Abstract
Bacterial cytoskeletal filamentous proteins, like their eukaryotic counterparts, are key regulators and central organizers of many cellular processes including morphogenesis, cell division, DNA segregation and movement. Such filaments often organize themselves into complex structures within the prokaryotic cell, driven by molecular crowding and cation association, to form bundles (ParM), rings, toroids and helical spirals (FtsZ) or interwoven sheets (MreB). The formation of complex structures is essential for bacterial cytoskeleton function. Here, we highlight the suprastructures of the prokaryotic cytoskeleton that have been observed by high resolution in vitro electron microscopy and set them in perspective with in vivo observations. We discuss the underlying physical principles that lead to complex structure formation.
Key words: bacterial cytoskeletal filaments, suprastructures, toroids, bundles, helices, molecular crowding, cations, counterion mediated electrostatic attractive forces
Ions and water molecules screen charged molecules from each other within a cell. However, the biological polymers that constitute the cytoskeleton associate with counterions that can result in strong electrostatic interactions between the like-charged filaments.1,2 In addition, the interior of a cell is about 40% full of various macromolecules, termed molecular crowding. Molecular crowding has been known for almost half a century to have profound effects on the thermodynamics and kinetics of cellular processes and can enhance the native state of proteins.3,4 Due to the excluded volume effect, molecular crowding leads to short range forces between cytoskeletal proteins and shifts the equilibrium from single filaments to more complex supramolecular structures.5 The complex eukaryotic cytoskeleton is tightly regulated by a large number of actin- or microtubule-associated proteins. In contrast, bacterial actins and microtubules have few associated regulatory proteins. This simplifies the quest for finding the basic physical principles that govern the formation of filamentous suprastructures.
Suprastructures of the bacterial cystoskeleton have been directly visualized by high resolution in vitro electron microscopy. We present three examples:
Cell division protein FtsZ, a microtubule homolog, assembles at the midcell into a ring of multiple filaments called the Z-ring. The ring forms on the inside of the cytoplasmatic membrane and meditates membrane constriction, resulting in the production of two newborn cells. The Z-ring donut or toroid-like appearance has only been observed at low resolution (0.2 µm) by fluorescent light microscopy. In addition FtsZ also forms dynamic helical like assemblies6,7 which transform