Abstract
Multistep synthesis in the laboratory typically requires numerous reaction vessels, each containing a different set of reactants. In contrast, cells are capable of performing highly efficient and selective multistep biosynthesis under mild conditions with all reactants simultaneously present in solution. If the latter approach could be applied in the laboratory, it may improve the ease, speed, and efficiency of multistep reaction sequences. Here we show that a DNA mechanical device— a DNA walker moving along a DNA track— can be used to perform a series of amine acylation reactions in a single solution without any external intervention. The multistep products generated by this primitive ribosome mimetic are programmed by the sequence of the DNA track, are unrelated to the structure of DNA, and are formed with speeds and overall yields significantly greater than those previously achieved by multistep DNA-templated small-molecule synthesis.
The exquisite selectivity and efficiency of many biosynthetic pathways rely on the use of protein or nucleic acid templates to modulate the effective molarities of substrates. For combinatorial biosyntheses including mRNA-templated peptide synthesis during ribosomal translation1-2 as well as pathways that generate polyketides and non-ribosomal peptides,3-4 this strategy enables the sequence of building blocks in a biosynthetic product to be determined by the sequence of mRNA nucleotides or enzyme modules in the template, enabling different multistep reaction products to be generated selectively from a single set of substrates with no external intervention.
In the absence of enzymes, laboratory reaction sequences to generate multistep synthetic products generally proceed in multiple reaction vessels, such that each vessel exposes a different set of substrates to a different set of reaction conditions. In principle, performing multistep reaction sequences in a single solution programmed by a template could significantly increase the ease, speed, and overall efficiency of multistep syntheses. We previously engineered a DNA template that undergoes a series of secondary structure changes when subjected to a schedule of increasing temperatures.5 These changes expose hybridization sites for DNA-templated synthesis, an approach to controlling chemical reactivity in which reactions between DNA-linked reagents are triggered by DNA hybridization, enabling a multistep reaction sequence to take place in one solution. While successful, the generality of this approach is significantly limited by requiring a different arrangement of substrates for each step and requiring large changes in temperature during the reaction sequence (for example, 4 °C to 72 °C).5
Recent advances in engineering DNA-based devices6-9 have resulted in nanometer-scale DNA machines that are capable of autonomously changing their physical location over time.10-16 For example, Mao and coworkers recently reported a DNA “nanowalker” that moves unidirectionally along a DNA track from station to station.12 We envisioned that the ability of a DNA walker to translocate could be integrated with DNA-templated synthesis to enable a specific chemical reaction to take place upon arrival of the walker at each station. Because the reaction product remains linked to the walker and serves as the starting material for the next reaction, this strategy results in a progressively more advanced synthetic reaction product as the walker moves along its track. Here we report the development of an autonomous DNA walker that performs a series of amine acylation reactions as it moves from station to station along a DNA track. The resulting machine mediates multistep synthesis of an oligoamide in a single isothermal solution programmed by the sequence of a DNA track, conceptually resembling an artificial ribosome. The system can generate multistep products in overall yields that are much higher than those of previous DNA-templated small-molecule syntheses,5,18 and in total reaction times of only a few hours. This work also represents one of the first DNA devices that manipulates the covalent structure of non-nucleic acid molecules,17,19-20 and thus expands the functional scope of DNA-based devices.
