Abstract
Frequently, primary hepatocytes are used as an in vitro model for the liver in vivo. However, the culture conditions reported vary considerably, with associated variability in performance. In this study, we characterized the differentiation character of primary human hepatocytes cultured using a highly defined, serum-free two-dimensional sandwich system, one that configures hepatocytes with collagen I as the substratum together with a dilute extracellular matrix (Matrigel) overlay combined with a defined serum-free medium containing nanomolar levels of dexamethasone. Gap junctional communication, indicated by immunochemical detection of connexin 32 protein, was markedly enhanced in hepatocytes cultured in the Matrigel sandwich configuration. Whole genome expression profiling enabled direct comparison of liver tissues to hepatocytes and to the hepatoma-derived cell lines, HepG2 and Huh7. PANTHER database analyses were used to identify biological processes that were comparatively overrepresented among probe sets expressed in the in vitro systems. The robustness of the primary hepatocyte cultures was reflected by the extent of unchanged expression character when compared directly to liver, with more than 77% of the probe sets unchanged in all overrepresented categories, representing such genes as C/EBPα, HNF4α, CYP2D6, and ABCB1. In contrast, HepG2 and Huh7 cells were unchanged from the liver tissues for fewer than 48% and 55% of these probe sets, respectively. Further, hierarchical clustering of the hepatocytes, but not the cell lines, shifted from donor-specific to treatment-specific when the probe sets were filtered to focus on phenobarbital-inducible genes, indicative of the highly differentiated nature of the hepatocytes when cultured in a highly defined 2-dimensional sandwich system.
Keywords: DNA microarray, human hepatocytes, in vitro hepatic model
INTRODUCTION
Primary hepatocytes are often used as an in vitro system to model biological processes that occur in the liver in vivo. In part due to rapid loss of hepatocyte differentiation status reported for most culture conditions, a variety of culture methodologies have been explored, with varying success, to better maintain hepatic character and function. In the 2-dimensional sandwich system, primary hepatocytes are plated on collagen-coated dishes and then overlaid with either collagen or Matrigel™, a commercially available extracellular matrix (ECM) material derived from the Engelbreth-Holm-Swarm sarcoma. The ECM includes such components as laminin, collagen IV, heparin sulfate proteoglycans, and entactin, components that comprise the extracellular milieu of the liver in vivo. Some reports have indicated that the sandwich culture model facilitates the preservation of certain liver characteristics, including cuboidal morphology of hepatocytes with features such as bile canaliculi, tight junctions, and gap junctions (Hoffmaster et al., 2004;LeCluyse et al., 1999;Moghe et al., 1996;Richert et al., 2002;Sidhu et al., 1993;Sidhu et al., 2004), expression of basolateral and canalicular domains of the plasma membrane such that polarized hepatic transport is retained (Annaert et al., 2005;Hoffmaster et al., 2004;Liu et al., 1999;Moghe et al., 1996), and expression and activity of drug metabolizing enzymes (De Smet et al., 2001;Kern et al., 1997;LeCluyse et al., 1999;Sidhu et al., 1993;Sidhu et al., 2004). In comparison to conventional monolayer cultures, the sandwich culture model appeared to enhance expression of liver-selective proteins such as albumin (De Smet et al., 2001;Moghe et al., 1996;Sidhu et al., 1993;Sidhu et al., 2004), transthyretin (Sidhu et al., 2004) and transferrin (Sidhu et al., 2004), and contributes to decreased levels of spontaneous apoptosis (De Smet et al., 2001) and oxidative stress (Richert et al., 2002).
Despite these reports, the specific conditions used by different investigators still vary considerably, and concerns remain that the sandwich culture model may not accurately represent the in vivo response to chemical challenge. For example, it was reported that the sandwich configuration was unable to rescue basal levels of cytochrome P450 (CYP) activity to that observed in freshly isolated hepatocytes (Richert et al., 2002) and that, while the expression of some CYP proteins, namely CYP1A1, 2B1/2, and 3A2 in the rat, increase with time in culture, others, such as CYP2E1, decrease until expression is completely lost (Farkas et al., 2005). Phase II biotransformation enzymes were reported to undergo a similar mixed fate in culture (Kern et al., 1997;Richert et al., 2002). Overall, losses of specific biotransformation function, together with other hepatic differentiation features, have limited the utility of these models with respect to their predictive potential. Thus, further optimization and validation of the sandwich culture model is required, especially for human, to establish a robust in vitro system for use in mechanistic and predictive studies of toxicology and drug metabolism.
In this investigation, we hypothesized that, based on previous development studies using rat hepatocytes (Sidhu et al., 1993;Sidhu et al., 2004), an optimized human 2-dimensional primary hepatocyte culture system could be deployed that robustly reflects the differentiation status and biotransformation functions of hepatocytes in vivo. To this end, we adapted the use of a dilute ECM overlay within a highly defined serum-free media, supplemented with physiological concentrations of a glucocorticoid, and used DNA microarray analysis to assess transcriptional expression patterns in culture across a series of ten independent human donors. In parallel, whole genome expression profiling was conducted directly in human liver tissue samples and in two commonly used established cell lines derived from human hepatomas, HepG2 and Huh7 cells. The data from the microarrays was subjected to expression analyses using the Protein ANalysis THrough Evolutionary Relationships (PANTHER) Classification system (www.pantherdb.org; Applied Biosystems, Foster City, CA) to identify overrepresented biological processes and molecular functions in each model. Using these methods, we illustrate that primary human hepatocytes cultured in a defined sandwich configuration represent an in vitro hepatic model that closely resembles an in vivo hepatic phenotype, with respect to differentiation, liver-specific markers and the ability to support functional responses to xenobiotic challenge.
MATERIALS AND METHODS
Cell culture
Primary human hepatocytes were obtained from the Liver Tissue Procurement and Distribution System (LTPDS) at the University of Pittsburgh, through NIH Contract #NO1-DK-9-2310. Available donor information is presented in Table 1. Donor organs not designated for transplantation were used to isolate hepatocytes according to a three step collagenase perfusion protocol (Strom et al., 1996). Preparations enriched for hepatocytes were received plated in collagen-coated T25 flasks. Upon arrival, the media was changed to William’s Media E supplemented with 1% penicillin/streptomycin, 10 mM HEPES, 20 μM glutamine, 25 nM dexamethasone, 10 nM insulin, 30 mM linoleic acid, 1 mg/ml BSA, 5ng/ml selenious acid, 5 μg/ml transferrin. Within 4–16 h, a 10 mg/ml stock solution of Matrigel™ (BD Biosciences, San Jose, CA), was added dropwise to the culture media and evenly distributed by gentle swirling diluted to a final concentration of 225 μg/ml. The media was subsequently changed every two days until cells were harvested for RNA extraction at 90 h (HH-G, HH-I) or 114 h (HH-A through HH-F, HH-H, HH-J), depending on the condition of the cells. Time course studies were also performed on hepatocytes from 3 donors cultured up to 258 h, however the extended time in contact with Matrigel™ did not result in any substantive changes in global gene expression profiles or on hierarchical clustering results relative to the 90 or 114 h culture periods. Huh-7 cells were maintained in minimal essential medium supplemented with 10% fetal bovine serum, 10 mM HEPES, 1% penicillin/streptomycin, and 0.1 mM nonessential amino acids. HepG2 cells were maintained under the same conditions as Huh-7 with one additional supplement to the media, 1 mM sodium pyruvate. All cells were maintained at 37°C under 5% CO2. All culturing materials were purchased from Invitrogen (Carlsbad, CA), unless otherwise noted.
Table 1.
Donor information for the ten human hepatocyte donors5.
| Donor identification | Age (years) | Gender | Ethnicity | Cause of death | Smoker/alcohol use |
|---|---|---|---|---|---|
| HH-A | 0.75 | M | C6 | Anoxia | No/No |
| HH-B | 57 | M | C | CVA/ICH7 | Yes (15 years)/Yes (5 drinks/week) |
| HH-C | 52 | M | C | N/A8 | N/A |
| HH-D | 61 | M | C | Gunshot wound | No/Yes (21 drinks/week) |
| HH-E | 63 | M | C | IVH9 | No/No |
| HH-F | 3 | F | C | Anoxia | No/No |
| HH-G | 73 | F | C | Head injury | No/No |
| HH-H | 29 | F | C | N/A | No/No |
| HH-I | 46 | F | C | Head injury | Yes (13 years)/Yes (3 drinks/week) |
| HH-J | 16 | F | C | N/A | No/No |
Donors were obtained from Dr. Stephen Strom at the University of Pittsburgh under the regulations of the Liver Tissue Procurement and Distribution System (LTPDS).
