Abstract
Frontotemporal Dementia (FTD) is the second major cause of dementia in persons under the age of 65 after Alzheimer’s disease (AD). FTD is clinically, pathologically and genetically heterogeneous and has been associated with mutations in different genes located on chromosomes 17, 9 and 3. In our study we report a novel heterozygous g.26218G>A variant in exon 6 of Charged Multivesicular body Protein 2B (CHMP2B), predicted to cause the amino acid change p.Ser187Asn, in one patient diagnosed with FTD. We were not able to determine the mode of inheritance of the mutation since we did not have access to the genetically informative family members of the proband; those who were screened did not carry the variant. We didn’t find this variant in 273 Caucasian controls while we did find it in 6 of 94 African American controls. Most of the mutations in CHMP2B which are considered pathogenic lead to partial deletion of the C-terminus region of CHMP2B protein. Based on previous reports and on our current data, missense mutations seem unlikely to be pathogenic. The pathogenicity of CHMP2B mutations requires further investigation.
Keywords: dementia, FTD, CHMP2B, gene, missense mutation
INTRODUCTION
Frontotemporal Dementia (FTD) is the second most common cause of dementia in persons under the age of 65, after Alzheimer’s disease (AD)1. Familial cases account for up to 40% of all FTD cases2 and the average onset age lies in the mid to late 50s3.
Mutations in two genes located on chromosome 17 (microtubule associated protein TAU [MAPT] and progranulin [PGRN]) have been shown to be the major cause of FTD. FTD has also been linked to chromosome 9 with mutations in the valnosin-containing protein (VCP)4 and the intraflagellar transport 74 homolog (IFT74)5 and to chromosome 3 with mutations in the charged multivesicular body protein 2B (CHMP2B)6.
In 1995 the case of a Danish family with autosomal-dominantly inherited dementia was linked to chromosome 37; the disorder was named chromosome-3 linked frontotemporal dementia (FTD3)8. With a mean onset age of 57 years, FTD3 presents with symmetrical frontal and temporal cortical atrophy and, clinically, with personality change (mainly social behavioral disinhibition), hyperorality, dyscalculia, a range of speech disturbances and dystonic postures9. FTD3 is a TDP-43 negative FTLD-U; neuronal inclusions (in the cytoplasm of neurons located either in the dentate gyrus or sparse in the frontal or other cortical areas9) are characterized by ubiquitin and/or p62, which are proteins of the ubiquitin proteosome system (UPS). Based on these observations a novel nomenclature has been suggested for FTD3: FTLD-UPS10. In 2005 Skibinski et al. reported a G>C transition in the acceptor splice site of exon 6 in CHMP2B causing the two aberrant transcripts CHMP2Bintron5 and CHMP2BΔ10 6. This mutation segregated with 11 affected family members6. Screening of all the open reading frames in the region linked to FTD3 in one affected member who carried the CHMP2B mutation of the Danish family6 did not reveal any other pathogenic variant11. Momeni et al. described a C>T variant in exon 6 causing the nonsense mutation p.R186X in two asymptomatic siblings of a familial case of FTD with an apparent autosomal dominant mode of inheritance12. Further an A>G mutation in exon 2 was reported: the variant, with unclear pathogenicity, was predicted to cause the non-synonymous change p.I29V13, 14. More recently a novel missense mutation was reported in a Belgian familial case of FTD: a C>T transition predictive of a 165 amino acids long C-terminus truncated protein (p.Q165X), for which functional studies suggested impairment of late endosomal trafficking15. The authors reported also a p.N143S missense mutation in a patient with corticobasal syndrome (CBS)15: for this mutation functional unpublished data revealed no association with enlarged or aberrant endosomal phenotype15. Another report suggested a link between CHMP2B and amyotrophic lateral sclerosis (ALS): two missense mutations were reported in two ALS patients (p.Q206H and p.I29V)16. A recent report showed the same mutation (G>C) reported by Skibinski et al., 2005 in one Danish patient with FTD and his half sibling (the patient is related to the Danish pedigree reported in Skibinski et al., 2005)17.
CHMP2B is located on chromosome 3p11.2. CHMP2B protein is composed of 213 amino acids and is a component of the heteromeric ESCRT-III (Endosomal Sorting Complex required for Transport III) complex. CHMP2B is involved in 1) the process of sorting and trafficking surface receptors or proteins into intraluminal vesicules (ILVs) for lysosomal degradation and 2) binding the Vps4 protein responsible for the dissociation of ESCRT components18, 9. CHMP2B is expressed in all major regions of the brain, including the hippocampus, frontal and temporal lobes and cerebellum6.