interchangeably into the Z-ring. We have recently shown in vitro that FtsZ, from different species (Escherichia coli or Mycobacteria tuberculosis), can be organized into supramolecular structures such as straight bundles, helices, rings or toroids as a function of counterions and molecular crowding.8,9 The transformation between the different configurations (straight ↔ helical ↔ circular) can be explained assuming that FtsZ subunits exist in two different structural conformations that are bistable (Fig. 1), an interpretation similar to that given for the multiple states seen in bacterial flagella.10 Local conditions may shift the bistable potential and induce a flip in curvature or pitch.
Figure 1.
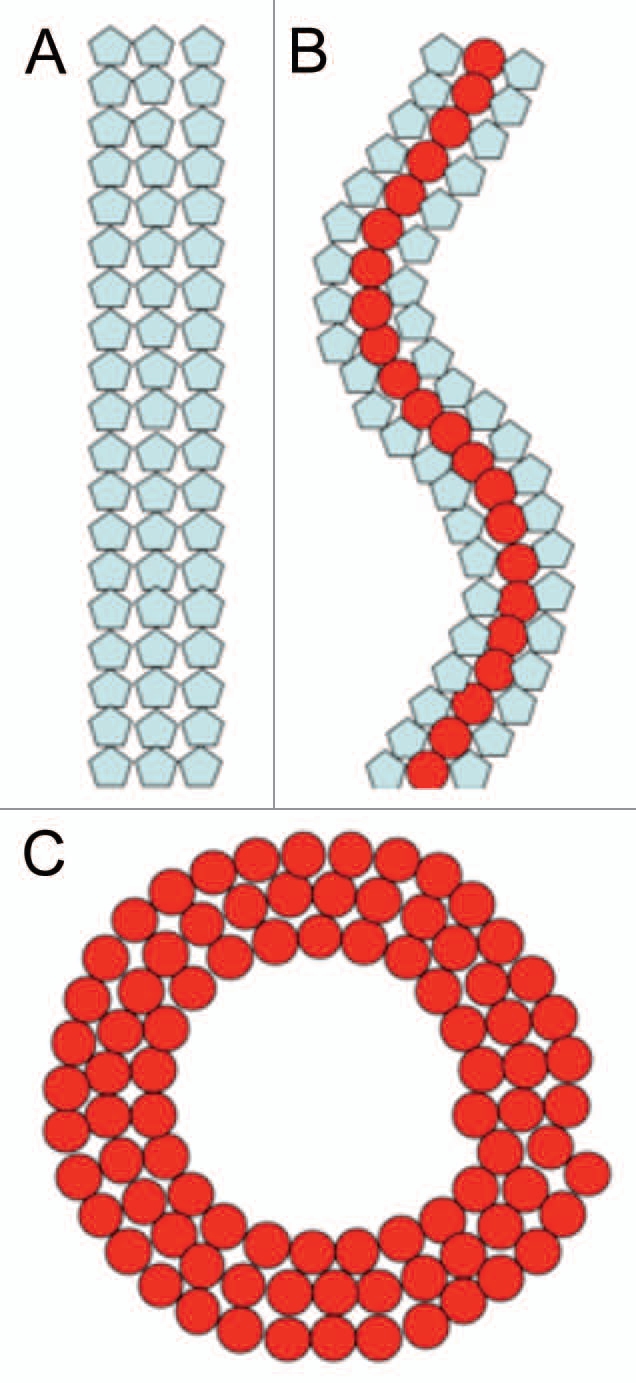
Cartoon of three different FtsZ suprastructures. (A) Straight filaments. (B) Helical filaments. (C) Toroids. Blue and red subunits illustrate two different conformations of the monomer. The transformation between the different supramolecular structures is caused by changes between the bistable conformations, due to a shift of local conditions (pH or ions). The inner diameter of rings and toroids is ∼200 nm, which could be an energy minimized state during Z-ring constriction.
ParM is a bacterial DNA segregation protein and actin homolog. In cryosectioned E. coli cells, ParM filaments were observed to consist of small filament bundles rather than individual filaments.11 We employed electron micrographs of in vitro ParM rafts, which are 2-D analogs of 3-D bundles, to identify the main molecular interfilament contacts within these suprastructures.12 Surprisingly the interface between filaments was similar for both parallel and antiparallel orientations suggesting that the distribution of filament polarity is random within a bundle (Fig. 2). Furthermore the interfilament interactions were not due to the interactions of specific residues but rather to long-range, counter ion mediated, electrostatic attractive forces. This bundle design has two advantages when bidirectionally segregating large DNA in the prokaryotic cell. The randomly oriented nature of the bundle allows DNA to be captured with equal efficiency at both ends of the bundle. Secondly the bundling of filaments greatly increases stiffness allowing the system to maneuver relatively large payloads, DNA plasmids.
Figure 2.
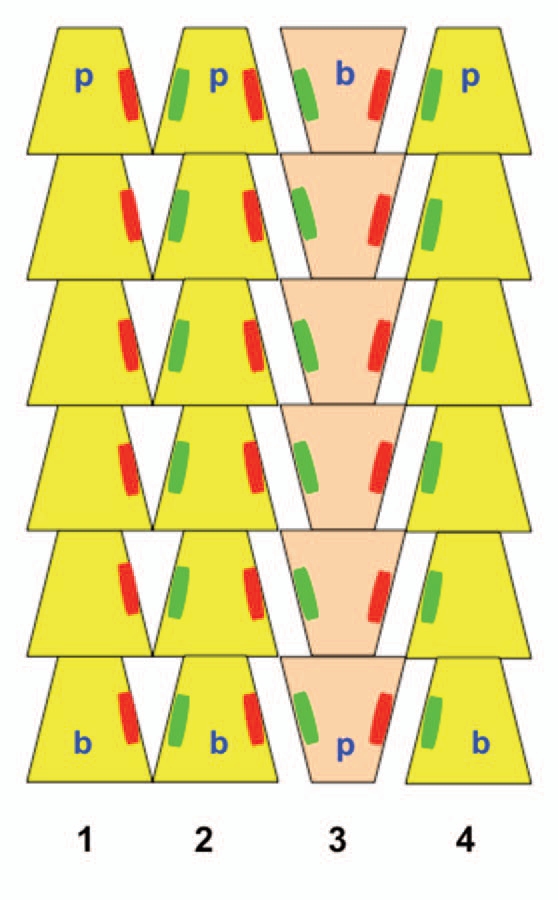
Schematic model of ParM bundles. Three filaments with their pointed ends (p) up are shown in yellow one filament with the barbed end (b) up is shown in tan. Parallel filaments within the bundle (filaments 1–2) share similar large areas of molecular interaction (illustrated as red and green patches) as filaments arranged anti-parallel (filaments 2–3 or 3–4).