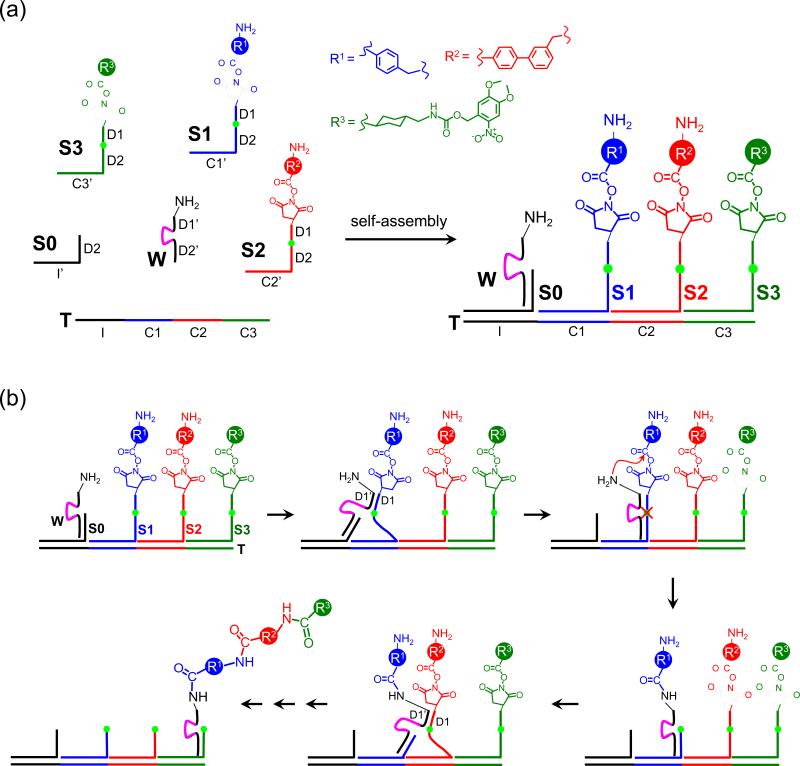
The DNA-based ribosome mimetic (“DNAsome”) system (Fig. 1a) consists of four sets of components. (i) The track (T), analogous to an mRNA, is a long single-stranded DNA molecule that contains an initiation site (I) followed by codon sites (C1, C2, C3, etc.) that each encodes a different substrate (S1, S2, S3, etc.). (ii) The substrates, analogous to aminoacylated tRNAs, each contain (from 5’ to 3’) a synthetic small-molecule building block, a docking region (D1) of constant sequence, two consecutive central RNA nucleotides, a second constant docking region (D2), and a unique 21-nucleotide anticodon complementary to a template codon C1, C2, or C3. In the case of the system described here, the small-molecule reagents are amino acid N-hydroxysuccinimidyl (NHS) esters that cannot undergo intramolecular cyclization due to the enforced separation of amine and ester groups. (iii) The initiator (S0) is a truncated substrate that contains only D2 followed by an anticodon complementary to site I of the template. (iv) The DNA walker (W) is a DNA 38-mer containing a 5’ region complementary to D2 (D2’), the 10-23 RNA-cleaving DNAzyme evolved by Joyce and coworkers,21 a region complementary to D1 (D1’), and a 3’ primary amine group. The interval between any two adjacent stations corresponds to two turns of B-form DNA, which defines each walking step as ~7 nm.
Figure 1.
Overview of the DNAsome system. (a) The system described in this work comprises six DNA or DNA-linked molecules. Three substrates (S1-S3) and an initiator (S0) can hybridize on a single-stranded DNA track (T). Each substrate has an amino acid NHS ester at its 5’ end and two ribonucleotides (green dot) in the middle of its DNA sequence. The DNA walker (W) contains a 3’ amine group and an RNA-cleaving DNAzyme (purple line) that can cleave the ribonucleotides in the substrates. (b) DNAsome-mediated multistep synthesis of a triamide product. All steps take place in a single solution under one set of reaction conditions without external intervention.
When all components except the walker and initiator are combined in one solution, the substrates and templates self-assemble into a double-stranded complex. Upon addition of S0 and W, the initiator and walker associate via D2:D2’ hybridization and localize to the track-substrate complex through S0:I hybridization (Fig. 1a). The template-bound walker then hybridizes with the nearest D1 docking region, which is found on the S1 substrate annealed to the first station (Fig. 1b). Hybridization between W and S1 induces the favorable translocation of W from S0 (which can hybridize only to the D2’ region of W) to S1 (which can hybridize to both the D1’ and D2’ regions of W). W-S1 hybridization also triggers DNA-templated acylation of the walker's amine group with the NHS ester of S1, resulting in transfer of the first amino acid building block from S1 to W. The 10-23 DNAzyme core in the walker cleaves the ribonucleotide linkage in S1, allowing the 5’ fragment of S1 to dissociate (Fig. 1b). The system now is identical to the W:S0:T starting state, except W has captured the first reaction product and has translocated from the initiation site to the first station.