Here we report a novel missense mutation in CHMP2B: the nucleotide change G>A (AGC>AAC), in exon 6, determines the p.S187N amino acid change in a patient diagnosed with FTD. Pathogenicity of mutations in CHMP2B is discussed.
MATERIAL AND METHODS
All experiments on human subjects were conducted in accordance with the Declaration of Helsinki and informed consent was obtained from all living individuals participating in this study under Protocol 02-N-0010 of the National Institute of Health (NIH). The Neuroscience IRB of the NIH and the institutional IRB at Texas Tech University approved the study.
We extracted genomic DNA from the peripheral blood of the patient and the available relatives using the Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) following standard protocol as recommended by manufacturer. We evaluated MAPT haplotype, ApoE genotyping and performed sequencing of the candidate genes TAU, PGRN and CHMP2B. Sequencing of purified PCR amplicons was carried out from both directions using the Big Dye Terminator kit (ABI, Fosters City, CA, USA) following standard protocol as recommended by manufacturer, run in 3730 DNA analyzer (ABI) and analyzed with the Sequencher 4.9 (Gene codes corporation, Ann Arbor, MI, USA).
RESULTS
Case report
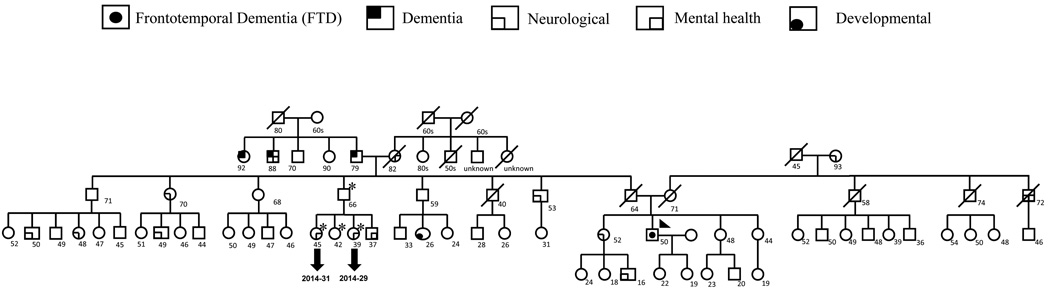
The proband is of English heritage from his father’s side and Irish/Swedish heritage from his mother’s side. As seen in the pedigree (Fig. 1), the proband’s paternal grandfather was affected by dementia in his 70s marked by bizarre dysexecutive behaviors (such as covering up lamps with blankets rather than turning them off), which was attributed at the time to “hardening of the arteries”. A sister and a brother of the paternal grandfather also suffered from dementia in their 70s, which was also attributed to “hardening of the arteries”. The paternal grandmother suffered from strokes in her 70s. The maternal grandmother had depression. The father of the proband died at the age of 62 from a myocardial infarction, whereas his mother is alive at age 70 with history of depression, but cognitively intact. Two siblings of the proband’s father suffer from depression and alcoholism and a sibling of the proband’s mother also suffers from alcoholism. A sibling of the proband has depression and one of her children has attention deficit disorder (ADD). The patient presented in this report developed depression in his early 40s and showed the first symptoms suggestive of FTD at the age of 50 (executive dysfunction leading to mistakes at work which caused him to be fired). Over time, his executive function worsened, in terms of action sequencing, multi-tasking and working memory. In parallel, he developed personality change, with loss of empathy, emotional disengagement from his family, and low tolerance to frustration. He developed a number of compulsive behaviors, including incidents of kleptomania, a tendency to collect useless items, compulsive overeating with stuffing of large quantities of food into his mouth, as well as repetitive purposeless behaviors, such as constant pacing. His motivation and energy decreased dramatically over time. His speech output decreased dramatically over time, eventually to the point of mutism other than occasional answers to yes/no questions. He did not develop significant gait disorder, other motor symptoms or fasciculations.
Figure 1. Pedigree of the proband.
The complete pedigree of the family of the proband is shown. Proband is identified by the arrow. Family members who have been screened for follow up are identified by asterisk. Characteristics of the index patient and family history are discussed in the manuscript.
His neurological exam was most interesting for paucity of spontaneous movement and inattention. He made poor eye contact with the examiner or the caregiver, but appeared euthymic. His speech was sparse, but grammatically and syntactically intact without evidence of dysarthria or apraxia. His profound executive dysfunction and inattention precluded most neurologic testing relying on co-operation. Saccadic breakdown of smooth pursuit was the only occulomotor abnormality noted. Motor exam showed paratonia, increased reflexes throughout (without fasciculations) and bilateral Babinski sign. He had positive glabellar and palmomental reflexes. His cerebellar exam was unremarkable; his gait exam showed decreased arm swing bilaterally.
