Summary
Cell surface glycans play important cellular functions and are synthesized by glycosyltransferases. Structure and function studies show that the donor sugar specificity of the invertebrate β1,4-N-acetyl-glactosaminyltransferase (β4GalNAc-T) and the vertebrate β1,4-galactosyltransferase I (β4Gal-T1) are related by a single amino acid residue change. Comparison of the catalytic domain crystal structures of the β4Gal-T1 and the α-polypeptidyl-GalNAc-T (αppGalNAc-T) shows that their protein structure and sequences are similar. Therefore, it seems that the invertebrate β4GalNAc-T and the catalytic domain of αppGalNAc-T might have emerged from a common primordial gene. When vertebrates emerged from invertebrates, the amino acid that determines the donor sugar specificity of the invertebrate β4GalNAc-T might have mutated, thus converting the enzyme to a β4Gal-T1 in vertebrates.
Introduction
Glycosyltransferases exist as a super family of enzymes and constitute 1 to 2% of the human genome [1]. These transferases have been thought to have evolved by gene duplication and divergence as evidenced from their high protein sequence and gene structure (exon/intron organization) similarity among the subfamily members [2–4]. They generally transfer a donor sugar from an activated donor substrate such as a nucleotide-diphosphate-sugar to an acceptor sugar, extending the glycan chain of glyco-proteins and glyco-lipids, one sugar moiety at a time. The complex glycans, synthesized by involving many glycosyltransferases, are known to have very important cellular functions [5]. Therefore, absence or lethal mutation in any glycosyltransferase gene and its corresponding protein might lead to a total absence of a sugar epitope, thus changing the cellular physiology. For example, in humans the αGal1-3βGal epitope is absent and has an anti-αGal IgG antibody against this epitope [6]. Sequencing of the human α1,3-galactosyltransferase gene that is responsible for the synthesis of this epitope has revealed that because of a number of lethal mutations, the active α3 galactosyltransferase enzyme can not be expressed in humans, thus explaining the absence of the epitope [7]. Similarly, sequencing of the human CMP-N-acetylneuraminic acid hydroxylase gene showed an inactivating mutation that is responsible for the lack of the N-glycolylneuraminic acid (Neu5Gc) in humans; Neu5Gc is otherwise widely found in most mammalian tissues [8]. The loss of Neu5Gc and access of its precursor N- acetylneuraminic acid (Neu5Ac) has been implicated in the evolution of their binding proteins, Siglecs, and in the evolutionary changes in the human lineage [9]. Similarly, other glycosyltransferases are known to have been involved in the evolution and diversification of species [10].
Crystal structure of bovine β4Gal-T1 revealed that a single amino acid, Tyr289, determines the donor sugar specificity
In vertebrate cells, the most prevalent epitope, LacNAc (βGal1-4βGlcNAc), is recognized by asialoglycoprotein receptors [11]. Furthermore, a LacNAc moiety with a terminal sialic acid linked in different configurations is involved in immunological and cell signaling functions [12–14]. Thus, the galactose moiety in the LacNAc epitope seems to have played an important role in vertebrate evolution. In the presence of manganese, the β1,4-galactosyltransferase I (β4Gal-T1) enzyme transfers the donor sugar, galactose (Gal), from UDP-Gal to an acceptor sugar β-N-acetylglucosamine (βGlcNAc) synthesizing LacNAc, βGal1-4βGalNAc [15]. From the enzyme kinetic studies it has been known that the β4Gal-T1 poorly transfers N-acetylgalactosmine (GalNAc) from UDP-GalNAc to the acceptor sugar GlcNAc [16,17]. In the crystal structure of the bovine β4Gal-T1•Mn2+•UDP-GalNAc complex, the side-chain hydroxyl group of the Tyr289 residue forms a hydrogen bond with the carbonyl oxygen atom of the N-acetyl group of the bound UDP-GalNAc molecule; due to the lack of enough space between them, the N-acetyl moiety is found in a unfavorable orientation (Figure 1a) [17]. It was reasoned that this steric hindrance could be responsible for the poor catalytic activity by the β4Gal-T1 enzyme with UDP-GalNAc as the donor substrate. Therefore, it was thought that a substitution of the Tyr289 residue with an amino acid with a less bulky side chain, such as Leu, Ile, or Asn, might not cause such steric hindrance and thereby would be expected to enhance the catalytic activity with UDP-GalNAc. Indeed, it was found that such substitution with any of these three amino acids enhanced the catalytic activity using UDP-GalNAc. The mutant enzyme with Leu substitution exhibited catalytic activity that was as efficient with the UDP-GalNAc as it was with the UDP-Gal. Therefore, the single amino acid, Tyr289, determines the donor sugar specificity of bovine β4Gal-T1 [17].
