Abstract
Rapid detection of antibody immunity in serum or plasma, whether to pathogenic antigens, tumor antigens, or autoimmune antigens, is critical for diagnosis, monitoring, and biomarker assessment of the immune response. Individual or multiplexed ELISAs that use purified recombinant proteins are dependent on a priori protein purification, a labor-intensive process that may take months to obtain proteins of sufficient purity and stability for serologic assays. We developed a programmable multiplexed immunoassay for the rapid monitoring of humoral immunity using the Luminex suspension bead array platform. In this approach, epitope-tagged antigens (GST- or FLAG-tagged) are expressed using in vitro transcription and translation, and captured onto anti-epitope-coupled Luminex SeroMap beads. The antigen-loaded beads are mixed, serum is added, and human IgG detected with standard secondary detection reagents. By coupling high-throughput DNA preparation of cDNA ORFs with antigen expression/capture, we demonstrate that 71/72 (98.6%) of GST-tagged proteins can be expressed and specifically detected on the bead ELISA. Detection of antibodies to the test viral antigen EBNA-1 in human sera is highly reproducible, with intra-assay variation of 3–8%, inter-assay variation of 5%, and with stability over 11 months. The specificity and limits of detection of the bead ELISAs for the tumor antigen p53 are comparable to both standard protein ELISAs and plate-based programmable (RAPID) ELISAs, and are also comparable to the detection of directly-conjugated p53 protein. Multiplexing a panel of analytes does not impair the sensitivity of antibody detection. Immunity to a panel of EBV-derived antigens (EBNA-1, EBNA-3A, EBNA-3B, and LMP-2) is specifically and differentially detected within healthy donor sera. This method allows for rapid conversion of ORFeome-derived cDNAs to a multiplexed bead ELISA to detect antibody immunity to both infectious and tumor antigens.
Keywords: Autoantibodies, Tumor Antigen, Biomarker, Luminex, Multiplex immunoassay, EBV
1. Introduction
The detection of the humoral immune response is essential for the diagnosis and prognosis of infectious disease and autoimmunity, and may also provide biomarkers for the detection of cancer (Coronella-Wood and Hersh, 2003; Chapman et al., 2007). Several proteomic multiplexed immunoassays have been developed to facilitate the detection of these antibodies (Robinson et al., 2003; Ramachandran et al., 2004; Anderson and LaBaer, 2005; Hudson et al., 2007). The slide-based assays, in particular, are excellent discovery tools for the detection of antibodies, but require specialized high-throughput equipment not generally found in routine immunology laboratories.
Confirmation of antibody detection requires assays that allow for the intermediate-throughput (hundreds) of serum screening on fewer antigens. The gold standard, ELISA, uses purified recombinant proteins adhered onto 96-well plastic plates and probed with sera, one antigen at a time. One intermediate step for the validation of antibody responses is a bead ELISA using the Luminex platform. The recent development of spectrally-distinguishable fluorescent beads (Luminex) (Kellar and Iannone, 2002) has resulted in the widespread use of antigen-coupled beads for monitoring antibodies in sera by flow cytometry. These bead arrays have been adapted for serologic screening of antigens and has been described for up to ten antigens for HIV (Opalka et al., 2004), HPV (Opalka et al., 2003; Dias et al., 2005), Epstein-Barr virus (Klutts et al., 2004; Binnicker et al., 2008; Gu et al., 2008), B. anthracis (Biagini et al., 2004; Biagini et al., 2005), Influenza (Drummond et al., 2008) and M. tuberculosis (Khan et al., 2008). When using purified antigens covalently linked on the beads, multiplexed serologic screening is both highly sensitive and efficient, but requires bacterial protein production and protein purification. Batch-to-batch and antigen-dependent variations in protein expression, purification, and stability limit the development of novel ELISA assays, and increase the risk of false-positive antibody detection (Schmetzer et al., 2005).
We have previously developed a method for in situ transcription/translation of immunogenic antigens as epitope-tagged proteins for in situ expression and capture on protein microarrays (NAPPA) (Ramachandran et al., 2004; Anderson et al., 2008). This results in highly reproducible and easily produced proteins for serologic screening, with comparable limits of detection of antibody to p53 antigen, for example, as standard protein ELISA. Using this approach, we have successfully expressed and captured over 15,000 different human, viral, and bacterial proteins for monitoring humoral immunity in both human and animal model systems. Slide-based protein microarrays allow for rapid screening of thousands of potentially immunogenic antigens. NAPPA expression and capture of tagged antigens for serum detection has been adapted for plate-based assays (RAPID ELISA) for confirmation of individual antigens (Ramachandran et al., 2008a).