Neuropsychological testing revealed that he was severely impaired on the Mattis Dementia Rating Scale-2. His performance on Wechler Adult Intelligence Scale-III showed moderately to severely impaired verbal and visuospatial skills and mildly impaired digit span. He showed borderline naming ability on Boston Naming Test. His memory on Wechler Memory Scale-III was severely impaired, as well as his executive functions (concept formation, reasoning and planning) on the Delis-Kaplan Executive Function System. His wife reported increase in apathy, disinhibition and executive dysfunction (on the Frontal Systems Behavior Scale) and increase in irritability and agitation on the UCLA Neuropsychiatric Inventory19, 20, 21, 22, 23, 24, 25. His brain MRI showed marked atrophy of the frontal and temporal lobes, more severe to the right hemisphere. The observed atrophy was progressive on serial MRIs. His fluoro-deoxy-glucose positron emission tomography (FDG-PET) showed reduction in cerebral glucose metabolism across the frontal and temporal cortex with involvement of the parietal cortex in both hemispheres, more severe to the right hemisphere.
Overall, the patient’s history, clinical exam, neuropsychological testing and neuroimaging studies are consistent with the diagnosis of Frontotemporal Dementia- Frontal Variant26.
Genetic screening
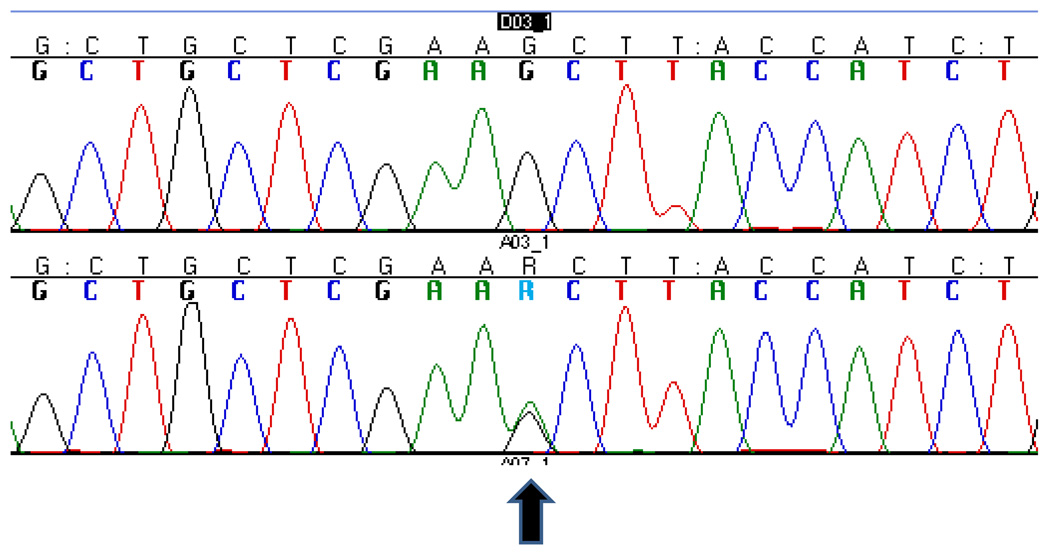
The complete results of the genetic screening of the proband are summarized in Table 1A. We identified a novel missense mutation: g.26218G>A (Fig.2) in exon 6 of CHMP2B in the C-terminus of the protein (p.S187N). This mutation was not found in 273 Caucasian neurologically normal controls (NDPT 098, NDPT 099, and NDPT 096: Coriell Cell Repositories, Camden, NJ, USA). In Momeni et al., 2006, 400 neurological normal controls were sequenced for CHMP2B exon 6 and this variant was not found in those samples12. However, we found the variant reported in this paper in 6 of 94 (6.4%) normal controls of African American ethnicity (Coriell Cell Repositories: NDPT 031). We were not able to determine the mode of inheritance of the mutation, since the father of the proband is deceased and it was not possible to collect blood samples from the mother. We were able to collect samples from an uncle and 3 cousins of the proband (Fig.1). Two of the cousins show neurological problems: one (2014–31) suffers from migraine and has abnormal brain imaging, the other (2014–29) only suffers from migraine. We did not detect the mutation reported for the index patient in any of his relatives.