Figure 1.
(a) The crystal structure of the bovine β4Gal-T1•Mn2+•UDP-GalNAc complex (pdb 1OQM) with a GlcNAc molecule modeled in the acceptor binding site. The UDP-GalNAc and GlcNAc molecules are shown in a ball and stick diagram. The side-chain hydroxyl group of Tyr289 residue forms a hydrogen bond (shown as black dotted line) with the carbonyl oxygen atom of the N-acetyl group of the GalNAc moiety. (b) Protein sequence comparison of the bovine β4Gal-T1 shown near the vicinity of the Y289 residues (black arrow), along with its homolog proteins from various species. Only in the vertebrate homolog proteins is the Y289 residue conversed as a Tyr residue, whereas in the invertebrates it is either Leu or Ile.
The β4Gal-T1 homolog in the invertebrates is β4GalNAc-T enzyme and has a mutation at the Tyr289 residue
In contrast to vertebrate cells that have LacNAc moiety invertebrate cells have mainly a LacdiNAc moiety where β-N-acetylgalactosamine (βGalNAc) is 1–4 linked to βGlcNAc [18]. Thus, it seems that in the invertebrate cells the vertebrate β4Gal-T1, that is responsible for the synthesis of LacNAc moiety, is replaced by a β1,4-N-acetyl-galactosaminyltransferase (β4GalNAc-T) that synthesize the LacdiNAc moiety. Indeed in several invertebrates a β4GalNAc-T enzyme has been found that is homologous to the vertebrate β4Gal-T1 protein [19–22]. These two enzymes not only exhibit reasonable protein sequence similarity but also similar gene structure [23]. However, the corresponding Y289 residue of bovine β4Gal-T1 in the invertebrate β4GalNAc-T proteins is found either as Leu or Ile residue, suggesting that this amino acid change may be responsible for the altered donor sugar, UDP-GalNAc, specificity of these enzymes (Figure 1b) [20,21].
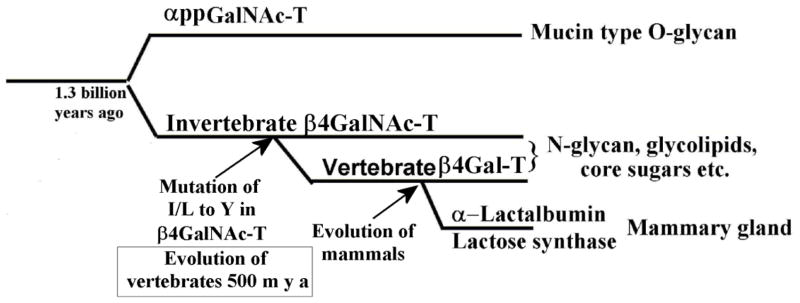
To provide support for this hypothesis, we have expressed Drosophila β4GalNAc-TA protein, a homolog protein of vertebrate β4Gal-T1, that transfers GalNAc from UDP-GalNAc to the acceptor substrate GlcNAc; it transfers Gal from UDP-Gal to GlcNAc very poorly [23]. When the Ile289 residue is mutated to a Tyr residue, the mutant enzyme loses the catalytic activity with UDP-GalNAc; instead, it exhibits high catalytic activity with UDP-Gal transferring Gal to GlcNAc, thus confirming that this single amino acid determines the sugar donor specificity of Drosophila β4GalNAc-TA enzyme [23]. It seems that when the vertebrates emerged from the invertebrates nearly 500 million years ago, the vertebrate β4Gal-T1 enzyme might have also emerged from invertebrate β4GalNAc-T, by the single amino acid mutation of Leu/Ile in the β4GalNAc-TA to Tyr, thus converting the β4GalNAc-T to β4Gal-T (Figure 2) [23].
Figure 2.
Since the donor sugar specificity of the invertebrate β4GalNAc-T and vertebrate β4Gal-T are related by a single amino acid change, it is possible that when the vertebrate emerged from the invertebrate 500 million years ago, this single amino acid change might have happened, thus converting the invertebrate β4GalNAc-T enzyme to a β4Gal-T enzyme in the vertebrates. Furthermore, since the catalytic domains of β4Gal-T1 and αpptGalNAc-T enzymes exhibit protein structure, sequence, and gene structure similarity, the vertebrate β4GalNAc-T and the αpptGalNAc-T might have evolved from a common primordial gene by gene duplication and, further, during evolution the αpptGalNAc-T might have picked up the lectin domain to define its acceptor specificity. Since α-lactabumin binds and inhibits but does not modulate the invertebrate β4GalNAc-T enzyme, its binding site existed before the evolution of vertebrates. Only during the evolution of mammals did α-lactabumin evolve to bind to this site in the vertebrate β4Gal-T1 and modulate its acceptor sugar specificity to synthesize lactose during lactation of mammals.