Caucasian
CVA/ICH; cerebrovascular accident/intracranial hemorrhage
N/A; not available
IVH; intraventricular hemorrhage
Immunohistochemistry
Following removal of culture media, the cells were rinsed with phosphate buffered saline (PBS), fixed in methanol for 5 min, rinsed with PBS, and blocked with 1X blocking buffer (Sigma-Aldrich, St. Louis, MO) for 1 h at room temperature. Anti-Connexin 32 monoclonal antibody (Zymed Laboratories, Inc., San Francisco, CA) was added to the cells diluted in blocking buffer (1:250), the cells were incubated at 4°C for 48 h. After rinsing with PBS 3X for 5 min each, the cells were incubated with secondary antibody, Alexa Fluor® 488 goat anti-mouse IgG (Molecular Probes, Invitrogen), diluted 1:400 in blocking buffer, for 45 min. After rinsing the cells with PBS, Immunofluorescence of CAR protein was observed with a Nikon inverted fluorescence microscope (Nikon USA, Melville, NY). Image capture was performed with SpotRT software and digital camera (Diagnostic Instruments, Sterling Heights, MI).
Phenobarbital treatment
After 66 or 90 hours in culture, selected cell cultures were treated with 500 μM PB for 24 h, an exposure shown previously to provide optimal induction of typical markers of response to PB without associated toxicity (Sidhu et al., 2004).
RNA isolation
For primary hepatocytes and hepatoma-derived cell lines, the media was aspirated, 1 ml of TRIzol® Reagent (Invitrogen) was added directly to each flask, and RNA was extracted according to the manufacturer’s instructions. Contaminating DNA was removed using DNA-free™ DNase Treatment & Removal Reagents (Ambion, Inc., Austin, TX) according to manufacturer’s instructions for rigorous DNase treatment. RNA concentrations were assessed with UV absorbance at 260 nm, using a SmartSpec 3000 Spectrophotometer (BioRad, Hercules, CA). Human liver RNA was obtained from flash-frozen human livers generously contributed by Dr. Kenneth Thummel from the University of Washington.
Quantitative Real-Time PCR
RNA was reverse transcribed into cDNA using the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA), following the manufacturer’s instructions. Gene expression was estimated using TaqMan® Assays-on-Demand™ Gene Expression systems (Applied Biosystems). Assays were prepared according to manufacturer’s recommendations for a 50 μl reaction volume (5 μl cDNA, 1X TaqMan® Universal PCR Master Mix, and 1X Assays-on-Demand™ Gene Expression Assay Mix containing unlabeled PCR primers and TaqMan® FAM™ dye-labeled MGB probe), divided into duplicate 25-μl reactions in a 96-well plate, and conducted using an Applied Biosystems 7300 Real-Time PCR System. Thermal cycling consisted of a UNG activation step for 2 minutes at 50°C and an initial denaturation step for 10 minutes at 95°C followed by 40 cycles of denaturation (15 seconds at 95°C) and annealing/extension (1 minute at 60°C). Gene products measured by QRT-PCR were albumin (ALB; Assay ID Hs00609411_m1) and α-fetoprotein (AFP; Assay ID Hs00173490_m1). Expression levels were estimated using the ΔΔCt method and normalized to 18S (Assay ID Hs99999901_s1) using pooled RNA from 6 human livers as a reference standard. ΔΔCt was transformed into fold-change with the following formula: fold change = 2−ΔΔCt.
Microarray protocol
A total of 27 RNA samples (5 μg) were hybridized to Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA) by Paradigm Array Labs, Inc (Icoria, Research Triangle Park, NC). The samples consisted of: three identical human liver RNA samples, each pooled from six individuals (3 arrays); one pool each of untreated and PB-treated HepG2 RNA, each from two cultures (2 arrays); one pool each of untreated and PB-treated Huh7 RNA, each from two cultures (2 arrays); and RNA from untreated and PB-treated human hepatocytes from ten independent donors (20 arrays). RNA quality was assessed at Paradigm Array Labs using an Agilent 2100 Bioanalyzer and NanoDrop® ND-1000 Spectrophotometer. Paradigm Array Labs performed amplification and labeling of cRNA, hybridization of the cRNA to the arrays, and scanning of the arrays, using Affymetrix reagent kits and instrumentation platforms.
Differentiation marker analysis
To estimate differentiation status in the in vitro models, prototypical differentiation markers were evaluated. Detection of each probe set on each array was assessed by the Microarray Suite version 5 (MAS5) algorithm in ArrayAssist 4.0 (Affymetrix), which uses a One-sided Wilcoxon’s signed rank test to assign a p-value for each probe set. This p-value is then used to make a ‘detection call’ to determine if a transcript is measurable (present), undetectable (absent), or uncertain (marginal) (for a more detailed explanation of the algorithm, see the Affymetrix Technical Note ‘Statistical Algorithms Reference Guide’, Part No. 701137, Rev. 3 at www.affymetrix.com). Only present probe sets were included in all subsequent analyses. The PLIER algorithm in the software package ArrayAssist 4.0 (Stratagene, La Jolla, CA) was used to summarize data from individual probes within a probe set (for further explanation of the algorithm, see the Affymetrix Technical Note ‘Guide to Probe Logarithmic Intensity Error (PLIER) Estimation’, Part No. 701903, Rev. 1 at www.affymetrix.com). Data were converted to signal log ratio format using ArrayAssist 4.0. For all gene-level analysis, the probe set determined as the most specific for the respective gene was chosen for subsequent analyses, unless otherwise noted (Dallas et al., 2005). Specificity was determined by the identifier at the end of the probe set ID; ‘_at’ indicates that that probe set is predicted to perfectly match only a single transcript, ‘_s_at’ may match multiple transcripts, and ‘_x_at’ indicates that one or more probes in this probe set are predicted as identical or highly similar to sequences other than the intended transcript (Affymetrix Technical Note ‘Design and Performance of the GeneChip® Human Genome U133 Plus 2.0 and Human Genome U133a 2.0 Arrays’, Part No. 701483, Rev. 2 at www.affymetrix.com). In this study, the probe sets used for ALB and AFP were 214837_at and 204694_at, respectively. When the signal log ratio resulted in a positive value, the fold change was calculated as: fold increase = 2Signal log ratio. When the signal log ratio resulted in a negative value, the fold change was calculated as: fold decrease = −(2(−Signal log ratio)).
Correlation analysis
The resulting data were then filtered from 54,675 probe sets to 39,485 probe sets by removing any probe sets detected as ‘Absent’ in all 27 arrays. To obtain an estimate of similarity in global gene expression between the arrays, ArrayAssist generated scatter plots and correlation coefficients for: the three comparisons between technical replicates (Liver A-Liver B; Liver A-Liver C; Liver B-Liver C), the six comparisons between the liver replicates and the two untreated hepatoma-derived cell lines (Liver A-HepG2; Liver A-Huh7; Liver B-HepG2; etc.), and the thirty comparisons between the liver replicates and the untreated hepatocytes from ten donors (Liver A-HH-A; Liver B-HH-A; Liver C-HH-A; Liver A-HH-B; Liver B-HH-B; etc.). Data were expressed as mean correlation ± SD, where SD represents deviation between the three measurements of correlation between a single in vitro sample and the three human liver replicates. One-way analysis of variance in combination with Tukey’s multiple comparison post-test was used to determine significance of the difference in correlation between Liver B (chosen as a representative liver sample) and each of the three groups of samples (technical replicates, cell lines, and hepatocytes).
Overrepresentation analysis
In order to identify biological trends, differentially expressed genes were subjected to gene ontology (GO) overrepresentation analysis. For each untreated array, a list was created consisting of all probe sets that were both present and expressed differentially in that array from the human liver tissues. Differential expression was defined as expression greater than a signal log ratio of 2 (4-fold increase from human liver) or less than a signal log ratio of −2 (4-fold decrease from human liver). Each list of differentially expressed probe sets was then submitted to the PANTHER Classification System to find overrepresented ontology categories (Thomas et al., 2003). For each ontology category, PANTHER calculates the number of genes identified in that category in both a list of differentially regulated genes and a reference list containing all the probe sets present on the Affymetrix array and compares these results using the binomial test to determine if there are more genes than expected in the differentially regulated list (Thomas et al., 2006). Overrepresentation is defined by p<0.05.