Two subsequent cycles of walker translocation, DNA-templated amine acylation, DNAzyme-catalyzed cleavage, and dissociation of the 5’ fragment of the expended substrate result in the walker resting at the final station on the template covalently linked to the final multistep reaction product (in this case, a triamide containing three amino acid building blocks in a specific T-programmed order). Because each step of this cycle occurs spontaneously under identical conditions, the entire three-step reaction sequence proceeds autonomously, requiring no changes in temperature or pH, and no intervention from the researcher.
A major challenge of any effort to perform a multistep reaction sequence in a single solution is to prevent reactive substrates from undergoing any of the many possible reactions other than those on the desired pathway. In the case of transforming amino acid building blocks into a specific oligoamide, each activated amino acid can only react with the nascent product, and not with each other. In nature, the ribosomal machinery modulates the effective molarity of aminoacylated tRNAs with respect to water or other nucleophiles to greatly increase the likelihood than an aminoacylated tRNA in the ribosome's A-site couples with the nascent peptide in the P-site before hydrolysis or errant coupling takes place.22-23 Like the ribosome, the DNAsome system displays multiple activated amino acids on a single track simultaneously. These amino acids each contain a free amine group that can potentially react with the aminoacyl NHS ester on an adjacent substrate station, leading to misordered products or precluding the formation of the desired final product. We hypothesized based on our previous observations5,24-25 that the double-stranded nature of the intervening track between adjacent substrates would effectively separate adjacent amino acids, decreasing their effective molarity and preventing side reactions. To test this hypothesis, we performed a model reaction in which a substrate containing a primary amine (S3a) and an NHS ester-linked substrate (S2a) were hybridized at adjacent stations of the same track (Supplementary Information Fig. S3). After incubation in 50 mM MOPS buffer, pH 7.5, with 10 mM Mg(OAc)2 for 16 hours, no significant product formation was observed, indicating that adjacently docked substrates are not prone to uncontrolled cross-reaction in the absence of the walker (Supplementary Information Fig. S3).
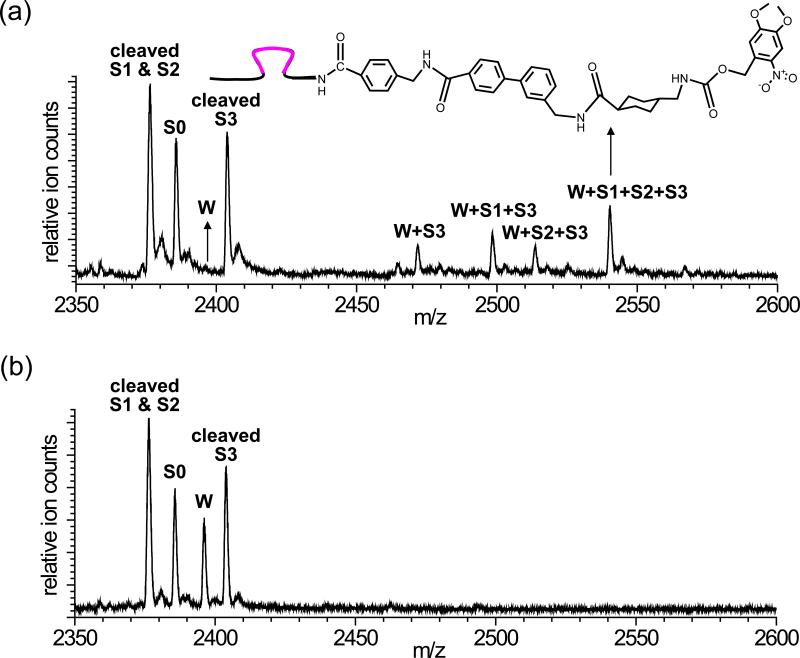
In a typical multistep synthesis experiment, T, S1, S2, and S3 (1.6 μM each) were combined at room temperature in aqueous buffer containing 5 mM MOPS and 10 mM Mg(OAc)2 at pH 5.0. In a separate tube, W and S0 (3.0 μM each) were combined in the same buffer. After 1 hour at room temperature, the two solutions were combined into 50 mM aqueous sodium MOPS buffer at pH 7.5 (1.0 μM final concentration of each of the six molecules) to initiate the walking and multistep synthesis process. After 8 hours at room temperature, the crude reaction was desalted by gel filtration then analyzed by high-resolution mass spectrometry (Figs. 2 and Supplementary Information Fig. S1). Following the conditions described above, W was completely consumed and the three-step desired triamide arising from reaction of W with S1, S2, and S3 represented the major product (Fig. 2). We estimated the overall yield of desired three-step product to be ~45% of detectable DNA-linked species by comparison with mass spectra of authentic product standards mixed in various ratios (Supplementary Information Fig. S2). This overall yield is the highest of any three-step DNA-templated small-molecule synthesis reported to date.5,18 Minor truncation products corresponding to the omission of a reaction (such as W+S1+S3) were also observed, typically in ~10-25% yield each. These results indicate that the DNAsome system is capable of mediating an efficient three-step synthesis over several hours with no intervention.