The invertebrate β4GalNAc-T enzymes have an α-lactalbumin binding site
Lysozyme and α-lactalbumin (α-LA) share high similarity between their protein sequences and gene structures but have completely different biological functions [24]. These proteins are thought to have evolved from a primordial gene by a gene duplication nearly 300 million years ago. α-LA is a calcium binding protein that is only expressed in the mammary gland during lactation. The evolution of mammals has been associated with the appearance of α-LA in the mammary gland where it plays a role in the synthesis of lactose, a hallmark of mammals. α-LA modulates β4Gal-T1 by altering its acceptor specificity from GlcNAc to glucose (Glc), thus synthesizing the disaccharide lactose, βGal1-4βGlc [25]. Also, α-LA is known to inhibit the transfer of Gal to a GlcNAc acceptor by β4Gal-T1. Interestingly, we have found that α-LA binds to the wild-type Drosophila β4GalNAc-TA enzyme and inhibits the catalytic activity of this protein but it does not alter its acceptor specificity from GlcNAc to Glc [23]. Earlier, a similar property was also observed for the C. elegans β4GalNAc-T enzyme [20]. These observations suggest that the α-LA binding site existed even before the appearance of the vertebrate β4Gal-T1 protein, and that during the evolution of mammals, this site was utilized by α-lactalbumin to modulate the acceptor specificity of the β4Gal-T1 in mammals (Figure 2). This is further evidenced from the fact that the α-LA can bind and modulate the acceptor specificity of a non-mammal vertebrate β4Gal-T1 enzyme, like the one from chicken [26].
Although the evolution of β1,4-galactosyltransferase appears to be coincidental with the appearence of vertebrates, the evolution of this enzyme played an important role in vertebrate development. For example, only in vertebates several sialyltransferases are found to synthesize varity of sialyated LacNAc containing glycans, and during the evolution of mammals α-LA was recruited for the synthesis of lactose. Similarly, different lectins had to be recruited to recognize these new glycoconjugates. Thus, the appearance of β1,4-galactosyltransferases in vertebrates created a boundary between vertebrate and invertebrate glycans and made it possible to expand the variety of glycoconjugates that were needed for vertebrate development.
The β4Gal-T1 and the catalytic domain of αppGalNAc-T enzymes show structural similarity
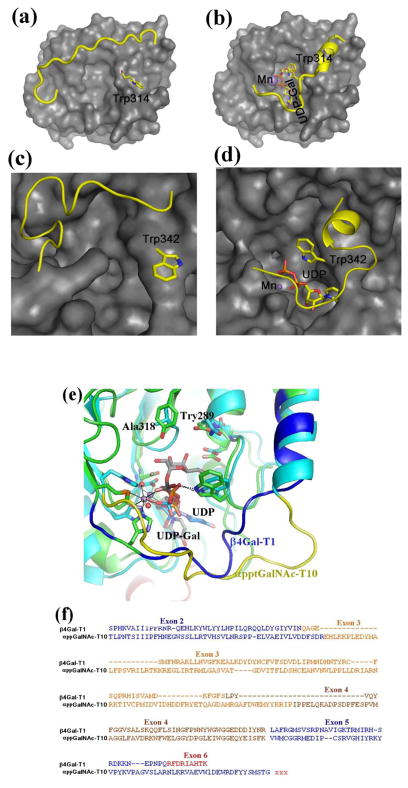
Having observed an evolutionary relationship between the vertebrate β4Gal-T1 and invertebrate β4GalNAc-T enzymes, it was important to find out if there are any other GalNAc-T enzymes that might be related to them. In all species, α-polypeptidyl-GalNAc-T (αppGalNAc-T) exists as a family of enzymes, and in the presence of manganese, they transfer GalNAc to the side-chain hydroxyl group of a Thr/Ser amino acid in a polypeptide acceptor [27]. In these transferases the presence of a lectin domain C-terminal to the catalytic domain and separated by a linker polypeptide had been predicted well before the crystal structure of αppGalNAc-T was available [28]. However, its structural relationship with the β4Gal-T1 was first shown by Kubota et al. [29] who determined the crystal structure of an αppGalNAc-T10•Mn2+•UDP-GalNAc complex, which showed that the binding of UDP-GalNAc to αppGalNAc-T10 molecules was similar to UDP-GalNAc binding to β4Gal-T1 enzyme [29]. The crystal structure studies on αppGalNAc-T2 revealed that upon the binding of manganese and the donor sugar substrate, the enzyme undergoes conformational changes involving separate short and long flexible loops, similar to the β4Gal-T1 enzyme (Figure 3a and b) [30]. Interestingly, in the short loop of these proteins, the side chain of a Trp residue undergoes conformational changes from outside to inside of the the catalytic pocket allowing it to interact with the β-phosphate oxygen atom of the bound UDP-sugar