Table 1.
| A. Summary of genotyping and sequences screening for patient 165 | ||
|---|---|---|
| ApoE | Sequencing | MAPT Haplotype |
| MAPT |
||
| E3/e3 | Exon 1, A>G (+8) from 5' exon 1, homozygous G (rs17650901) | H2/H2 |
| Intron 8, G>A (−26) from 5' of exon 9, homozygous A (rs62063850) | ||
| Exon 9, A>G, p.A227A (silent), homozygous G (rs1052553) | ||
| Exon 9, T>C, p.N255N (silent), homozygous C (rs17652121) | ||
| Intron 11, G>A (+34) from 3' of exon 11, homozygous A * | ||
| PGRN |
||
| Intron 3, G>A (+21) from 3' of exon 3 (rs9897526) | ||
| Intron 4, del/ins GTCA (−47−50) from 5' exon 5, heterozygous deletion (rs34424835) | ||
| Intron 5, G>A (+24) from 3' exon 5 (rs850713) | ||
| Intron 12, C>T (+78) from 3' exon 12 (rs5848) | ||
| CHMP2B |
||
| Exon 3, T>C, p.T104T (silent), homozygous C (rs11540913) | ||
| Exon 6, G>A, p.S187N * | ||
| B. Summary of C-truncation and missense mutations in CHMP2B | |||
|---|---|---|---|
| C-truncation mutations | Base change | Disease | Reference |
| CHMP2Bintron5 | g.26189G>C | FTD | 6 |
| CHMP2BΔ10 | g.26189G>C | FTD | 6 |
| p.Q165X | g.25950C>T | FTD | 15 |
| p.R186X | g.26214C>T | FTD | 12 |
| Missense mutations | |||
| p.I29V | g.13227A>G | FTD3; ALS | 13; 16 |
| p.N143S | g.25885A>G | CBD | 15 |
| p.D148Y | g.25899G>T | FTD | 6 |
| p.S187N | g.26218G>A | FTD | this report |
| p.Q206H | g.26276A>C | ALS | 16 |
Sequencing analysis resulted in the identification of single nuclear polymorphysms (SNPs) in microtubule associated protein TAU (MAPT), progranulin (PGRN) and Charged Multivesicular body Protein 2B (CHMP2B). Most of the SNPs are known; novel, non previously reported SNPs, are identified by an asterisk. The G>A variant in intron 11 in MAPT gene is probably tagging the H2/H1 inversion.
Figure 2. Sequencing electropherogram.
The normal sequence (top) is compared to the sequence of the patient carrying the g.26218C>T, p.S187N.
DISCUSSION
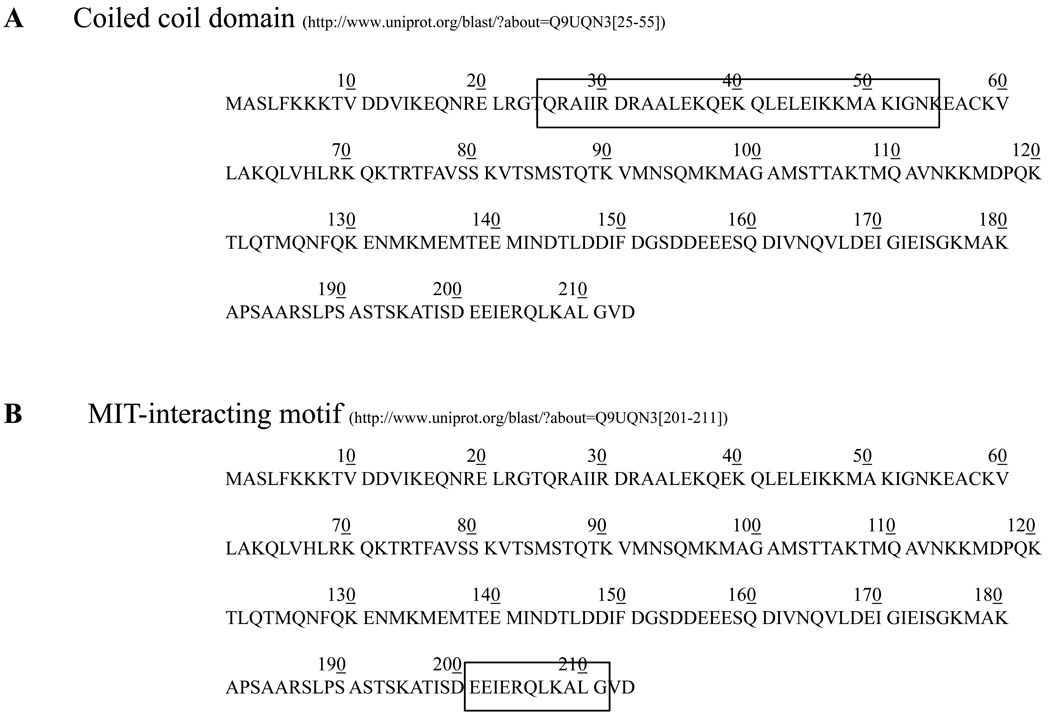
CHMP2B is part of the ESCRT-III complex which is directly involved in sorting the cargo proteins into ILVs18, 27. ESCRTs are highly conserved in all major taxa18. CHMP2B is characterized by 1) a coiled coil domain at the N-terminus (Fig. 3A28) and 2) an MIT (microtubule interacting and transport)-interacting region (MIR), at the C-terminus (Fig. 3B29). In the ESCRT-III complex, CHMP2B, together with CHMP2A and Vps24 (vacuolar protein sorting 24), binds the MIT domain of the hexameric protein Vps4 through its MIR domain30, 31. The interaction between these components determines 1) the active dissociation of ESCRTs from the endosomal membrane and 2) the formation and release of ILVs32, 33. Impairment of this machinery could determine the disruption of the endosomal trafficking leading, potentially, to 1) the lack of trophic support for the cell, 2) aberrant cellular signaling and 3) impairment of autophagy 9.