In vivo fluorescence microscopy studies of bacterial cells have shown that the bacterial shape-determining protein and actin homolog, MreB, forms cable-like structures that spiral around the periphery of the cell.13 Surprisingly, MreB from Thermatoga maritime appears in vitro to consist of complex, several µm long multilayered sheets of interwoven filaments in the presence of either ATP or GTP14 (Fig. 3). The crystalline order is highest in the presence of divalent cation Mg2+, which can be present at high millimolar levels in bacterial cells. This architecture, in agreement with recent rheological measurements on MreB cables,15 has superior mechanical properties compared to a single filament or a sheet with filaments aligned in parallel and could be an important feature for maintaining bacterial cell shape.
Figure 3.
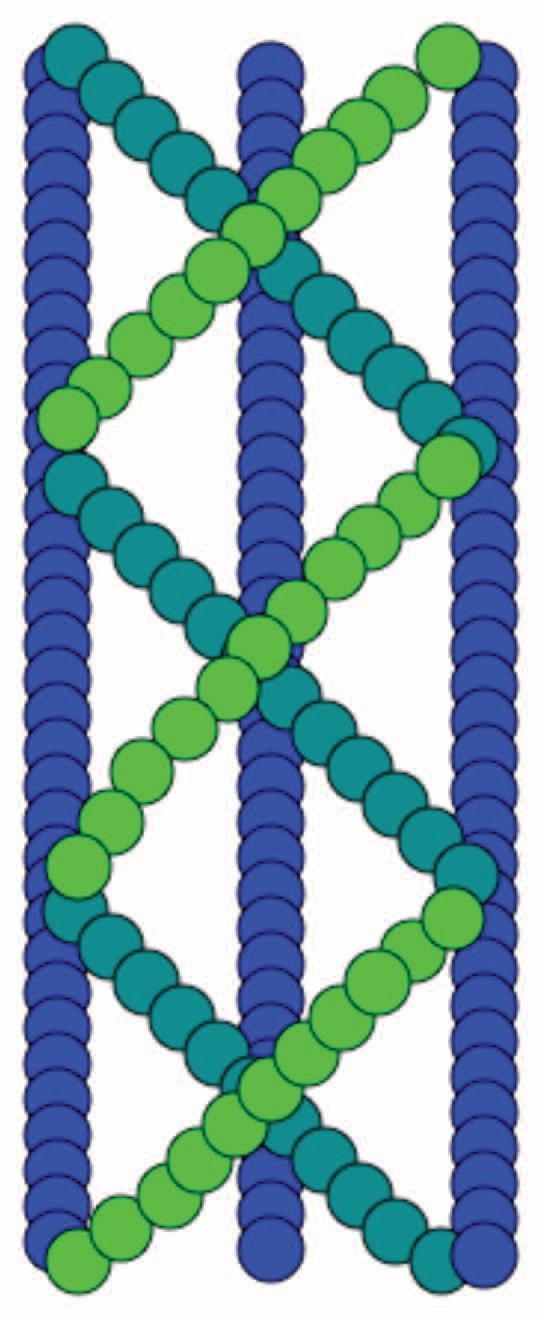
Schematic diagram of the molecular arrangement of interwoven MreB filaments within cables.
Collectively these findings suggest, that the filaments of the bacterial cytoskeleton can adopt specific supramolecular structures in response to the presence of both molecular crowding and cations that make them uniquely suited for the cellular processes in which they participate.
Addendum to: Popp D, Narita A, Maeda K, Fujisawa T, Ghoshdastider U, Iwasa M, et al. Filament structure, organization and dynamics in MreB sheets. J Biol Chem. 2010;285(21):15858–15865. doi: 10.1074/jbc.M109.095901. Popp D, Iwasa M, Erickson HP, Narita A, Maeda Y, Robinson RC. Suprastructures and dynamic properties of Mycobacterium tuberculosis FtsZ. J Biol Chem. 2010;285:11281–11289. doi: 10.1074/jbc.M109.084079. Popp D, Narita A, Iwasa M, Maeda Y, Robinson RC. Molecular mechanism of bundle formation by the bacterial actin ParM. Biochem Biophys Res Com. 2010;391:1598–1603. doi: 10.1016/j.bbrc.2009.12.078.