substrate molecule. The superposition of the crystal structures of the bovine β4Gal-T1 and the catalytic domain of αppGalNAc-T10 shows similarity in their crystal structures, protein sequences, and gene structures (Figure 3e and f). Interestingly, the Tyr289 residue that determines the donor sugar specificity of the bovine β4Gal-T1 is present as Ala in the αppGalNAc-Ts. Therefore, the catalytic domains of αppGalNAc-T and the vertebrate β4Gal-T1 seem to be related. Since vertebrate β4Gal-T1 emerged from the invertebrate β4GalNAc-T, the catalytic domain of the αppGalNAc-T and the invertebrate β4GalNAc-T might have evolved from a common primordial glycosyltransferase; and during the evolution the αppGalNAc-T acquired the lectin domain (Figure 2). Based on their DNA sequences, this gene duplication and divergence was predicted to have occurred 1.3 billion years ago [2].
Figure 3.
The crystal structure of the bovine β4Gal-T1 in the open (a) and closed (b) conformation. Upon the binding of manganese and UDP-Gal, the enzyme undergoes conformational changes involving a short and a long flexible loop. In the short loop, the side chain of the Trp314 residue moves from outside to inside the catalytic pocket to bind to the bound UDP-Gal molecule, while the long loop moves over the bound UDP-GalNAc molecule to cover it. The crystal structure of the human αppt-GalNAc-T2 is also found in an open (c) and a closed (d) conformation. Similar to β4Gal-T1, upon the binding of manganese and UDP-GalNAc molecule, this enzyme also undergoes conformational changes involving two flexible loops. Also, in the short loop, the Trp342 residue moves from outside to inside the catalytic pocket to bind to the bound UDP-GalNAc molecule, while the long flexible loop covers the bound UDP-GalNAc molecule.. (e) The superposition of the catalytic domains of bovine β4Gal-T1•Mn2+•UDP-Gal complex with the αppGalNAc-T10•Mn2+•UDP-GalNAc complex and (f) the corresponding protein sequence comparison. The Trp residue in the short flexible loops of these structures show good agreement, and the long flexible loop of β4Gal-T1 and αpptGalNAc-T10 are shown in blue and yellow, respectively. The Tyr289 residue in the bovine β4Gal-T that determines the donor sugar specificity is naturally present as Ala318 residue in αppGalNAc-T10 in order to accommodate the N-acetyl moiety of the donor sugar GalNAc, and it is conserved in all αppGalNAc-T enzymes. In the protein-protein sequence comparison (f), the amino acids are colored, based on their exons. Since the acceptor substrate for β4GalNAc-T1 is a sugar residue, while it is a linear peptide with a Thr/Ser amino acid for the αppGalNAc-T10, these enzymes are expected to have different acceptor binding sites. The additional amino acids in the exons 3 and 4 of αppGalNAc-T10 are found as a part of the acceptor substrate binding site in the αppGalNAc-T10 crystal structure.
An additional protein domain, as either a contiguous or a non-contiguous polypeptide, has been observed in many other glycosyltransferases, and such domains have been defined as “add-on” domains [31,32]. For example, in the lactose synthase complex, α-LA alters the acceptor sugar specificity of β4Gal-T1 to Glc, thus acting as a non-contiguous “add-on” domain, similar to the lectin domain in the αppGalNAc-Ts (Figure 4). Recently, the crystal structure of α1,6-fucosyltransferase-8 revealed that this enzyme also has at its C-terminal end a SH3-like “add-on” domain that is usually found in proteins that interact with other proteins and mediate assembly of specific protein complexes, typically via binding to proline-rich peptides in their respective binding partner [31]. Interestingly, the N-terminal tandem-repeat, an “add-on” domain in O-βGlcNAc-T, is known to impart acceptor substrate specificity of the enzyme [33]. Thus, it seems that during evolution these enzymes have acquired these “add-on” domains to impart new functions [32].
Figure 4.