Figure 3. Functional domains in CHMP2B.
A. The coiled coil domain is located close to the N-terminus of CHMP2B, between aminoacids 25 and 55. B. The microtubule-interacting and transport (MIT)-interacting region (MIR) is located at the C-terminus of CHMP2B, between aminoacids 201–211. This domain is responsible for the binding of CHMP2B to Vps4, the hexameric AAA + ATPase that disassembles the ESCRT complex prior formation of ILVs.
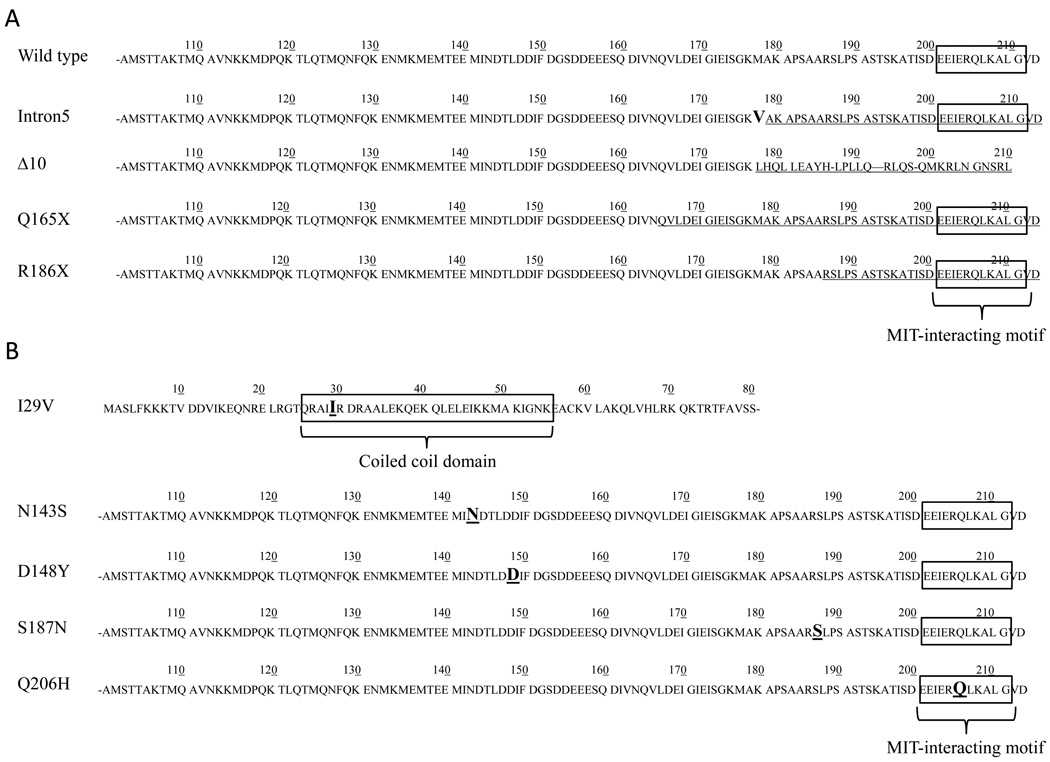
During the past five years several mutations in CHMP2B have been reported. These mutations can be divided into two groups: 1) the ones leading to C-terminus truncated proteins and 2) the missense mutations (table 1B). Researchers investigated both kinds of mutations in functional studies to evaluate their pathogenicity. While missense mutations have not been related to specific pathogenic pathways, the C-terminus truncation mutations have been associated with abnormal phenotypes of the late endosomes6, 9, 15. Impaired trafficking of multivesicular bodies (MVBs) to lysosomes would cause cytoplasmic accumulation of vesicles: such a phenotype has shown to lead to neurodegeneration in mice34, 35. Investigation of the ESCRT machinery showed that depletion of ESCRT subunits causes abnormal morphology of the MVBs. For example, cells depleted of Tsg101 (subunit of ESCRT-I) and Vps24 (subunit of ESCRT-III) showed p62 positive structures in the cytoplasm of HeLa cells, which is similar phenotype of cells expressing CHMP2Bintron5 mutants36. These studies show that depletion or dysfunction of different subunits of the ESCRT complex can affect late endosomal trafficking and cause protein or vesicles accumulation. However the ubiquitin- and p62-positive inclusions observed in the Danish FTD patient brains with the CHMP2B mutation occur at low frequency when compared with other cases of FTLD-U and are observed mostly in the hippocampus, which is not a site of neurodegenerative pathology in this disease37. Recently, a study suggested the implication of CHMP2Bintron5 in the misregulation of Toll-like receptor (TLR), due to abnormal sorting in the endocytic pathway; this would affect the TLR pathway, which may lead to neurodegeneration38.
C-terminus truncation mutations
Mutations leading to the variants CHMP2Bintron5, CHMP2BΔ10, p.Q165X and p.R186X (Fig. 4A) cause loss of the Vsp4 binding domain. CHMP2Bintron5 and p.Q165X mutations cause aberrant cytoplasmic phenotype if compared to cells transfected with wild-type CHMP2B leading to the assumption that these mutations could cause FTD6, 9 15. On the other hand, interestingly, functional analysis of CHMP2BΔ10 showed no real implication of CHMP2BΔ10 in neurodegeneration39 suggesting the possibility that not all reported CHMP2B mutations are pathogenic38. Further, the p.R186X mutation found in two asymptomatic members of an FTD family raises questions about the pathogenicity or penetrance of the C-terminus truncating mutations in CHMP2B. Based on the present clinical data, p.R186X could be 1) a non pathogenic CHMP2B C-terminus truncating mutation, 2) a pathogenic mutation with variable penetrance or 3) a pathogenic mutation causing variable age of onset in FTD.