Regulation of individual probe sets in GO categories
To determine how the overrepresented categories were regulated in each in vitro model, a GO category-specific list for each category of interest was compiled containing those probe sets identified as differentially expressed in any of the three in vitro models. For each untreated array, all of the probe sets in each GO category-specific list detected as present on that array were scored as belonging to one of three regulation groups: ‘Not changed’ (−2 < signal log ratio < 2), ‘Increased’ (signal log ratio > 2), or ‘Decreased’ (signal log ratio < −2). For HepG2 and Huh7 cells, data in each regulation group are expressed as the percentage of probe sets detected as present. For the human hepatocytes, data in each regulation group are expressed as the mean percentage ± SD of probe sets detected as present, summarized from the ten individual hepatocyte donors. For the human hepatocytes, one-way analysis of variance in combination with Tukey’s multiple comparison post-test was used to determine significance of the difference between regulation groups in each GO category.
Gene-level analysis of liver-specific functions
To verify the biological relevance of the overrepresentation analysis, genes with known biological roles in three liver-specific ontology categories were assessed. Data are expressed as fold change, as described under ‘Differentiation marker analysis’. The following probe sets were used: C/EBPα, 204039_at; HNF4α, 230914_at; NFE2L2, 201146_at; NR1I2, 207202_s_at; NR1I3, 207007_at; PPARα, 206870_at; PPARδ, 37152_at; RXRα, 202449_s_at; TCF1, 210515_at; CYP1A2, 207609_s_at; CYP2B6, 206755_at; CYP2C9, 217558_at; CYP2D6, 207498_s_at; CYP2E1, 209975_at; CYP3A4, 205999_x_at; FMO5, 205776_at; EPHX1, 202017_at; EPHX2, 209368_at; GSTA1, 203924_at; GPX1, 200736_s_at; ABCB1, 209993_at; ABCB11, 208288_at; ABCC1, 202804_at; ABCC2, 206155_at; ABCC4, 203196_at; SLC22A1, 207201_s_at; SLC22A3, 205421_at; SLC22A7, 231398_at; and SLCO2B1, 211557_x_at.
Gene-level analysis of a complex biological response
To assess the ability of each in vitro hepatic model to perform a complex biological response to chemical treatment, three marker genes were analyzed for transcriptional induction in response to PB. Data are reported as fold change as described under ‘Differentiation marker analysis’.
Hierarchical clustering
To obtain a global view of the response of each in vitro model system to PB, hierarchical clustering was done on two different datasets. Signal log ratio was obtained as described in ‘Differentiation marker analysis’. First, the 27 arrays were subjected to hierarchical clustering analysis using the 39,485 probe sets found to be present or marginal in at least one array as the dataset. Second, both the 27 arrays and PB-inducible probe sets were subjected to clustering analysis using a dataset consisting only of PB-inducible probe sets. PB-inducible probe sets were defined in each pair of samples (untreated and PB-treated) by defining the baseline array as the untreated sample and identifying any probe sets increased at least 4-fold in the treated sample. The PB-inducible dataset consisted of 54 probe sets, representing 31 unique genes that were both inducible in at least two pairs of samples and present in at least half of the arrays (13 arrays). All hierarchical clustering was performed using a Euclidean distance measurement and an average linkage.
Protein isolation
For each human hepatocyte and cell line sample, protein was isolated from the organic phase of the TRIzol® Reagent remaining after RNA isolation according to the protocol provided by the manufacturer. Protein concentrations were determined according to the Bradford method, using a commercially available kit (BioRad Protein Assay) and bovine serum albumin as the standard. S9 fractions were derived from human liver tissue samples as described previously (Hassett et al., 1997).
Western blotting
Protein samples (S9 fractions, 20 μg; protein isolated from TRIzol® reagent, 40 μg) were separated by electrophoresis on a precast Ready Gel (10% Tris-HCl, BioRad) and transferred to Immun-Blot PVDF membrane (BioRad). The PVDF membrane was subjected to a blocking procedure by incubation in 5% non-fat dry milk in TBS-Tween 20 (0.1%) for 1 h. Primary antibodies were diluted in blocking buffer as follows: albumin, 1:7500 (monoclonal antibody #A6684; Sigma-Aldrich); β actin, 1:3000 (monoclonal antibody #A5441; Sigma-Aldrich); CYP2B6, 1:650 (monoclonal antibody clone 49-10-20); CYP2C9/18, 1:1000 (monoclonal antibody clone 592-2-5); CYP3A4, 1:1000 (monoclonal antibody 275-1-2); and, RXRα, 1:1000 (polyclonal antibody D-20 Santa Cruz Biotechnology, Santa Cruz, CA). Incubation in primary antibody was either at 2 h at room temperature (for detection of albumin, β actin, CYP2C9/18, and CYP3A4) or overnight at 4°C (for detection of CYP2B6 and RXRα). Antibodies for CYP2B6, CYP3A4, and CYP2C9/18 were obtained through Dr. Harry C. Gelboin at the NCI/NIH. The PVDF membranes were then washed 4 times for 5 min each, followed by incubation in the appropriate secondary antibody, either goat anti-rabbit IgG-HRP (Santa Cruz, #sc-2004;1:3000 for detection of RXRα,) or goat anti-mouse IgG-HRP (Santa Cruz, #sc-2005; 1:3000 for detection of β actin and CYP2B6; 1:5000 for detection of CYP2C9/18 and CYP3A4; 1:20,000 for detection of albumin). Subsequently, the membranes were washed 4 times for 5 min prior to autoradiographic detection of signals by chemiluminescence using a commercially available kit (Lumi-light, Roche Diagnostic, Indianapolis, IN).
RESULTS
Immunohistochemistry
Visualized by phase contrast microscopy, primary hepatocytes cultured in both monolayer (on collagen) and in Matrigel sandwich configurations (Figure 1A and C, respectively) exhibited morphology indicative of a highly differentiated phenotype, comprised of striking cuboidal cellular architecture with hepatocytes arranged in closely-associated networks. Hepatocytes in a matrigel sandwich culture in particular exhibited enhanced gap junction formation, as assessed with immunohistochemical analyses of connexin 32 protein expression (Figure 1D vs Figure 1B).
Figure 1.
Detection of connexin 32 using immunohistochemistry in primary human hepatocytes maintained in monolayer and sandwich culture. Hepatocytes were cultured on collagen-coated flasks and were either maintained as a monolayer (A and B) or overlaid with a dilute layer of Matrigel (C and D). After fixation, cells were subjected to immunohistochemical analysis for connexin 32 using an Alexa Fluor secondary antibody conjugated with a FITC molecule. Images were taken as bright field (A and C) or with a FITC-specific filter (B and D). Magnification ×200.
Differentiation marker analysis
Albumin (ALB), a liver-selective protein, is generally used as a marker of the differentiation status of in vitro liver models (Ben Ze’ev et al., 1988;De Smet et al., 2001;Farkas et al., 2005;Moghe et al., 1996;Sidhu et al., 2004). Although a slight decrease from the liver was suggested, ALB expression was maintained in all in vitro hepatic models within the boundaries defined as unchanged from the liver (less than 4-fold decreased) (Figure 2A). An inverse marker of hepatic differentiation status is AFP, the fetal homologue of albumin. Because synthesis of AFP is turned off shortly after birth, its detection indicates loss of the differentiated liver phenotype and reversion to a more fetal status. Among the seven human hepatocyte donors where AFP was reliably detected, expression of AFP appeared modestly elevated, 4.6 ± 1.2(SD)-fold, from the liver, slightly above the 4-fold cutoff selected for differential expression (Figure 2C). In striking contrast, HepG2 and Huh7 cells exhibited markedly enhanced expression of AFP, with levels increased > 600- and 1000-fold, respectively, from levels detected directly in liver tissues.
Figure 2.
Albumin (ALB), a differentiation marker, and α-fetoprotein (AFP), a dedifferentiation marker, estimate the differentiation status of the in vitro hepatic models. Data are expressed as fold change (A and C). These markers were verified by QRT-PCR, using the ΔΔCt method for analysis where 18S is the reference gene and the human liver is the reference sample. Data are expressed as fold change, defined by 2−ΔΔCt (B and D). Differential expression is defined as greater than four-fold change from the human liver (dotted lines).