Figure 2.
Analysis of reaction products generated by the DNAsome system. (a) Mass spectroscopy analysis of the three-step DNAsome-mediated reaction sequence. See the text for a detailed description of reaction conditions. (b) Mass spectrometry analysis of the experiment in (a) repeated using substrates lacking amino acid NHS esters. See the Supplementary Information for all expected and observed masses.
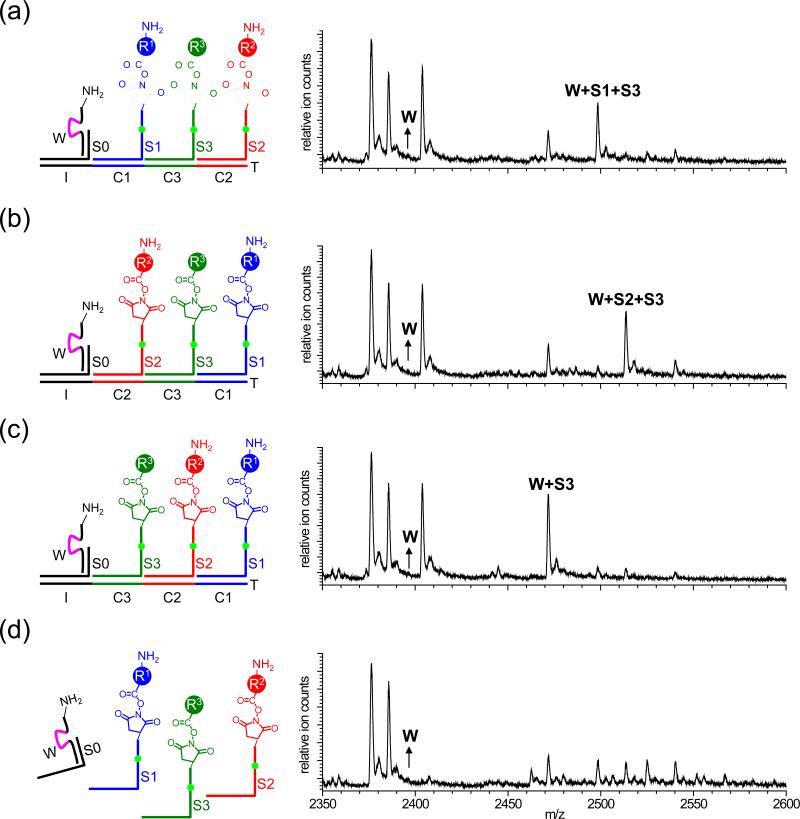
To confirm that multistep synthesis mediated by the DNAsome proceeds in an ordered manner programmed by the nucleotide sequence of the track (in the above example, I-C1-C2-C3), we repeated the above experiment using three different DNA tracks, or omitting T altogether. Since S3 lacks an amine group, it effectively serves as a translation terminator. When the experiment described above was repeated using an I-C1-C3-C2 track, W was again entirely consumed and the predominant product observed by mass spectrometry was consistent with the reaction of W+S1+S3 (Fig. 3a). The absence of products containing S2 is consistent with the ordered, sequence-dependent nature of DNAsome-mediated multistep synthesis. Likewise, using an I-C2-C3-C1 track resulted in the formation of a predominant product consistent with the reaction of W+S2+S3 (Fig. 3b), and using an I-C3-C2-C1 track yielded only W+S3 product (Fig. 3c). Finally, omission of the track altogether resulted in the formation of a complex mixture of minor products consistent with the uncontrolled random intermolecular reaction of W, S1, S2, and S3 (Fig. 3d). Taken together, these results establish that artificial translation mediated by the DNAsome proceeds in a manner programmed by the order of codons on the DNA track.