Figure 4. Primary protein structure of CHMP2B displaying mutations.
A. All the known mutations leading to a C-terminus truncation phenotype are shown and compared to the wild type protein sequence of CHMP2B (top). The C-terminus protein sequence that is lost, due to mutation, is underlined. Truncation determines the loss of the MIT-interacting region. B. All the known missense mutations are shown. Each amino acid change is bold and underlined. In the case of p.I29V the missense mutation is within the coiled coil domain, while in p.Q206H missense mutation the amino acid change appears in the MIT-interacting region. These are the two only missense mutations that are located within a functional domain. The remaining three mutations are in a non functional region of the protein.
Missense mutations
In this report we present a novel missense mutation, p.S187N. This mutation was not found in a total of 273 Caucasian and, in a previous study, in a total of 400 Caucasian neurologically normal controls12. We found this variant in 6.4% of African American controls. This variant is found more frequently in the African American than in the Caucasian population. It could be a non pathogenic polymorphism. Other missense mutations reported earlier, p.D148Y and p.N143S, were neither associated with pathogenicity nor with aberrant endosomal phenotype6, 15, 38, while the variant leading to p.I29V was found in only one normal control 13, 14. For p.Q206H, which appears to be within the MIR domain (Fig. 4B), no functional studies are available. Taken together these data do not support a direct pathogenetic role of CHMP2B missense mutations in FTD. In the case reported in this manuscript, beside the proband, no other family members carried the mutation. Unfortunately, we were unable to obtain DNA from the proband’s parents., therefore, we cannot conclude whether this mutation segregates with the disease. Pathological studies will show if the proband’s brain pathology is consistent with CHMP2B mutation-type FTD.
Overall, the pathogenicity of CHMP2B mutations requires further proof. As of today, based on the fact that mutations causing C-terminus truncation of the protein contribute to the impairment of endosomal functions and trafficking and have a detrimental effect on the normal function of the protein6, 15, mutations in CHMP2B are considered a rare cause of familial FTD. Further screening for CHMP2B mutations is needed to come to a concrete conclusion on the pathogenic role of CHMP2B variants in FTD.