To better estimate the variability of ALB and AFP expression, quantitative RT-PCR (QRT-PCR) was used as a verification of the microarray results. The data derived from both QRT-PCR and microarray analyses indicated that ALB expression was unchanged in the primary cultured hepatocytes relative to liver tissue (Figure 2B). Although the well-documented compression of signal effect is apparent (Hamadeh et al., 2002;Rockett et al., 2004), AFP expression in the different culture systems measured by QRT-PCR closely mimicked the microarray estimation; with the fold enhancement of AFP detected by QRT-PCR even greater than that detected by microarray analysis (59,000 ± 9000- and 130,000 ± 80,000-fold increased in HepG2 and Huh7 cells vs. 600-and 1000-fold increased; Figure 2C–D). Among the five hepatocyte donors for which QRT-PCR analysis was performed, AFP expression was 4.6 ± 4.5-fold higher than the liver, reflecting variability between cultures established from different donors. Thus, both techniques detect AFP expression in hepatocytes as nearly unchanged from the liver; while expression in the hepatoma-derived cell lines is dramatically increased relative to liver. Additionally, verification of the microarray results with a second method of mRNA quantification provides confidence that microarray analysis is an appropriate method for estimation of mRNA expression.
Correlation analysis
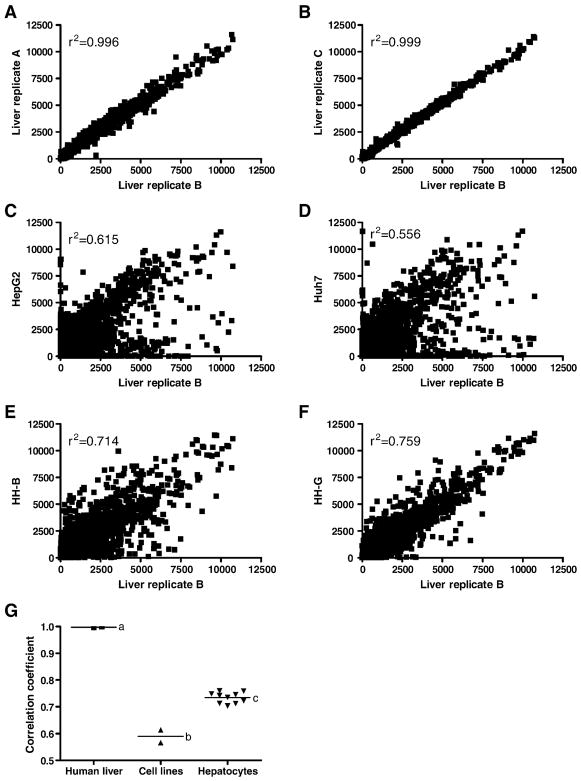
The pooled human liver replicates exhibited higher correlation with the primary human hepatocytes than with the hepatoma cell lines, although the highest correlation between two samples was apparent for the technical replicates (p<0.001, Figure 3G). Correlation between the three technical replicates was quite high; correlation coefficients in all combinations were greater than 0.996 (Figure 3A–B; all scatter plots are shown using liver replicate B as a representative liver). The six comparisons between the human liver replicates and the hepatoma cell lines revealed very poor correlation in gene expression profiles (HepG2, 0.614 ± 0.007; Huh7, 0.566 ± 0.006; Figure 3C–D). For the thirty comparisons between the human liver replicates and the ten human hepatocyte donors, the correlation to human liver among donors ranged from 0.76 ± 0.01 in donor HH-G to 0.70 ± 0.01 in HH-B (Figure 3E–F). The variability noted between the liver and the in vitro models is likely due to a combination of the absence of certain cell types in the purified hepatoycte preparations that are otherwise present in whole liver tissue, such as Kupffer cells and hepatic stellate cells, and interindividual differences between the hepatocyte donors and the human liver donors.
Figure 3.
Correlation of gene expression profiles between human liver and either technical replicates, hepatoma-derived cell lines, or primary human hepatocytes. Scatter plots are shown for correlation of a representative liver pool (Liver replicate B) to: two technical replicates, Liver replicates A (A) and C (B); HepG2 cells (C); Huh7 cells (D); two human hepatocyte donors, HH-B (E) and HH-G (F). G summarizes the mean correlation of the individual HepG2, Huh7, and human hepatocytes arrays to the human liver pools and the correlation of technical replicates to a representative array of pooled human liver (liver replicate B). Different letters (a, b, c) indicate significantly different correlation of that group to the human liver pool (p<0.001; one-way ANOVA with Tukey’s multiple comparison post-test).
Overrepresentation analysis
In an effort to examine biological trends among these basal level expression changes in the in vitro models, we used over-representation analysis with the PANTHER Classification system to identify biological processes and molecular pathways fundamental to drug metabolism among the differentially expressed probe sets in all of the models (Table 2). For example, two types of reactions catalyzed by biotransformation enzymes were identified as overrepresented: ‘oxidoreductase’ (hepatocytes, p=3.86×10−3; HepG2, p=5.31×10−2) and ‘transferase’ (HepG2, p=1.01×10−2; Huh7, p=7.03×10−4). ‘Lipid, fatty acid, and steroid metabolism’ was identified in all three models (HepG2, p=1.22×10−8; Huh7, p=1.24×10−7; hepatocytes, p=5.78×10−4). Additionally, drug metabolism, alternatively referred to as ‘detoxification’ (HepG2, p=3.44×10−2, Huh7, p=1.10×10−3), is dependent upon signaling pathways, which are carried out by ‘signaling molecules’ (hepatocytes, p=1.12×10−4) that often depend upon modifications such as ‘protein phosphorylation’ (HepG2, p=1.22×10−3).
Table 2.
Statistically overrepresented gene ontology (GO) categories among the differentially expressed genes in either HepG2 cells, Huh7 cells, or human hepatocytes.10
| Panther ID | Panther category | Significant children in this branch | HepG2 | Huh7 | Hepatocytes |
|---|---|---|---|---|---|
| BIOLOGICAL PROCESS ANNOTATIONS | p-value11 | ||||
| BP00019 | Lipid, fatty acid and steroid metabolism | 1.22×10−8 | 1.24×10−7 | 5.78×10−4 | |
| BP00295 | Steroid metabolism | 2.21×10−5 | 9.60×10−6 | 9.86×10−6 | |
| BP00020 | Fatty acid metabolism | - 12 | 7.69×10−4 | - | |
| BP00063 | Protein modification | 3.10×10−2 | - | - | |
| BP00064 | Protein phosphorylation | 1.22×10−3 | - | - | |
| BP00148 | Immunity and defense | 1.04×10−3 | 7.92×10−4 | 5.96×10−7 | |
| BP00153 | Complement-mediated immunity | 9.36×10−7 | 8.28×10−11 | - | |
| BP00180 | Detoxification | 3.44×10−2 | 1.10×10−3 | - | |
| BP00178 | Stress response | 2.10×10−2 | - | - | |
| BP00203 | Cell cycle | 1.76×10−11 | 4.42×10−9 | - | |
| BP00282 | Mitosis | 4.46×10−2 | 4.09×10−3 | - | |
| BP00035 | DNA replication | 1.38×10−8 | 3.39×10−6 | - | |
| BP00224 | Cell proliferation and differentiation | 2.32×10−2 | 2.96×10−2 | - | |
| MOLECULAR FUNCTION ANNOTATIONS | |||||
| MF00016 | Signaling molecule | - | - | 1.12×10−4 | |
| MF00107 | Kinase | 5.63×10−6 | - | - | |
| MF00108 | Protein kinase | 2.28×10−5 | - | - | |
| MF00123 | Oxidoreductase | - | - | 3.86×10−3 | |
| MF00124 | Oxygenase | - | - | 1.02×10−3 | |
| MF00131 | Transferase | 1.01×10−2 | 7.03×10−4 | - | |
| MF00133 | Methyltransferase | - | 1.47×10−4 | - | |
Signal intensity values in the untreated arrays were converted to signal log ratio format as described in Figure 2. PANTHER used the binomial test to determine biological processes or molecular functions overrepresented among probe sets differentially expressed (signal log ratio >2 or signal log ratio < −2) in each in vitro model. P values for each category were calculated from the binomial test statistic.