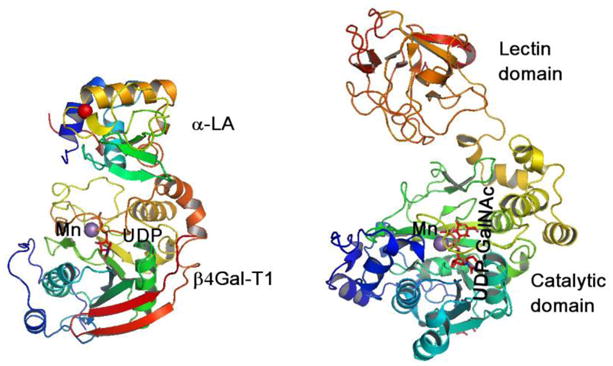
In the lactose synthase complex (left), α-LA modulates the sugar acceptor specificity of the β4Gal-T1 enzyme from GlcNAc to Glc, while in the αpptGalNAc-Ts (right), although the catalytic domain alone exhibits catalytic activity, the lectin domain defines acceptor substrate specificity to the enzyme. Thus, in these proteins α-LA and the lectin domain are considered as “add-on” domains. There are many other transferases found where such a domain brings about substrate specificity to the enzyme.
Evolution of β4Gal-T7 enzyme
In all vertebrates, the β1,4-galactosyltransferase family has seven members, β4Gal-T1 to -T7, and they all transfer Gal from UDP-Gal to different acceptor sugar substrates [4]. Interestingly, β4Gal-T7 differs from the rest by its gene structure [34]. However, all the known β4Gal-T7 protein homologs in invertebrates and vertebrates have the conserved Phe/Tyr residue as in β4Gal-T1 [34, 36–40]. The β4Gal-T7 is involved in the synthesis of the tetrasaccharide linker sequence of proteoglycans for heparin/heparan sulfate or chondroitin sulfate or dermatan sulfates, which are widely found on the cell surface and in the extracellular matrix of various tissues and are known to play important roles in several cellular functions. β4Gal-T7 is an important enzyme for the species viability [38]; mutation in the human β4Gal-T7 gene has been linked to Ehlers-Danlos syndrome [41,42]. Although recent crystal structure of the Drosophila β4Gal-T7 shows that the overall structure of this enzyme is similar to β4Gal-T1, with a similar catalytic pocket, its evolutionary relationship with β4Gal-T1 is still not clear [43].
Though high protein sequence similarity between the vertebrate β4Gal-T1 and invertebrate β4GalNAc-T enzymes suggests that these proteins are related by evolution, the difference in their donor sugar specificity is mainly due to a single amino acid change. Furthermore, superposition of the three-dimensional structure of the catalytic domain of β4Gal-T1 with the corresponding catalytic domain of ppGalNAc-T reveals a similarity in their protein sequence and gene structure, suggesting that these proteins are also evolutionarily related. In addition to protein sequence and structure similarity in their gene structure, exon/intron organization is also observed among the proteins that have been thought to have evolved from a common primordial gene. This is in contrast to the observation of a common structural motif among the glycosyltransferases that have DxD metal binding motif and use UDP-sugar as donor substrates without any similarity in their gene structure. Thus, the structural and functional studies of these enzymes have provided deeper insight into the evolution of these glycosyltransferases.
