [DOI] [PubMed] [Google Scholar]
- 139.Albertson DG, Ylstra B, Segraves R, Collins C, Dairkee SH, Kowbel D, Kuo WL, Gray JW, Pinkel D. Quantitative mapping of amplicon structure by array CGH identifies CYP24 as a candidate oncogene. Nat Genet. 2000;25:144–146. doi: 10.1038/75985. [DOI] [PubMed] [Google Scholar]
- 140.Kutsenko AS, Gizatullin RZ, Al-Amin AN, Wang F, Kvasha SM, Podowski RM, Matushkin YG, Gyanchandani A, Muravenko OV, Levitsky VG, Kolchanov NA, Protopopov AI, Kashuba VI, Kisselev LL, Wasserman W, Wahlestedt C, Zabarovsky ER. NotI flanking sequences: a tool for gene discovery and verification of the human genome. Nucleic Acids Res. 2002;30:3163–3170. doi: 10.1093/nar/gkf428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ahuja N, Mohan AL, Li Q, Stolker JM, Herman JG, Hamilton SR, Baylin SB, Issa JP. Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res. 1997;57:3370–3374. [PubMed] [Google Scholar]
- 142.Weisenberger DJ, Siegmund KD, Campan M, Young J, Long TI, Faasse MA, Kang GH, Widschwendter M, Weener D, Buchanan D, Koh H, Simms L, Barker M, Leggett B, Levine J, Kim M, French AJ, Thibodeau SN, Jass J, Haile R, Laird PW. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 143.Issa JP, Shen L, Toyota M. CIMP, at last. Gastroenterology. 2005;129:1121–1124. doi: 10.1053/j.gastro.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 144.Issa JP. Colon cancer: it's CIN or CIMP. Clin Cancer Res. 2008;14:5939–5940. doi: 10.1158/1078-0432.CCR-08-1596. [DOI] [PubMed] [Google Scholar]
- 145.Perucho M, Suzuki K. Reply to Commentary. New insights into a controversial topic: the methylation-cancer connection. Gastroenterology. 2007;132:2070–2071. doi: 10.1053/j.gastro.2007.03.089. [DOI] [PubMed] [Google Scholar]
- 146.Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden JM, Bollen M, Esteller M, Di Croce L, de Launoit Y, Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 147.Schlesinger Y, Straussman R, Keshet I, Farkash S, Hecht M, Zimmerman J, Eden E, Yakhini Z, Ben-Shushan E, Reubinoff BE, Bergman Y, Simon I, Cedar H. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat Genet. 2007;39:232–236. doi: 10.1038/ng1950. [DOI] [PubMed] [Google Scholar]
- 148.Widschwendter M, Fiegl H, Egle D, Mueller-Holzner E, Spizzo G, Marth C, Weisenberger DJ, Campan M, Young J, Jacobs I, Laird PW. Epigenetic stem cell signature in cancer. Nat Genet. 2007;39:157–158. doi: 10.1038/ng1941. [DOI] [PubMed] [Google Scholar]
- 149.Kane MF, Loda M, Gaida GM, Lipman J, Mishra R, Goldman H, Jessup JM, Kolodner R. Methylation of the hMLH1 promoter correlates with lack of expression of hMLH1 in sporadic colon tumors and mismatch repair-defective human tumor cell lines. Cancer Res. 1997;57:808–811. [PubMed] [Google Scholar]
- 150.Veigl ML, Kasturi L, Olechnowicz J, Ma AH, Lutterbaugh JD, Periyasamy S, Li GM, Drummond J, Modrich PL, Sedwick WD, Markowitz SD. Biallelic inactivation of hMLH1 by epigenetic gene silencing, a novel mechanism causing human MSI cancers. Proc Natl Acad Sci U S A. 1998;95:8698–8702. doi: 10.1073/pnas.95.15.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]