Significance is defined as p<0.05
–; p>0.05
Regulation of individual probe sets in the liver-specific GO categories
In the six overrepresented GO categories noted above as involved in drug metabolism, the individual probe sets in the primary hepatocyte samples were overwhelmingly unchanged from the liver, while no distinct pattern of regulation was evident among the hepatoma-derived cell lines (Figure 4A–F). Among these six categories, the average number of probe sets in the hepatocytes that were unchanged from the liver was 83.7 ± 3.7%. This observation is also consistent in each of the ten individual hepatocyte donors, as can be seen in two representative ontology categories, ‘oxidoreductase’ and ‘phosphorylation’ (Figure 4G–H). Within each donor, the percentages of probe sets that are scored as unchanged, increased, or decreased emphasize that, while there are slight differences between donors in the regulation of individual probe sets, more probe sets in the hepatocytes are unchanged from the liver than are increased or decreased (p<0.001). In HepG2 and Huh7 cells, on the other hand, exhibited an average of only 42.5 ± 3.1% and 39.2 ± 9.2% unchanged probe sets compared with the liver across the six ontology categories, respectively.
Figure 4.
Regulation of the individual probe sets in overrepresented GO categories in the in vitro hepatic models. For each array, the probe sets in each GO category with a detection call of Present were scored as either ‘not changed’ (−2 < signal log ratio < 2), ‘increased’ (signal log ratio > 2), or ‘decreased’ (signal log ratio < −2). Data for the human hepatocytes are expressed as mean ± SD, summarizing results from 10 human hepatocyte donors. Data for the hepatoma-derived cell lines are expressed as results from a single array. The list for ‘Oxidoreductase’ (A) contains 270 probe sets (186 genes); ‘Transferase’ (B) contains 329 probe sets (221 genes); ‘Lipid, fatty acid, and steroid metabolism’ (C) contains 339 probe sets (239 genes); ‘Detoxification’ (D) contains 35 probe sets (29 genes); ‘Signaling molecule’ (E) contains 285 probe sets (207 genes); and ‘Phosphorylation’ (F) contains 230 probe sets (155 genes). Data in G and H are shown as individual results from the ten hepatocyte donors in two representative GO categories. * indicates that the ‘not changed’ regulation group has significantly more probe sets than the ‘increased’ or ‘decreased’ groups (p<0.001; one-way ANOVA with Tukey’s multiple comparison post-test).
Gene-level analysis of liver-specific functions
Expression of individual genes with known biological roles in the overrepresented ontology categories supports the hypothesis that in vitro hepatocytes in a sandwich culture are functionally representative of the in vivo liver, in contrast to the established cell lines. As a case in point, liver-enriched transcription factors, including hepatocyte nuclear factor (HNF) family members, CAAT/enhancer binding protein α (C/EBPα), and members of the nuclear hormone receptor superfamily, are central to maintaining the hepatic phenotype. These transcription factors were tightly regulated in all ten hepatocyte donors, as expression of all nine genes is maintained at levels less than 4-fold changed from the liver (Figure 5A). Although expression of these transcription factors were also generally regulated in the established cell lines, there were notable exceptions NR1I2 (pregnane X receptor, or PXR) and NR1I3 (constitutive androstane receptor, or CAR) were > 6- and 42-fold decreased in HepG2 cells, respectively, and neither was reliably detected in Huh7 cells (Figure 5B). A third nuclear hormone receptor, the retinoid X receptor-α (RXRα), was also decreased in Huh7 cells in comparison to the liver (5.7-fold).
Figure 5.
Gene-level expression analysis of selected liver-specific categories in human hepatocyte donors and hepatoma-derived cell lines. Distribution of fold change from the liver in the ten hepatocyte donors are shown for genes encoding select transcription factors (A), drug metabolizing enzymes (C), and drug transporters (E). The fold change for the same genes in HepG2 cells and Huh7 cells are reported in B, D and F. Differential expression is defined as greater than four-fold change from the human liver (dotted lines). * indicates that the measured probe set is detected as absent in at least one human hepatocyte donor (PPARA: Absent in two donors; TCF1: two donors; CYP1A2: one donor; ABCB11: one donor; ABCC1: one donor). ** indicates that that probe set is detected as absent in Huh7 cells (NR1I2, NR1I3, CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP3A4, ABCB11, SLC22A1, SLC22A7). *** indicates that that probe set is detected as absent in HepG2 (CYP1A2, CYP2B6, CYP2C9, CYP2D6, CYP2E1, ABCB11, SLC22A1, SLC22A7, SLCO2B1).
Drug metabolizing enzymes, such as the various CYPs, flavin-containing monooxygenase 5 (FMO5), epoxide hydrolases (EPHXs), glutathione-s-transferase (GSTA1), and gluatathione peroxidase (GPX1), catalyze the ‘oxidoreductase’ and ‘transferase’ activities identified by the overrepresentation analysis and their expression status yields important insight regarding the metabolic functional capacity of in vitro hepatic models. As expected from reports of decreased basal activity of certain CYP enzymes in sandwich culture (Farkas et al., 2005;Richert et al., 2002), expression of certain enzymes appears to be decreased in culture from the liver; particularly affected are CYP1A2 and CYP2E1 (Figure 5C). However, the remaining enzymes are expressed at levels comparable to or slightly increased from the human liver in the majority of the hepatocyte donors. In contrast, expression of the CYP450 isoforms is generally lost or dramatically decreased in comparison to the liver in both of the hepatoma cell lines that were subjected to analysis (Figure 5D).
Drug transporters, a third functional group of genes whose products perform important roles in ‘detoxification’, have been reported previously to exhibit polarized expression in sandwich culture of hepatocytes. Our results support these findings, as transporters, such as P-glycoprotein (ABCB1), the bile salt export pump (ABCB11), multidrug resistance-associated protein family members (MRPs, or the ABCC family), organic cation and anion transporters (OCTs and OATs, or solute carrier family SLC22A), and organic anion transport proteins (OATP, or solute carrier family SLCO), were expressed at levels unchanged from the liver in nearly all of the human hepatocyte donors (Figure 5E). In the established cell lines, however, expression of select transporters is lost, consistent with the view that these cell models do not maintain polarized membranes reminiscent of the liver.
Gene-level analysis of a complex biological response
Hepatocytes, but not hepatoma-derived cell lines, retain the ability to respond to toxic insult by induction of biotransformation enzymes, a critical feature of an appropriate in vitro model of the liver. Among the ten hepatocyte donors, considerable interindividual variability was apparent in the basal expression levels of three PB-inducible target genes; CYP2B6, CYP3A4, and CYP2C9 (Figures 6A–C). Most notably, basal expression of CYP2B6 ranged in signal intensity from 142 to nearly 2900 fluorescence units; in comparison, the highest signal strength scored after induction by PB was approximately 4100 units. However, CYP2B6, CYP3A4, and CYP2C9 were expressed basally at levels unchanged from the human liver in a majority of the donors, and, overall, PB treatment induced a greater fold change from the liver than treatment with vehicle (p<0.01; Figure 6D–F). In the established cell lines, only CYP3A4 was reliably detected at the basal level, albeit more than 45-fold decreased from the human liver. After chemical treatment, the only genes that may be described as induced in either cell line were CYP2B6 and CYP3A4 in Huh7 cells, which were detected at levels estimated as comparable to and more than 45-fold decreased from basal human liver expression, respectively (data not shown). Basal levels of CYP2B6, CYP3A4, and CYP2C9 proteins were clearly detectable in the human liver pool, but not in the human hepatocyte and established cell line samples (Figure 6G and data not shown). This apparent discrepancy is likely due to the nature of the human liver tissues used for western blotting which, as S9 fractions, are enriched for microsomal content, while the human hepatocyte and cell line samples consisted of whole cell extracts. The different protein sources derived from the human liver tissues vs. in vitro models further explains the absence of RXRα and β actin selectively in the human liver tissues. After PB treatment, protein expression of the three target genes was clearly elevated in the human hepatocytes, but not the cell lines samples, further supporting the use of primary hepatocytes for studies of drug metabolism.
Figure 6.