Acknowledgments
We thank Dr. Nathan Sharon from Weizmann Institute of Science, Israel, for encouraging us to write this review and for his valuable comments on the manuscript. The writing of this review was funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the view or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government. This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The cccarbohydrate-active EnZymes database (CAZy): An expert resource for glycogenomics. Nucleic Acids Res. 2008;37:D233–238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2**.Kaneko M, Nishihara S, Narimatsu H, Saitou N. The evolutionary history of glycosyltransferase genes. Trends Glycosci Gycotechnol. 2001;13:147–155. Molecular evolutionary analysis on the glycosyltransferases that are involved in N- or O-glycan synthesis. The phylogeneitc trees showed that these glycosyltransferases not only have evolved through gene duplications but also evolved more slowly than other genes. Interestingly they have predicted that the β4Gal-T and αppGalNAc-T might have evolved from the same primordial gene. [Google Scholar]
- 3.Togayachi A, Sato T, Narimatsu H. Comprehensive enzymatic characterization of glycosyltransferases with a beta3GT or beta4GT motif. Methods Enzymol. 2006;416:91–102. doi: 10.1016/S0076-6879(06)16006-1. [DOI] [PubMed] [Google Scholar]
- 4.Hennet T. The galactosyltransferase family. Cell Mol Life Sci. 2002;59:1081–1095. doi: 10.1007/s00018-002-8489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5*.Varki A, Lowe JB. Essentials of Glycobiology. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2009. Biological role of glycans; pp. 75–88. A book on topics in glycobiology. [Google Scholar]
- 6.Galili U, Macher BA, Buehler J, Shohet SB. Human natural anti-α-galactosyl IgG. II. The specific recognition of α(1–3)-linked galactose residues. J Exp Med. 1985;162:573–582. doi: 10.1084/jem.162.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larsen RD, Rivera-Marrero CA, Ernst LK, et al. Frameshift and nonsense mutations in a human genomic sequence homologous to a murine UDP-Gal:β-D-Gal(1,4)-D-GlcNAc α(1,3)-galactosyltransferase cDNA. J Biol Chem. 1990:2657055–7061. [PubMed] [Google Scholar]
- 8.Varki A. Multiple changes in sialic acid biology during human evolution. Glycoconj J. 2009;26:231–245. doi: 10.1007/s10719-008-9183-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9**.Varki A. Uniquely human evolution of sialic acid genetics and biology. Proc Natl Aca Sci U S A. 2010;107:8939–8946. doi: 10.1073/pnas.0914634107. This review discusses the evolution of the proteins that are involved in the binding of sialic acid and its implications in human evolution. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turcot-Dubois AL, Le Moullac-Vaidye B, Despiau S, Roubinet F, Bovin N, Le Pendu J, Blancher A. Long-term evolution of the CAZY glycosyltransferase 6 (ABO) gene family from fishes to mammals—a birth-and-death evolution model. Glycobiology. 2007;17:516–528. doi: 10.1093/glycob/cwm016. [DOI] [PubMed] [Google Scholar]
- 11.Ashwell G, Morell AG. Adv. Enzymol. Relat. Areas. Mol Biol. 1974;41:99–128. doi: 10.1002/9780470122860.ch3. [DOI] [PubMed] [Google Scholar]
- 12.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 13.Liu FT, Rabinovich GA. Galectins: regulators of acute and chronic inflammation. Ann N Y Acad Sci. 2010;1183:158–182. doi: 10.1111/j.1749-6632.2009.05131.x. [DOI] [PubMed] [Google Scholar]
- 14.Vasta GR. Roles of galectins in infection. Nat Rev Microbiol. 2009;7:424–438. doi: 10.1038/nrmicro2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brew K, Vanaman TC, Hill RL. The role of alpha-lactalbumin and the A protein in lactose synthetase: a unique mechanism for the control of a biological reaction. Proc Natl Acad Sci U S A. 1968;59:491–497. doi: 10.1073/pnas.59.2.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Do KY, Do SI, Cummings RD. Alpha-lactalbumin induces bovine milk beta 1,4-galactosyltransferase to utilize UDP-GalNAc. J Biol Chem. 1995;270:18447–18451. doi: 10.1074/jbc.270.31.18447. [DOI] [PubMed] [Google Scholar]