Here, we have developed a bead-based multiplexed antigen programmable array for the detection of antibodies in sera. DNA preparation and in vitro expression has been adapted for high-throughput antigen production, exploiting ORFeome-derived gene sets. Anti-GST or anti-FLAG antibodies are coupled onto Luminex beads, and epitope-tagged proteins are expressed in vitro and bound to individual beads. Individual antigens are then pooled and antibodies within sera are detected with standard bead-based flow cytometry. Using well-validated sera from cancer patients and healthy donors, we demonstrate that the bead ELISA has comparable limits of detection for antibodies to EBNA-1 and p53 antigens as standard protein ELISA and RAPID. This bead-based assay allows for multiplexed detection of antibodies within serum for rapid detection of humoral immunity to both viral and tumor antigens.
2. Material and methods
2.1. Patient Sera
Breast cancer patient sera used in these analyses were obtained from the Lurie Breast Cancer Tissue and Blood Repository and the Specialized Research Program in Breast Cancer at the Dana-Farber Cancer Institute. Sera derived from breast cancer patients were obtained at the time of presentation with invasive breast cancer. Control sera were from healthy women undergoing blood donation. Written consent was obtained from all subjects under institutional review board approval.
2.2. Plasmid repository and high-throughput DNA preparation
Sequence-verified, full-length cDNA expression plasmids in flexible donor vector systems were obtained from the Harvard Institute of Proteomics and are publicly available (http://plasmid.med.harvard.edu/PLASMID/). These were converted to the T7-based mammalian expression vector pANT7_GST using LR recombinase (Invitrogen, Carlsbad, CA). The high-throughput preparation of high-quality supercoiled DNA for cell-free protein expression was performed as described (Ramachandran et al., 2008b). Briefly, expression plasmids were transformed into E.coli DH5α and grown in 1.5 mL terrific broth and ampicillin (100 µg/mL). DNA was purified with the NucleoPrepII anion exchange resin (Macherey-Nagel Inc., Bethlehem, PA) using a Biomek FX (Beckman Coulter, Inc., Fullerton, CA) automated laboratory workstation. Automated addition of all solutions was accomplished using a Matrix WellMate (Thermo Scientific, Hudson, NH) rapid bulk liquid-dispensing instrument. Purified DNA was precipitated by addition of 0.6 volumes isopropanol, followed by centrifugation at 5000 rcf for 30 minutes. The DNA pellet was washed with 200 µL of 80% ethanol, centrifuged at 5000 rcf for 15 minutes, dried, and resuspended in dH2O. For p21, p53 and EBV antigens, larger quantities of DNA were prepared using standard Nucleobond preparation methods (Macherey-Nagel Inc., Bethlehem, PA).
2.3. Antibody and protein coupling to carboxylated Luminex microspheres
Anti-GST antisera (GE Healthcare, Piscataway, NJ) or anti-FLAG antisera (Sigma-Aldrich, St. Louis, MO) was dialyzed into PBS to remove sodium azide and coupled to SeroMAP carboxylated microspheres (Luminex Corporation, Austin, TX) per manufacturer’s recommendations at the antibody to microsphere ratios indicated. Antibody coupling was confirmed with R-phycoerythrin (PE)-conjugated donkey anti-goat IgG antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Recombinant p53 protein (BD Biosciences, San Jose, CA) was coupled to SeroMAP microspheres at a ratio of 5 µg protein to 1 million beads. Bead coupling was confirmed with p53 monoclonal antibody (Sigma-Aldrich, St. Louis, MO) and goat anti-mouse IgG-PE antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). Beads from adjacent color regions (#34 to 44) were coupled with antibody. Beads #35–42 were multiplexed for detection by monoclonal antibodies.
2.4. Bead ELISA
Proteins were expressed using T7 reticulocyte lysate (Promega Corporation, Madison, WI) per manufacturer’s recommendations using 500 ng DNA. After expression, the in vitro transcription/translation (IVTT) product was incubated at room temperature (rt) for 2 hours while rotating with 5000 microspheres per reaction diluted to 100 microspheres/µL in PBS-1% BSA. After washing, protein-bound microspheres were pooled together and added to the appropriate wells in a 96-well filter plate (Millipore Corporation, Billerica, MA). Microspheres were washed again and incubated for 30 minutes at rt with serum samples diluted 1:80 unless otherwise indicated. After washing, biotin-conjugated goat anti-human IgG antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA) diluted to 4 µg/mL was incubated with the microspheres. Streptavidin-R-PE (Molecular Probes, Inc., Eugene, OR) diluted to 4 µg/mL was used to for detection. FLAG- and GST-protein tags were detected with anti-FLAG (Sigma-Aldrich, St. Louis, MO) and anti-GST (Cell Signaling Technology, Danvers, MA) monoclonal antibodies. Antigens were detected with monoclonal antibodies to PCNA (Cell Signaling Technology, Danvers, MA), BCL2 (Sigma-Aldrich, St. Louis, MO), CDK4 (Cell Signaling Technology, Danvers, MA and Sigma-Aldrich, St. Louis, MO), and p53 (Sigma-Aldrich, St. Louis, MO).