ACKNOWLEDGEMENTS
PM molecular genetics work is funded by the office of the Dean of the School of Medicine, department of Internal Medicine, at Texas Tech Health Sciences Center and a grant from South Plains Foundation. This study was supported by the intramural programs of The National Institutes of Health (NIH) / The National Institute on Aging (NIA)/ The National Institute of Neurological Disorders and Stroke (NINDS, JG and EDH), NINDS grant 1K99NS060766-01 (EDH), and the Litwin-Zucker Center for Research on Alzheimer’s Disease and Memory Disorders (EDH and PD). We would like to thank Eric M. Wassermann for clinical evaluations, Anne Leopold, Michael Tierney, and Karen DeTucci for patient testing, and the NINDS nurses for patient care.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Ratnavalli E, Brayne C, Dawson K, et al. The prevalence of frontotemporal dementia. Neurology. 2002;58(11):1615–1621. doi: 10.1212/wnl.58.11.1615. [DOI] [PubMed] [Google Scholar]
- 2.Snowden JS, Neary D, Mann DM. Frontotemporal dementia. Br J Psychiatry. 2002;180:140–143. doi: 10.1192/bjp.180.2.140. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher A, Friedrich P, Diehl-Schmid J, et al. No association of chromatin-modifying protein 2B with sporadic frontotemporal dementia. Neurobiol Aging. 2007;28(11):1789–1790. doi: 10.1016/j.neurobiolaging.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 4.Watts GD, Wymer J, Kovach MJ, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36(4):377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 5.Momeni P, Schymick J, Jain S, et al. Analysis of IFT74 as a candidate gene for chromosome 9p-linked ALS-FTD. BMC Neurol. 2006;6:44. doi: 10.1186/1471-2377-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skibinski G, Parkinson NJ, Brown JM, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nat Genet. 2005;37(8):806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 7.Brown J, Ashworth A, Gydesen S, et al. Familial non-specific dementia maps to chromosome 3. Hum Mol Genet. 1995;4(9):1625–1628. doi: 10.1093/hmg/4.9.1625. [DOI] [PubMed] [Google Scholar]
- 8.Gydesen S, Brown JM, Brun A, et al. Chromosome 3 linked frontotemporal dementia (FTD-3) Neurology. 2002;59(10):1585–1594. doi: 10.1212/01.wnl.0000034763.54161.1f. [DOI] [PubMed] [Google Scholar]
- 9.Urwin H, Ghazi-Noori S, Collinge J, et al. The role of CHMP2B in frontotemporal dementia. Biochem Soc Trans. 2009;37(Pt 1):208–212. doi: 10.1042/BST0370208. [DOI] [PubMed] [Google Scholar]
- 10.Mackenzie IR, Neumann M, Bigio EH, et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117(1):15–18. doi: 10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Momeni P, Bell J, Duckworth J, et al. Sequence analysis of all identified open reading frames on the frontal temporal dementia haplotype on chromosome 3 fails to identify unique coding variants except in CHMP2B. Neurosci Lett. 2006;410(2):77–79. doi: 10.1016/j.neulet.2006.06.065. [DOI] [PubMed] [Google Scholar]
- 12.Momeni P, Rogaeva E, Van Deerlin V, et al. Genetic variability in CHMP2B and frontotemporal dementia. Neurodegener Dis. 2006;3(3):129–133. doi: 10.1159/000094771. [DOI] [PubMed] [Google Scholar]
- 13.Rizzu P, van Mil SE, Anar B, et al. CHMP2B mutations are not a cause of dementia in Dutch patients with familial and sporadic frontotemporal dementia. Am J Med Genet B Neuropsychiatr Genet. 2006;141B(8):944–946. doi: 10.1002/ajmg.b.30410. [DOI] [PubMed] [Google Scholar]
- 14.Cannon A, Baker M, Boeve B, et al. CHMP2B mutations are not a common cause of frontotemporal lobar degeneration. Neurosci Lett. 2006;398(1–2):83–84. doi: 10.1016/j.neulet.2005.12.056. [DOI] [PubMed] [Google Scholar]
- 15.van der Zee J, Urwin H, Engelborghs S, et al. CHMP2B C-truncating mutations in frontotemporal lobar degeneration are associated with an aberrant endosomal phenotype in vitro. Hum Mol Genet. 2008;17(2):313–322. doi: 10.1093/hmg/ddm309. [DOI] [PubMed] [Google Scholar]
- 16.Parkinson N, Ince PG, Smith MO, et al. MRC Proteomics in ALS Study FReJA Consortium. Neurology. 2006;67(6):1074–1077. doi: 10.1212/01.wnl.0000231510.89311.8b. [DOI] [PubMed] [Google Scholar]