Gene-level analysis of a complex biological response in human hepatocytes. Data are reported as linear signal intensity values for each untreated and PB-treated array for three target genes: CYP2B6 (A), CYP3A4 (B), and CYP2C9 (C). * indicates that the PB-treated signal intensity in a donor is more than four-fold increased from the untreated signal intensity. Data in D, E, and F are expressed as fold change from the human liver (p<0.01; paired t-test). Differential expression is defined as greater than four-fold change from the human liver (dotted lines). Induction of protein in response to PB treatment is shown for selected donors and hepatoma-derived cells lines (G).
Hierarchical clustering analysis
When subjected to hierarchical clustering analysis, PB-treatment caused hepatocytes, but not hepatoma-derived cell lines, to cluster by treatment rather than by sample type. When clustering of the samples was performed using all 39,485 probe sets that remained after removal of those probe sets absent in all 27 arrays, the resulting dendrogram was divided into two major branches: a first branch containing the liver replicates and primary hepatocytes, and a second containing the hepatoma-derived cell lines (Figure 7A). These clustering results indicate that the gene expression patterns occurring in the liver are more similar to hepatocytes than to the hepatoma cell lines, and that, among the human hepatocytes, interindividual variation plays a crucial role in gene expression patterning, as eight of the ten untreated samples clustered most closely with the PB-treated sample from the same donor.
Figure 7.
Hierarchical clustering of the human liver replicates, hepatoma-derived cell lines, and human hepatocytes. Hierarchical clustering of the 27 arrays was performed using a dataset consisting of 39,485 probe sets (A), followed by clustering of both the 27 arrays and the PB-inducible probe sets, which were found to be both PB-responsive in at least 2 pairs of samples (untreated and PB-treated) and present in 13 or more of the arrays (B). A cluster of probe sets with a distinct expression pattern across the 27 arrays is magnified in C. All clustering was done using a Euclidean distance measure and an average linkage.
Clustering of the samples was performed a second time, including only probe sets increased more than 4-fold in at least two PB-treated samples. The dendrogram was again divided into the same two main branches described above, emphasizing that the liver replicates are more similar to the hepatocytes than the cell lines. However, the branch containing the liver pools and hepatocytes was now further divided into two treatment-specific arms; one containing the liver pools, eight of ten untreated samples, and one PB-treated sample, and the second containing nine PB-treated and two untreated hepatocyte samples (Figure 7B). Contrary to the hepatocytes, the clustering pattern of the hepatoma cell lines was not altered in this PB-inducible dataset to reflect treatment.
Hierarchical clustering was also used to group the PB-inducible probe sets on the basis of similarity in expression pattern across the 27 samples. One branch of the dendrogram stood out with a distinct expression pattern across the different types of samples (magnified in Figure 7C). This cluster of probe sets was characterized by little change or a slight increase in expression in the untreated hepatocytes, clearly increased expression in the PB-treated hepatocytes, and decreased expression in the hepatoma-derived cell lines, both treated and untreated. The majority of the probe sets in this cluster are specific for cytochrome P450 family members, specifically, members of the CYP2A, 3A, and 2B families. Three additional non-CYP genes were identified to exhibit this similar overall expression pattern: aminolevulinate, delta-, synthase 1 (ALAS1), leucine rich repeat containing 54 (LRRC54), and one probe set that is not annotated. Overall, hierarchical clustering confirmed the themes presented thus far in this study; that hepatocytes possess gene expression profiles that are more representative of human liver than the hepatoma cell lines, and that measurement of gene expression character provides novel insight into functional capacity, as hepatocytes, but not hepatoma-derived cell lines, respond robustly to inducer challenge.
DISCUSSION
To better assess the utility of primary hepatocytes in an ECM sandwich culture configuration as a model for drug metabolism studies, microarray technology was used to gain a global view of the biological processes and molecular functions altered in two commonly used human in vitro hepatic models from the in vivo phenotype. Although it is true that our culture conditions could be described as 3D, in that the hepatocytes are sandwiched between a substratum of collagen and an overlay of matrigel, the cellular milieu largely consists of a 2D surface provided in the context of a culture plate. Several investigators have devised more complex devices for the culture of cells, for example hollow fiber bioreactors that enable packed cells to share a 3D space, within an environment that may also provide dynamic fluidics forces. In our conceptual framework, packed cells within a bioreactor scheme are more adeptly labeled as 3D cultures. Throughout our manuscript, we have largely referred to our culture scheme as a ‘sandwich’ approach, a term that does accurately describe the culture situation; with 2D being used on occasion to reflect that the cells are plated on a flat surface. Our results show that primary hepatocytes cultured using highly defined conditions maintained basal expression of genes involved in liver-specific functions and facilitated a complex biological response to chemical treatment. Hepatoma-derived cell lines were unable to demonstrate either of these features.
Genes examined in this study were chosen based on inclusion in the overrepresented ontology categories identified by the PANTHER classification system, an unbiased method of identifying biological trends among large lists of transcriptional changes (Currie et al., 2005;Thomas et al., 2003;Thomas et al., 2006). Although many of the overrepresented ontology categories described functions integral to drug metabolism, such as ‘oxidoreductase’, ‘transferase’, ‘lipid, fatty acid, and steroid metabolism’, and ‘detoxification’ (Table 2), the prominent pattern of regulation of the individual probe sets within each of these categories was unchanged expression between the hepatocytes and the liver (Figure 4). Interestingly, liver-enriched transcription factors, such as nuclear receptors, were shown to be tightly regulated in all hepatocyte donors (Figure 5A). Conservation of nuclear receptor expression supports the functionality of hepatocytes in sandwich culture to predict drug metabolism, as these receptors have been shown to regulate batteries of genes involved in response to chemical stress (Wu et al., 2004;Zhang et al., 2002;Zhang et al., 2004). Many of the genes regulated by nuclear receptors are drug metabolizing enzymes and drug transporters (Assem et al., 2004;Rosenfeld et al., 2003;Ueda et al., 2002), and under the described culture conditions, were expressed at levels unchanged from the human liver in the majority of the human hepatocyte donors (Figure 5B–C). Again, conserved expression of drug metabolizing enzymes and drug transporters supports the functionality of this culture model for prediction of both drug metabolism and drug disposition.
Throughout this manuscript, differential expression has been defined on the basis of fold change. It is well documented that using fold change alone as a method to identify differentially expressed probe sets does not produce optimal results, as this method has a high probability of identifying false positives (Baldi et al., 2001;Jeffery et al., 2006;McClintick et al., 2006). However, other methods of detecting differential change (t-statistic, ANOVA, significance analysis of microarrays) require technical replicates in order to assess statistical significance. When we considered our experimental goal, which was to maximize the number of donors in order to address interindividual variability, together with the high level of reproducibility afforded by the Affymetrix arrays, we thought that it was more important to include as many biological replicates as possible at the expense of technical replicates that would allow significance analysis. In order to limit the number of false positives anticipated to result from using a fold change method to define differential expression, the fold change cutoff was increased above the more commonly used 2-fold cutoff. The rationale for this decision was based on results which suggested that small variations in mRNA levels (less than 2-fold) are less accurately reflected by microarray analysis than by more quantitative methods such as qRT-PCR (Dallas et al., 2005). Thus, a 4-fold cutoff is intended to minimize the number of false positives, and, even though we are likely losing some true positives, we decided to focus only on dramatic gene expression changes in the in vitro models from the human liver. Interestingly, analyses performed using a lowered 2-fold cutoff did not alter the conclusion that human hepatocytes maintained in sandwich culture exhibit a gene expression profile more similar to the in vivo human liver than hepatoma-derived cell lines with respect to ontology categories involved in drug metabolism. Over-representation analysis using all probe sets changed more than 2-fold in each sample identifies nearly the same ontology categories, although there are a few differences in significance at this cutoff level; ‘lipid, fatty acid, and steroid metabolism’ and ‘signaling molecule’ were no longer identified as significant in Huh7 cells and the hepatocytes, nor was ‘protein phosphorylation’ considered significant in HepG2 cells. Of the probe sets in the six categories of interest, more probe sets remained unchanged from the human liver than were increased or decreased in the hepatocytes (p<0.001), similar to the results using the 4-fold cutoff. Also, the percentage of probes unchanged from the liver across these six categories is higher in the hepatocytes than in the hepatoma-derived cell lines at a 2 fold cutoff (65.6 ± 5.1% vs. 40.7 ± 3.7% and 41.0 ± 4.1%; p<0.001), in agreement with the conclusions drawn at the 4-fold cutoff level.