Figure 3.
Mass spectroscopy analysis of reactions identical to the one shown in Figure 2a, but using different DNA tracks (a-c) or with no DNA track (d). The DNA tracks are as follows: (a) I-C1-C3-C2; (b) I-C2-C3-C1; (c) I-C3-C2-C1; (d) no DNA track. See the Supplementary Information for all expected and observed masses.
In this work we integrated a DNA mechanical device and DNA-templated synthesis into an system that is capable of mediating in a single solution sequence-programmed, autonomous, multistep ordered synthesis of oligoamides without natural enzymes. Because the device is powered by the favorable energetics of RNA cleavage, no changes in temperature, pH, or other reaction conditions are needed over the course of the multistep synthesis. The system described here extends the capabilities of DNA-templated synthesis in significant ways. While requiring the use of self-cleaving reagents that cannot react intramolecularly,18 the multistep syntheses presented here are more efficient (~45% overall yield for three steps) and proceed more quickly (starting materials to three-step product within several hours) compared with existing methods for sequential multistep DNA-templated syntheses.5,18 These results also raise the possibility of using a similar strategy to rapidly generate libraries of synthetic small molecules constructed through multiple consecutive DNA-templated synthesis steps in a single solution.26-27 Finally, the system described here demonstrates how the ordered synthesis of products unrelated to nucleic acids and the avoidance of undesired side reactions can take place in a manner directed and catalyzed entirely by nucleic acids, and therefore has conceptual relevance to early translation systems.
Supplementary Material
Acknowledgements
This work was supported by the Howard Hughes Medical Institute and NIH/NIGMS (R01GM065865). We thank Christoph Dumelin and Yiyun Chen for insightful discussions and experimental assistance. We are grateful to Yinghua Shen for assistance with mass spectrometry.
Footnotes
Additional Information
Supplementary information accompanies this paper at www.nature.com/naturenanotechnology. Reprints and permission information is available online at http://npg.nature.com/reprintsandpermissions/. Correspondence and requests for materials should be addressed to D.R.L.
Competing Financial Interests
D.L. is a consultant for Ensemble Discovery, a company that uses DNA-templated synthesis for industrial applications.
References
- 1.Hanes J, Pluckthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl Acad. Sci. USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zahnd C, Amstutz P, Pluckthun A. Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods. 2007;4:269–279. doi: 10.1038/nmeth1003. [DOI] [PubMed] [Google Scholar]
- 3.Walsh CT. Polyketide and nonribosomal peptide antibiotics: Modularity and versatility. Science. 2004;303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 4.Sattely ES, Fischbach MA, Walsh CT. Total biosynthesis: in vitro reconstitution of polyketide and nonribosomal peptide pathways. Nat. Prod. Rep. 2008;25:757–793. doi: 10.1039/b801747f. [DOI] [PubMed] [Google Scholar]
- 5.Snyder TM, Liu DR. Ordered multistep synthesis in a single solution directed by DNA templates. Angew. Chem. Int. Ed. 2005;44:7379–7382. doi: 10.1002/anie.200502879. [DOI] [PubMed] [Google Scholar]
- 6.Seeman NC. From genes to machines: DNA nanomechanical devices. Trends Biochem Sci. 2005;30:119–125. doi: 10.1016/j.tibs.2005.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bath J, Turberfield AJ. DNA nanomachines. Nature Nanotech. 2007;2:275–284. doi: 10.1038/nnano.2007.104. [DOI] [PubMed] [Google Scholar]