- 17**.Ramakrishnan B, Qasba PK. Structure-based design of beta-1,4-Galactosyl-transferase (beta 4Gal-T1) with equally efficient N-Acetylgalactosaminyltransferase activity. J Biol Chem. 2002;277:20833–20840. doi: 10.1074/jbc.M111183200. Crystal structure and the enzyme kinetic studies showed that the in bovine β4Gal-T1 enzyme a single amino acid residue (Tyr289), found in the vicinity of the sugar donor-binding site that is not involved in any direct interactions with the donor sugar, determines the donor sugar specificity of the enzyme. The first successful structure based design of a glycosyltransferase to transfer unnatural substrates. [DOI] [PubMed] [Google Scholar]
- 18.Betenbaugh MJ, Tomiya N, Narang S, Hsu JT, Lee YC. Biosynthesis of human-type N-glycans in heterologous systems. Curr Opin Struct Biol. 2004:14601–606. doi: 10.1016/j.sbi.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 19.Haines N, Irvine KD. Functional analysis of Drosophila β1,4-N-Acetlygalactosaminyltransferases. Glycobiology. 2004;15:335–346. doi: 10.1093/glycob/cwi017. [DOI] [PubMed] [Google Scholar]
- 20.Kawar ZS, van Die I, Cummings RD. Molecular cloning and enzymatic characterization of a UDP-GalNAc:GlcNAc-R β1,4-N-Acetylgalactosaminyltransferase from Caenorhabditis elegans. J Biol Chem. 2002;277:34924–34932. doi: 10.1074/jbc.M206112200. [DOI] [PubMed] [Google Scholar]
- 21.Vadaie N, Jarvis DL. Molecular cloning and functional characterization of a lepidopteran insect β4-N-Acetylgalactosaminyltransferase with broad substrate specificity, a functional role in glycoprotein biosynthesis, and a potential functional role in glycolipid biosynthesis. J Biol Chem. 2004;279:33501–33518. doi: 10.1074/jbc.M404925200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakker H, Van Tetering A, Agterberg M, Smit AB, Van den Eijnden DH, Van Die I. Deletion of two exons from the Lymnaea stagnalis beta1—>4-N-acetylglucosaminyltransferase gene elevates the kinetic efficiency of the encoded enzyme for both UDP-sugar donor and acceptor substrates. J Biol Chem. 1997;272:18580–18585. doi: 10.1074/jbc.272.30.18580. [DOI] [PubMed] [Google Scholar]
- 23**.Ramakrishnan B, Qasba PK. Role of a single amino acid in the evolution of glycans of invertebrates and vertebrates. J Mol Biol. 2007;365:570–576. doi: 10.1016/j.jmb.2006.10.034. This demonstrated that the donor-sugar specificity of Drosophila β4GalNAc-TA and mammalian β4Gal-T1 are related by a single amino acid change. Furthermore, based on the binding of α-lactalbumin, it has been proposed that during the evolution of vertebrates from invertebrates, this single amino acids change must have taken place to convert the invertebrate β4GalNAc-T enzyme to a β4Gal-T enzyme in vertebrates. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24**.Qasba PK, Kumar S. Molecular divergence of lysozymes and alpha-lactalbumin. Crit Rev Biochem Mol Biol. 1997;32:255–306. doi: 10.3109/10409239709082574. Review on the evolutionary relationship between the lysozyme and α-lactalbumin and demonstrating that these proteins not only have similarity in their protein sequence but also in their gene structures. [DOI] [PubMed] [Google Scholar]
- 25.Brodbeck U, Denton WL, Tanahashi N, Ebner KE. The isolation and identification of the B protein of lactose synthetase as alpha-lactalbumin. J Biol Chem. 1967;242:1391–1397. [PubMed] [Google Scholar]
- 26.Shaper NL, Meurer JA, Joziasse DH, Chou TD, Smith EJ, Schnaar RL, Shaper JH. The chicken genome contains two functional nonallelic β1,4-Galactosyltransferase genes. Chromosomal assignment to syntenic regions tracks fate of the two gene lineages in the human genome. J Biol Chem. 1997;272:31389–31399. doi: 10.1074/jbc.272.50.31389. [DOI] [PubMed] [Google Scholar]
- 27.Ten Hagen KG, Fritz TA, Tabak LA. All in the family: the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology. 2003;13:1R–16R. doi: 10.1093/glycob/cwg007. [DOI] [PubMed] [Google Scholar]
- 28.Imberty A, Piller V, Piller F, Breton C. Fold recognition and molecular modeling of a lectin-like domain in UDP-GalNac:polypeptide N-acetylgalactosaminyltransferases. Protein Eng. 1997;10:1353–1356. doi: 10.1093/protein/10.12.1353. [DOI] [PubMed] [Google Scholar]
- 29**.Kubota T, Shiba T, Sugioka S, Furukawa S, Sawaki H, Kato R, Wakatsuki S, Narimatsu H. Structural basis of carbohydrate transfer activity by human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase (pp-GalNAc-T10) J Mol Biol. 2006;359:708–727. doi: 10.1016/j.jmb.2006.03.061. The x-ray crystal structure of αppGalNAc-T10 in complex with UDP-GalNAc shows that the binding of UDP-GalNAc to αppGalNAc-T10 enzyme is similar to the binding of UDP-GalNAc to β4Gal-T1. [DOI] [PubMed] [Google Scholar]