A second endpoint, complex biological responsiveness to chemical treatment, indicated that large-scale transcriptional analysis can be used to assess an in vitro functional response. For example, gene-level analysis of classical PB-inducible markers, CYP2B6, CYP3A4 and CYP2C9, showed that the induction response occurred at both the mRNA and protein levels in the hepatocyte donors but not in the hepatoma derived cell lines (Figure 6), which are often used as representative models of hepatocyte biology. Hierarchical clustering of the samples also confirmed that the primary hepatocytes retain induction responsiveness to chemical treatment, since treatment-specific clustering was observed only in the hepatocyte samples (Figure 7B), not in the hepatoma cell lines. Since induction of the CYP2B family members by PB treatment has historically been the most difficult P450 response to retain in culture (LeCluyse et al., 1999;Sidhu et al., 2004), these results suggest that, functionally, the primary human hepatocytes can replicate the metabolic capacity of the in vivo human liver.
Similar large-scale transcriptional analyses to compare in vivo and in vitro basal gene expression profiles in rodent models have been conducted, and offer similar results to those presented here. For example, one study established that the global expression profiles between in vivo rat liver and primary rat hepatocytes exhibited a correlation coefficient of 0.8023 (Jessen et al., 2003), while another reported a correlation of 0.68 (Boess et al., 2003). Both studies are in agreement with our findings, where correlation coefficients ranged from 0.76 ± 0.01 to 0.70 ± 0.01 among the ten human donors (Figure 3G). Hierarchical clustering analysis is a further area of agreement, with Boess et al. (2003) finding that rat liver and primary hepatocytes cluster on the same arm of the dendrogram compared to rat liver cell lines that cluster on a separate arm. Our conclusions are further corroborated by reports that primary human hepatocytes in sandwich culture maintain expression of drug metabolizing enzymes similar to that in human liver while HepG2 cells exhibit dramatically decreased levels of these enzymes (Rodriguez-Antona et al., 2002;Tuschl et al., 2006;Wilkening et al., 2003).
As anticipated, interindividual variability among the ten donors was noted throughout the study. Hierarchical clustering analysis most greatly illustrated the contribution of interindividual variability among donors, since samples clustered according to donor rather than treatment when all present probe sets were included in the dataset (Figure 7A). Also noted in the clustering analysis was that both untreated and PB-treated samples from donors HH-D and HH-F clustered on the PB-treated arm of the dendrogram (Figure 7B), which was consistent both with the minimal induction response upon chemical treatment and with the higher basal expression of CYP2B6 and CYP3A4 in these two donors when compared to the liver replicates and other donors (Figure 6). These results suggest that, even after almost 5 days in culture, the hepatocytes from these two donors were nearly maximally induced, perhaps because of exposure to environmental factors such as food or drugs (Parkinson, 1996). Thus, these hepatocytes were not capable of being further induced by treatment with PB, a trend noted by several other studies (Kostrubsky et al., 1999;McCune et al., 2000). A second discrepancy noted was that the induction response in the PB-treated HH-E sample was minimal, such that this sample clustered more closely with the untreated samples; specifically, with the untreated HH-E sample. Other minor variations included a decrease in albumin expression in a single donor (HH-C, decreased 6.8-fold from liver expression) and undetectable expression of certain transcription factors (TCF1 in HH-C and HH-J; PPARA in HH-E), drug metabolizing enzymes (CYP1A2 in HH-J), and drug transporters (ABCB11 in HH-E). Further, albumin protein was undetectable in HH-J (Figure 6G). Loss of liver-specific gene and protein expression in hepatocytes from these three donors is likely an indication of loss of differentiation in these samples.
Despite the noted discrepancies, overall, gene expression trends were clearly identified among donors. One such trend was that the regulation of probe sets in each overrepresented ontology category was largely unchanged from the liver in all ten donors. Similarly, as indicated above, expression of liver-enriched transcription factors was tightly regulated in the sandwich culture model, such that, when present, none were more than 4-fold changed in any of the donors. This trend also was true for all the drug transporters that were analyzed, with only a single gene (SLC22A7) decreased more than 4-fold in a single donor (HH-I). These results suggest that, while interindividual differences will contribute to differences in metabolic capacity in vitro, primary hepatocytes cultured in a sandwich model exhibit conserved biological trends from preparation to preparation. These results argue that even with the use of hepatocytes from multiple donors, general trends in response to a chemical or treatment are likely still discernable, despite minor inter-individual differences that inherently will exist.
In conclusion, in this study we conducted a global, unbiased view of the biological processes and molecular functions altered at the transcriptional level in two commonly used in vitro models of the human liver. Overall, the results support the utility of primary human hepatocytes cultured in a defined ECM sandwich configuration as a robust model of the in vivo hepatic phenotype, as a striking similarity exists in the expression profiles for genes involved in hallmark categories of liver function, such as drug metabolism, between in vitro hepatocytes in an ECM sandwich culture model and the liver. Further, this model maintains cellular responses to PB, a unique indicator of a well-differentiated hepatocyte. Conversely, the results obtained underscore caution when attempting to derive similar biological information from hepatoma-derived cell lines, as the expression levels and profiles of genes involved in liver-specific functions are poorly well-maintained in these systems.
Acknowledgments
This study was supported by a Toxicogenomics Research Consortium grant from the National Institutes of Environmental Health Sciences, U19 ES11387 and a grant from the National Institute of General Medical Sciences, GM066411.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Annaert PP, Brouwer KL. Assessment of drug interactions in hepatobiliary transport using rhodamine 123 in sandwich-cultured rat hepatocytes. Drug Metab Dispos. 2005;33:388–394. doi: 10.1124/dmd.104.001669. [DOI] [PubMed] [Google Scholar]
- Assem M, Schuetz EG, Leggas M, Sun D, Yasuda K, Reid G, Zelcer N, Adachi M, Strom S, Evans RM, Moore DD, Borst P, Schuetz JD. Interactions between hepatic Mrp4 and Sult2a as revealed by the constitutive androstane receptor and Mrp4 knockout mice. J Biol Chem. 2004;279:22250–22257. doi: 10.1074/jbc.M314111200. [DOI] [PubMed] [Google Scholar]
- Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t -test and statistical inferences of gene changes. Bioinformatics. 2001;17:509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- Ben Ze’ev A, Robinson GS, Bucher NL, Farmer SR. Cell-cell and cell-matrix interactions differentially regulate the expression of hepatic and cytoskeletal genes in primary cultures of rat hepatocytes. Proc Natl Acad Sci USA. 1988;85:2161–2165. doi: 10.1073/pnas.85.7.2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boess F, Kamber M, Romer S, Gasser R, Muller D, Albertini S, Suter L. Gene expression in two hepatic cell lines, cultured primary hepatocytes, and liver slices compared to the in vivo liver gene expression in rats: possible implications for toxicogenomics use of in vitro systems. Toxicol Sci. 2003;73:386–402. doi: 10.1093/toxsci/kfg064. [DOI] [PubMed] [Google Scholar]