- 8.Liedl T, Sobey TL, Simmel FC. DNA-based nanodevices. Nano Today. 2007;2:36–41. [Google Scholar]
- 9.Beissenhirtz MK, Willner I. DNA-based machines. Org. Biomol. Chem. 2006;4:3392–3401. doi: 10.1039/b607033g. [DOI] [PubMed] [Google Scholar]
- 10.Yin P, Yan H, Daniell XG, Turberfield AJ, Reif JH. A unidirectional DNA walker that moves autonomously along a track. Angew. Chem. Int. Ed. 2004;43:4906–4911. doi: 10.1002/anie.200460522. [DOI] [PubMed] [Google Scholar]
- 11.Bath J, Green SJ, Turberfield AJ. A free-running DNA motor powered by a nicking enzyme. Angew. Chem. Int. Ed. 2005;44:4358–4361. doi: 10.1002/anie.200501262. [DOI] [PubMed] [Google Scholar]
- 12.Tian Y, He Y, Chen Y, Yin P, Mao CD. Molecular devices - A DNAzyme that walks processively and autonomously along a one-dimensional track. Angew. Chem. Int. Ed. 2005;44:4355–4358. doi: 10.1002/anie.200500703. [DOI] [PubMed] [Google Scholar]
- 13.Yin P, Choi HMT, Calvert CR, Pierce NA. Programming biomolecular self-assembly pathways. Nature. 2008;451:318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 14.Omabegho T, Sha R, Seeman NC. A Bipedal DNA Brownian Motor with Coordinated Legs. Science. 2009;324:67–71. doi: 10.1126/science.1170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu HZ, Chao J, Xiao SJ, Seeman NC. A proximity-based programmable DNA nanoscale assembly line. Nature. 2010;465:202–205. doi: 10.1038/nature09026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lund K, et al. Molecular robots guided by prescriptive landscapes. Nature. 2010;465:206–210. doi: 10.1038/nature09012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chhabra R, Sharma J, Liu Y, Yan H. Addressable molecular tweezers for DNA-templated coupling reactions. Nano Lett. 2006;6:978–983. doi: 10.1021/nl060212f. [DOI] [PubMed] [Google Scholar]
- 18.Gartner ZJ, Kanan MW, Liu DR. Multistep small-molecule synthesis programmed by DNA templates. J. Am. Chem. Soc. 2002;124:10304–10306. doi: 10.1021/ja027307d. [DOI] [PubMed] [Google Scholar]
- 19.Chen Y, Mao C. Reprogramming DNA-directed reactions on the basis of a DNA conformational change. J. Am. Chem. Soc. 2004;126:13240–13241. doi: 10.1021/ja045718j. [DOI] [PubMed] [Google Scholar]
- 20.Voigt NV, et al. Single-molecule chemical reactions on DNA origami. Nature Nanotech. 2010;5:200–203. doi: 10.1038/nnano.2010.5. [DOI] [PubMed] [Google Scholar]
- 21.Santoro SW, Joyce GF. A general purpose RNA-cleaving DNA enzyme. Proc. Natl Acad. Sci. USA. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 23.Trobro S, Aqvist J. Mechanism of peptide bond synthesis on the ribosome. Proc. Natl Acad. Sci. USA. 2005;102:12395–12400. doi: 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Snyder TM, Tse BN, Liu DR. Effects of template sequence and secondary structure on DNA-templated reactivity. J. Am. Chem. Soc. 2008;130:1392–1401. doi: 10.1021/ja076780u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gartner ZJ, Liu DR. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J. Am. Chem. Soc. 2001;123:6961–6963. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li XY, Liu DR. DNA-Templated organic synthesis: Nature's strategy for controlling chemical reactivity applied to synthetic molecules. Angew. Chem. Int. Ed. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- 27.Tse BN, Snyder TM, Shen YH, Liu DR. Translation of DNA into a Library of 13 000 Synthetic Small-Molecule Macrocycles Suitable for in Vitro Selection. J. Am. Chem. Soc. 2008;130:15611–15626. doi: 10.1021/ja805649f. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.