2.5. P53 antibody ELISA
The p53 protein ELISA assay was performed per manufacturer’s recommendations (p53 ELISAPLUS Autoantibody Kit, Calbiochem, Gibbstown, NJ) (Anderson et al., 2008). Calibrators, controls, and serum samples were added at 100 µl each in duplicate. The calibrators were used neat, 1:1.5, 1:2, 1:3, 1:4, and 1:6 to obtain a calibration curve. Serum samples were diluted 1:100 and incubated for 1 hour at rt and bound IgG was detected by absorbance at 450 nm using a VICTOR3TM 1420 Multilabel Plate Reader running Wallac 1420 software (Perkin Elmer, Waltham, MA).
2.6. RAPID ELISA
GST 96-well detection plates coated with anti-GST antibody (GE Healthcare, Piscataway, NJ) were blocked overnight at 4°C with 5% milk in 0.2% Tween20 in PBS (PBST-0.2%). GST fusion proteins were expressed using the T7 reticulocyte lysate (Promega, Madison, WI) per manufacturer’s recommendations using 500 ng DNA. After expression, the IVTT product was transferred to the anti-GST plates for 2 hours at rt while shaking at 800 rpm, washed, and incubated with serum samples diluted 1:300. HRP-conjugated goat anti-human IgG antibody was diluted at 1:1000 (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). SuperSignal ELISA Femto Max Sensitivity (Pierce, Rockford, IL) was used for detection.
2.7. Statistical Analysis
Protein ELISA analyses for antibodies in sera were performed in duplicate, and positive sera were confirmed on repeat experiments. Values are plotted as mean values. P53 positive sera were previously defined on RAPID ELISA and on standard ELISA as those that have fold change of 3 or more when compared to the p53 negative sera and control sera. We assumed that majority of sera was p53 negative and computed fold changes as ratio of the serum signal to the median signal of all sera assayed (Anderson et al., 2008).
For the bead ELISA, measurements were performed on a Luminex 200 using Luminex IS 2.3 software, counting 50 events for each analysis, and median fluorescence intensity (MFI) was measured. Intra-assay variability was determined using fluorescent measurements of EBNA-1 IgG antibody detection of the same sera repeated 7 times on the same day for 5 days. These signals were averaged, and the coefficient of variation (CV, (SD/mean)×100%) was calculated. Inter-assay variability was determined using fluorescent measurements of anti-EBNA-1 IgG detection of the same sera repeated on 5 different days. These signals were also averaged and used to calculate the CV.
3. Results
3.1. Schematic of the bead ELISA
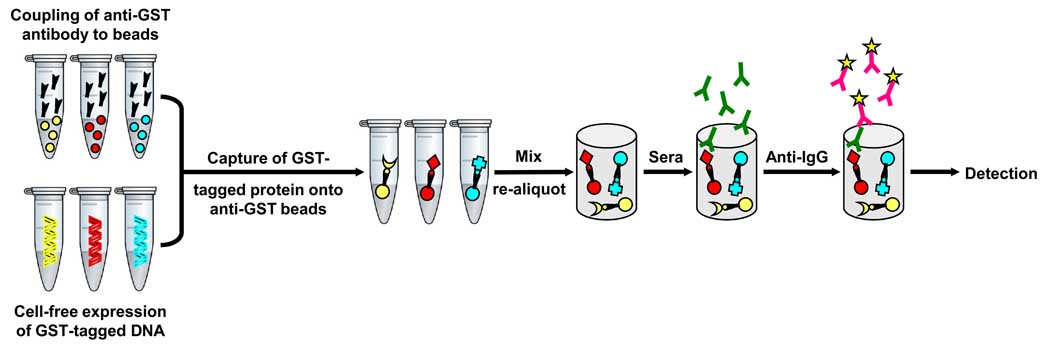
The bead ELISA for the multiplexed detection of antibodies in sera uses anti-GST antibodies that are coupled to different Luminex beads, each containing different mixtures of red and infrared fluorophores. The cDNA ORFs are expressed as C-terminal GST fusion proteins using IVTT. The individual protein antigens are then captured via their GST tags to anti-GST-coupled beads. This results in “addressable” recombinant proteins that are non-covalently attached to different fluorescent beads (Figure 1). The individual protein-bound beads are then pooled and incubated with individual sera for the detection of bound IgG antibodies.