- Currie RA, Orphanides G, Moggs JG. Mapping molecular responses to xenoestrogens through Gene Ontology and pathway analysis of toxicogenomic data. Reprod Toxicol. 2005;20:433–440. doi: 10.1016/j.reprotox.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Dallas PB, Gottardo NG, Firth MJ, Beesley AH, Hoffmann K, Terry PA, Freitas JR, Boag JM, Cummings AJ, Kees UR. Gene expression levels assessed by oligonucleotide microarray analysis and quantitative real-time RT-PCR -- how well do they correlate? BMC Genomics. 2005;6:59. doi: 10.1186/1471-2164-6-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet K, Loyer P, Gilot D, Vercruysse A, Rogiers V, Guguen-Guillouzo C. Effects of epidermal growth factor on CYP inducibility by xenobiotics, DNA replication, and caspase activations in collagen I gel sandwich cultures of rat hepatocytes. Biochem Pharmacol. 2001;61:1293–1303. doi: 10.1016/s0006-2952(01)00612-8. [DOI] [PubMed] [Google Scholar]
- Farkas D, Tannenbaum SR. Characterization of chemically induced hepatotoxicity in collagen sandwiches of rat hepatocytes. Toxicol Sci. 2005;85:927–934. doi: 10.1093/toxsci/kfi145. [DOI] [PubMed] [Google Scholar]
- Hamadeh HK, Bushel PR, Jayadev S, Martin K, DiSorbo O, Sieber S, Bennett L, Tennant R, Stoll R, Barrett JC, Blanchard K, Paules RS, Afshari CA. Gene expression analysis reveals chemical-specific profiles. Toxicol Sci. 2002;67:219–231. doi: 10.1093/toxsci/67.2.219. [DOI] [PubMed] [Google Scholar]
- Hassett C, Lin J, Carty CL, Laurenzana EM, Omiecinski CJ. Human hepatic microsomal epoxide hydrolase: comparative analysis of polymorphic expression. Arch Biochem Biophys. 1997;337:275–283. doi: 10.1006/abbi.1996.9794. [DOI] [PubMed] [Google Scholar]
- Hoffmaster KA, Turncliff RZ, LeCluyse EL, Kim RB, Meier PJ, Brouwer KL. P-glycoprotein expression, localization, and function in sandwich-cultured primary rat and human hepatocytes: relevance to the hepatobiliary disposition of a model opioid peptide. Pharm Res. 2004;21:1294–1302. doi: 10.1023/b:pham.0000033018.97745.0d. [DOI] [PubMed] [Google Scholar]
- Jeffery IB, Higgins DG, Culhane AC. Comparison and evaluation of methods for generating differentially expressed gene lists from microarray data. BMC Bioinformatics. 2006;7:359. doi: 10.1186/1471-2105-7-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen BA, Mullins JS, De Peyster A, Stevens GJ. Assessment of hepatocytes and liver slices as in vitro test systems to predict in vivo gene expression. Toxicol Sci. 2003;75:208–222. doi: 10.1093/toxsci/kfg172. [DOI] [PubMed] [Google Scholar]
- Kern A, Bader A, Pichlmayr R, Sewing KF. Drug metabolism in hepatocyte sandwich cultures of rats and humans. Biochem Pharmacol. 1997;54:761–772. doi: 10.1016/s0006-2952(97)00204-9. [DOI] [PubMed] [Google Scholar]
- Kostrubsky VE, Ramachandran V, Venkataramanan R, Dorko K, Esplen JE, Zhang S, Sinclair JF, Wrighton SA, Strom SC. The use of human hepatocyte cultures to study the induction of cytochrome P-450. Drug Metab Dispos. 1999;27:887–894. [PubMed] [Google Scholar]
- LeCluyse E, Bullock P, Madan A, Carroll K, Parkinson A. Influence of extracellular matrix overlay and medium formulation on the induction of cytochrome P-450 2B enzymes in primary cultures of rat hepatocytes. Drug Metab Dispos. 1999;27:909–915. [PubMed] [Google Scholar]
- Liu X, Chism JP, LeCluyse EL, Brouwer KR, Brouwer KL. Correlation of biliary excretion in sandwich-cultured rat hepatocytes and in vivo in rats. Drug Metab Dispos. 1999;27:637–644. [PubMed] [Google Scholar]
- McClintick JN, Edenberg HJ. Effects of filtering by Present call on analysis of microarray experiments. BMC Bioinformatics. 2006;7:49. doi: 10.1186/1471-2105-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCune JS, Hawke RL, LeCluyse EL, Gillenwater HH, Hamilton G, Ritchie J, Lindley C. In vivo and in vitro induction of human cytochrome P4503A4 by dexamethasone. Clin Pharmacol Ther. 2000;68:356–366. doi: 10.1067/mcp.2000.110215. [DOI] [PubMed] [Google Scholar]
- Moghe PV, Berthiaume F, Ezzell RM, Toner M, Tompkins RG, Yarmush ML. Culture matrix configuration and composition in the maintenance of hepatocyte polarity and function. Biomaterials. 1996;17:373–385. doi: 10.1016/0142-9612(96)85576-1. [DOI] [PubMed] [Google Scholar]
- Parkinson A. An overview of current cytochrome P450 technology for assessing the safety and efficacy of new materials. Toxicol Pathol. 1996;24:48–57. [PubMed] [Google Scholar]
- Richert L, Binda D, Hamilton G, Viollon-Abadie C, Alexandre E, Bigot-Lasserre D, Bars R, Coassolo P, LeCluyse E. Evaluation of the effect of culture configuration on morphology, survival time, antioxidant status and metabolic capacities of cultured rat hepatocytes. Toxicol In Vitro. 2002;16:89–99. doi: 10.1016/s0887-2333(01)00099-6. [DOI] [PubMed] [Google Scholar]
- Rockett JC, Hellmann GM. Confirming microarray data--is it really necessary? Genomics. 2004;83:541–549. doi: 10.1016/j.ygeno.2003.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Antona C, Donato MT, Boobis A, Edwards RJ, Watts PS, Castell JV, Gomez-Lechon MJ. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32:505–520. doi: 10.1080/00498250210128675. [DOI] [PubMed] [Google Scholar]
- Rosenfeld JM, Vargas R, Jr, Xie W, Evans RM. Genetic profiling defines the xenobiotic gene network controlled by the nuclear receptor pregnane X receptor. Mol Endocrinol. 2003;17:1268–1282. doi: 10.1210/me.2002-0421. [DOI] [PubMed] [Google Scholar]
- Sidhu JS, Farin FM, Omiecinski CJ. Influence of extracellular matrix overlay on phenobarbital-mediated induction of CYP2B1, 2B2, and 3A1 genes in primary adult rat hepatocyte culture. Arch Biochem Biophys. 1993;301:103–113. doi: 10.1006/abbi.1993.1121. [DOI] [PubMed] [Google Scholar]
- Sidhu JS, Liu F, Omiecinski CJ. Phenobarbital responsiveness as a uniquely sensitive indicator of hepatocyte differentiation status: requirement of dexamethasone and extracellular matrix in establishing the functional integrity of cultured primary rat hepatocytes. Exp Cell Res. 2004;292:252–264. doi: 10.1016/j.yexcr.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Strom SC, Pisarov LA, Dorko K, Thompson MT, Schuetz JD, Schuetz EG. Use of human hepatocytes to study P450 gene induction. Methods Enzymol. 1996;272:388–401. doi: 10.1016/s0076-6879(96)72044-x. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas PD, Kejariwal A, Guo N, Mi H, Campbell MJ, Muruganujan A, Lazareva-Ulitsky B. Applications for protein sequence-function evolution data: mRNA/protein expression analysis and coding SNP scoring tools. Nucleic Acids Res. 2006;34:W645–W650. doi: 10.1093/nar/gkl229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl G, Mueller SO. Effects of cell culture conditions on primary rat hepatocytes-cell morphology and differential gene expression. Toxicology. 2006;218:205–215. doi: 10.1016/j.tox.2005.10.017. [DOI] [PubMed] [Google Scholar]
- Ueda A, Hamadeh HK, Webb HK, Yamamoto Y, Sueyoshi T, Afshari CA, Lehmann JM, Negishi M. Diverse roles of the nuclear orphan receptor CAR in regulating hepatic genes in response to phenobarbital. Mol Pharmacol. 2002;61:1–6. doi: 10.1124/mol.61.1.1. [DOI] [PubMed] [Google Scholar]
- Wilkening S, Stahl F, Bader A. Comparison of primary human hepatocytes and hepatoma cell line Hepg2 with regard to their biotransformation properties. Drug Metab Dispos. 2003;31:1035–1042. doi: 10.1124/dmd.31.8.1035. [DOI] [PubMed] [Google Scholar]
- Wu Y, Zhang X, Bardag-Gorce F, Robel RC, Aguilo J, Chen L, Zeng Y, Hwang K, French SW, Lu SC, Wan YJ. Retinoid X receptor alpha regulates glutathione homeostasis and xenobiotic detoxification processes in mouse liver. Mol Pharmacol. 2004;65:550–557. doi: 10.1124/mol.65.3.550. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Chua SS, Wei P, Moore DD. Modulation of acetaminophen-induced hepatotoxicity by the xenobiotic receptor CAR. Science. 2002;298:422–424. doi: 10.1126/science.1073502. [DOI] [PubMed] [Google Scholar]
- Zhang J, Huang W, Qatanani M, Evans RM, Moore DD. The constitutive androstane receptor and pregnane X receptor function coordinately to prevent bile acid-induced hepatotoxicity. J Biol Chem. 2004;279:49517–49522. doi: 10.1074/jbc.M409041200. [DOI] [PubMed] [Google Scholar]