- 17.Lindquist SG, Braedgaard H, Svenstrup K, et al. FReJA Consortium, Frontotemporal dementia linked to chromosome 3 (FTD-3)--current concepts and the detection of a previously unknown branch of the Danish FTD-3 family. Eur J Neurol. 2008;15(7):667–670. doi: 10.1111/j.1468-1331.2008.02144.x. [DOI] [PubMed] [Google Scholar]
- 18.Williams RL, Urbé S. The emerging shape of the ESCRT machinery. Nat Rev Mol Cell Biol. 2007;8(5):355–368. doi: 10.1038/nrm2162. [DOI] [PubMed] [Google Scholar]
- 19.Wechsler D. Wechsler Memory Scale Revised. San Antonio, TX: Harcourt Brace Jovanovich, Inc; 1987. [Google Scholar]
- 20.Wechsler D. WAIS-III. Administration and scoring manual. San Antonio, TX: The Psychological Corporation; 1997. [Google Scholar]
- 21.Kaplan H, Goodglass H, Weintraub S. Boston Naming Test. Philadelphia: Lea & Febiger; 1983. [Google Scholar]
- 22.Mattis S. Mental Status examination for organic mental syndrome in the elderly patient. In: Bellack L, Karusu TB, editors. Geriatric Psychiatry. New York: Grune & Stratton; 1976. pp. 77–121. [Google Scholar]
- 23.Delis DC, Kaplan E, Kramer JH. The Delis-Kaplan Executive Function System: Examiner's Manual. San Antonio, TX: The Psychological Corporation; 2001. [Google Scholar]
- 24.Cummings JL, Mega M, Gray K, et al. The Neuropsychiatric Inventory: comprehensive assessment of psychopathology in dementia. Neurology. 1994;44(12):2308–2314. doi: 10.1212/wnl.44.12.2308. [DOI] [PubMed] [Google Scholar]
- 25.Grace J, Malloy P. Frontal Systems Behavior Scale (FrSBe): Professional Manual. Lutz, FL: Psychological Assessment Resources; 2001. [Google Scholar]
- 26.Neary D, Snowden JS, Gustafson L, et al. Benson, Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 27.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3(12):893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 28.Q9UQN3[25-55, Charged multivesicular body protein 2b, Homo sapiens (Human) [Accessed July 2009]. Available at http://www.uniprot.org/blast/?about=Q9UQN3[25-55.
- 29.Q9UQN3[201-211, Charged multivesicular body protein 2b, Homo sapiens (Human) [Accessed July 2009]. Available at http://www.uniprot.org/blast/?about=Q9UQN3[201-211.
- 30.Nickerson DP, West M, Odorizzi G. Did2 coordinates Vps4-mediated dissociation of ESCRT-III from endosomes. J Cell Biol. 2006;175(5):715–720. doi: 10.1083/jcb.200606113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang HT, Connell JW, Brown SE, et al. A systematic analysis of human CHMP protein interactions: additional MIT domain-containing proteins bind to multiple components of the human ESCRT III complex. Genomics. 2006;88(3):333–346. doi: 10.1016/j.ygeno.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Hurley JH, Yang D. MIT domainia. Dev Cell. 2008;14(1):6–8. doi: 10.1016/j.devcel.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 33.Obita T, Saksena S, Ghazi-Tabatabai S, et al. Structural Basis for Selective Recognition of Escrt-III by the Aaa ATPase Vps4. Nature. 2007;449:735–739. doi: 10.1038/nature06171. [DOI] [PubMed] [Google Scholar]
- 34.Hara T, Nakamura K, Matsui M, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441(7095):885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 35.Komatsu M, Waguri S, Chiba T, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 36.Filimonenko M, Stuffers S, Raiborg C, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179(3):485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holm IE, Englund E, Mackenzie IR, et al. A reassessment of the neuropathology of frontotemporal dementia linked to chromosome 3. J Neuropathol Exp Neurol. 2007;66(10):884–891. doi: 10.1097/nen.0b013e3181567f02. [DOI] [PubMed] [Google Scholar]
- 38.Lee JA, Beigneux A, Ahmad ST, et al. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17(18):1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 39.Ahmad ST, Sweeney ST, Lee JA, et al. Genetic screen identifies serpin5 as a regulator of the toll pathway and CHMP2B toxicity associated with frontotemporal dementia. Proc Natl Acad Sci U S A. 2009;106(29):12168–12173. doi: 10.1073/pnas.0903134106. [DOI] [PMC free article] [PubMed] [Google Scholar]