- 30*.Fritz TA, Raman J, Tabak LA. Dynamic association between the catalytic and lectin domains of human UDP-GalNAc:polypeptide alpha-N-acetylgalactosaminyltransferase-2. J Biol Chem. 2006;281:8613–8619. doi: 10.1074/jbc.M513590200. The x-ray crystal structure and enzyme kinetics studies on human αppGalNAc-T2. This study showed that the catalytic domain of the enzyme alone exhibits catalytic activity. [DOI] [PubMed] [Google Scholar]
- 31.Ihara H, Ikeda Y, Toma S, Wang X, Suzuki T, Gu J, Miyoshi E, Tsukihara T, Honke K, Matsumoto A, et al. Crystal structure of mammalian alpha1,6-fucosyltransferase. FUT8 Glycobiology. 2007;17:455–466. doi: 10.1093/glycob/cwl079. [DOI] [PubMed] [Google Scholar]
- 32.Qasba PK, Ramakrishnan B. Letter to the Glyco-Forum: Catalytic domains of glycosyltransferases with ‘add-on’ domains. Glycobiology. 2007;17:7G–9G. doi: 10.1093/glycob/cwm013. [DOI] [PubMed] [Google Scholar]
- 33.Martinez-Fleites C, He Y, Davies GJ. Structural analyses of enzymes involved in the O-GlcNAc modification. Biochim Biophys Acta. 2010;1800:122–133. doi: 10.1016/j.bbagen.2009.07.019. [DOI] [PubMed] [Google Scholar]
- 34.Almeida R, Levery SB, Mandel U, Kresse H, Schwientek T, Bennett EP, Clausen H. Cloning and expression of a proteoglycan UDP-galactose:beta-xylose beta1,4-galactosyltransferase I. A seventh member of the human beta4-galactosyltransferase gene family. J Biol Chem. 1999;274:26165–26171. doi: 10.1074/jbc.274.37.26165. [DOI] [PubMed] [Google Scholar]
- 35.Stolz A, Haines N, Pich A, Irvine KD, Hokke CH, Deelder AM, Gerardy-Schahn R, Wuhrer M, Bakker H. Distinct contributions of beta 4GalNAcTA and beta 4GalNAcTB to Drosophila glycosphingolipid biosynthesis. Glycoconj J. 2008;25:167–175. doi: 10.1007/s10719-007-9069-5. [DOI] [PubMed] [Google Scholar]
- 36.Vadaie N, Hulinsky RS, Jarvis JD. Identification and characterization of a Drosophila melanogaster ortholog of human beta1,4-galactosyltransferase VII. Glycobiology. 2002;12:589–597. doi: 10.1093/glycob/cwf074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakamura Y, Haines N, Chen J, Okajima T, Furukawa K, Urano T, Stanley P, Irvine KD, Furukawa K. Identification of a Drosophila gene encoding xylosylprotein beta4-galactosyltransferase that is essential for the synthesis of glycosaminoglycans and for morphogenesis. J Biol Chem. 2002;277:46280–46288. doi: 10.1074/jbc.M203873200. [DOI] [PubMed] [Google Scholar]
- 38.Takemae H, Ueda R, Okubo R, Nakato H, Izumi S, Saigo K, Nishihara S. Proteoglycan UDP-galactose:beta-xylose beta 1,4-galactosyltransferase I is essential for viability in Drosophila melanogaster. J Biol Chem. 2003;278:15571–15578. doi: 10.1074/jbc.M301123200. [DOI] [PubMed] [Google Scholar]
- 39.Herman T, Horvitz HR. Three proteins involved in Caenorhabditis elegans vulval invagination are similar to components of a glycosylation pathway. Proc Natl Acad Sci USA. 1999;96:974–979. doi: 10.1073/pnas.96.3.974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okajima T, Yoshida K, Kondo T, Furukawa K. Human homolog of Caenorhabditis elegans sqv-3 gene is galactosyltransferase I involved in the biosynthesis of the glycosaminoglycan-protein linkage region of proteoglycans. J Biol Chem. 1999;274:22915–22918. doi: 10.1074/jbc.274.33.22915. [DOI] [PubMed] [Google Scholar]
- 41.Okajima T, Fukumoto S, Furukawa K, Urano T. Molecular basis for the progeroid variant of Ehlers-Danlos syndrome. Identification and characterization of two mutations in galactosyltransferase I gene. J Biol Chem. 1999;274:28841–28844. doi: 10.1074/jbc.274.41.28841. [DOI] [PubMed] [Google Scholar]
- 42.Seidler DG, Faiyaz-Ul-Haque M, Hansen U, Yip GW, Zaidi SH, Teebi AS, Kiesel L, Götte M. Defective glycosylation of decorin and biglycan, altered collagen structure, and abnormal phenotype of the skin fibroblasts of an Ehlers-Danlos syndrome patient carrying the novel Arg270Cys substitution in galactosyltransferase I (beta4GalT-7) J Mol Med. 2006;84:583–594. doi: 10.1007/s00109-006-0046-4. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrishnan B, Qasba PK. Crystal structure of the catalytic domain of Drosophila β1,4-galactosyltransferase-7. J Biol Chem. 2010;285:15619–15626. doi: 10.1074/jbc.M109.099564. [DOI] [PMC free article] [PubMed] [Google Scholar]