Footnotes
Previously published online: www.landesbioscience.com/journals/cib/article/12340
References
- 1.Ise N. When, why, and how does like like like? Proc Jpn Acad Ser B. 2008;83:192–198. doi: 10.2183/pjab/83.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wong CL, Pollack L. Electrostatics of strongly charged biological polymers: ion-mediated interactions and self-organization in nucleic acids and proteins. Annu Rev Phys Chem. 2010;61:171–189. doi: 10.1146/annurev.physchem.58.032806.104436. [DOI] [PubMed] [Google Scholar]
- 3.Minton AP. Influence of macromolecular crowding upon the stability and state of association of proteins: predictions and observations. J Pharm Sci. 2005;94:1668–1675. doi: 10.1002/jps.20417. [DOI] [PubMed] [Google Scholar]
- 4.Minton AP. Implications of macromolecular crowding for protein assembly. Curr Opin Struct Biol. 2000;10:34–39. doi: 10.1016/s0959-440x(99)00045-7. [DOI] [PubMed] [Google Scholar]
- 5.Popp D, Gov N, Iwasa M, Maeda Y. Effect of short-range forces on the length distribution of fibrous cytoskeletal proteins. Biopolymers. 2008;89:711–721. doi: 10.1002/bip.20999. [DOI] [PubMed] [Google Scholar]
- 6.van den Ent F, Amos L, Löwe J. Bacteria ancestry of actin and tubulin. Curr Opin Microbiol. 2000;4:634–638. doi: 10.1016/s1369-5274(01)00262-4. [DOI] [PubMed] [Google Scholar]
- 7.Peters PC, Migocki MD, Thoni C, Harry EJ. A new assembly pathway for the cytokinetic Z ring from a dynamic helical structure in vegetatively growing cells of Bacillus subtilis. Mol Microbiol. 2007;64:487–499. doi: 10.1111/j.1365-2958.2007.05673.x. [DOI] [PubMed] [Google Scholar]
- 8.Popp D, Iwasa M, Narita A, Erickson HP, Maeda Y. FtsZ condensates: an in vitro electron microscopy study. Biopolymers. 2009;91:340–350. doi: 10.1002/bip.21136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Popp D, Iwasa M, Erickson HP, Narita A, Maeda Y, Robinson RC. Suprastructures and dynamic properties of Mycobacterium tuberculosis FtsZ. J Biol Chem. 2010;285:11281–11289. doi: 10.1074/jbc.M109.084079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wakabayashi K, Mitsui T. A phenomenological theory of polymorphism of Salmonella flagella I. A model. J Phys Sc Jpn. 1972;33:175–182. [Google Scholar]
- 11.Salje J, Zuber B, Löwe J. Electron microscopy of E. coli reveals filament bundles involved in plasmid DNA segregation. Science. 2009;232:509–512. doi: 10.1126/science.1164346. [DOI] [PubMed] [Google Scholar]
- 12.Popp D, Narita A, Iwasa M, Maeda Y, Robinson RC. Molecular mechanism of bundle formation by the bacterial actin ParM. Biochem Biophys Res Com. 2010;391:1598–1603. doi: 10.1016/j.bbrc.2009.12.078. [DOI] [PubMed] [Google Scholar]
- 13.Jones LJ, Carballido-Lopez R, Errington J. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell. 2001;104:913–922. doi: 10.1016/s0092-8674(01)00287-2. [DOI] [PubMed] [Google Scholar]
- 14.Popp D, Narita A, Maeda K, Fujisawa T, Ghoshdastider U, Iwasa M, et al. Filament structure, organization and dynamics in MreB sheets. J Biol Chem. 2010 doi: 10.1074/jbc.M109.095901. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esue O, Wirtz D, Tseng Y. GTPase activity, structure and mechanical properties of filaments assembled from bacterial cytoskeleton protein MreB. J Bacteriol. 2006;188:968–976. doi: 10.1128/JB.188.3.968-976.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]