Figure 1. Schematic of the bead ELISA for the detection of antibodies in serum.
The cDNAs encoding full-length antigens are individually expressed as GST-tagged fusion proteins using rabbit reticulocyte lysate. Recombinant proteins are captured onto anti-GST-coupled Luminex microspheres. The beads displaying different antigens are pooled and re-aliquoted prior to addition of human serum so that there are 5000 of each color/antigen bead per serum. Human IgG is detected with PE-conjugated secondary anti-IgG antibodies.
3.2. Detection of recombinant GST-tagged and FLAG-tagged proteins by the bead ELISA
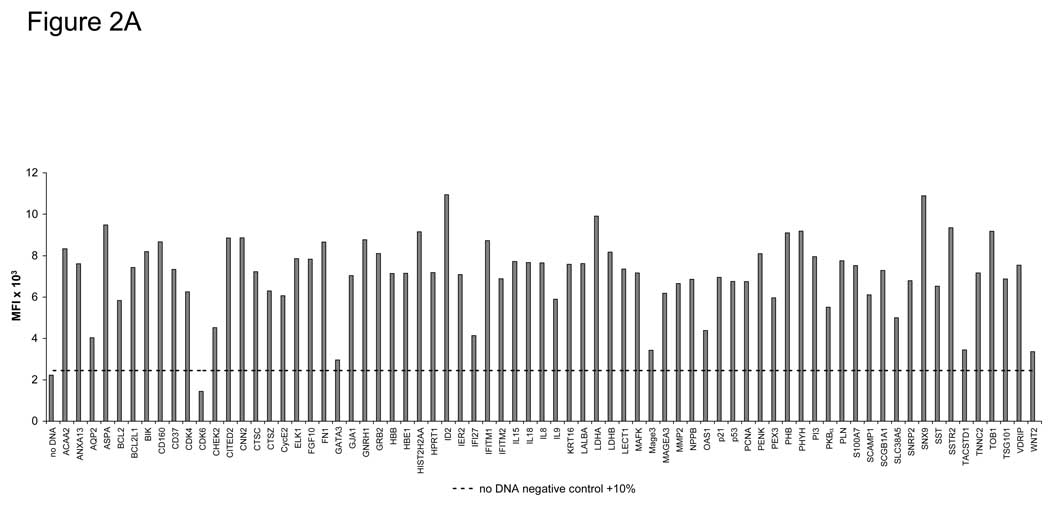
To determine if the programmable bead array can be adapted for multiple-antigen detection, we selected 72 human tumor antigen clones from our PlasmID repository (http://plasmid.med.harvard.edu/PLASMID/) for high-throughput DNA preparation (Table 1). The 72 tumor antigens were expressed as C-terminal GST fusion proteins in duplicate. The success of protein expression and capture on the bead ELISA was measured by probing with an anti-GST antibody that recognizes a C-terminal GST epitope on every antigen (Figure 2A). Expression ranged from 1446–10947 MFI (mean=7102). We determined that 71 of 72 antigens (98.6%) had detectable protein signal over background, defined as 10% above the average of non-antigen containing beads (dotted line). Using this expression method, we have expressed over 10,000 human and infectious disease antigens in slide microarray format and have demonstrated overall expression of >90% of fusion proteins, including membrane proteins (Ramachandran et al., 2008b).
Table 1.
List of Human Tumor Antigens.
| ACAA2 | HBB | p21 |
| ANXA13 | HBE1 | p53 |
| AQP2 | HIST2H2AA | PCNA |
| ASPA | HPRT1 | PENK |
| BCL2 | ID2 | PEX3 |
| BCL2L1 | IER2 | PHB |
| BIK | IFI27 | PHYH |
| CD160 | IFITM1 | PI3 |
| CD37 | IFITM2 | PKBα |
| CDK4 | IL15 | PLN |
| CDK6 | IL18 | S100A7 |
| CHEK2 | IL8 | SCAMP1 |
| CITED2 | IL9 | SCGB1A1 |
| CNN2 | KRT16 | SLC38A5 |
| CTSC | LALBA | SNRP2 |
| CTSZ | LDHA | SNX9 |
| CycE2 | LDHB | SST |
| ELK1 | LECT1 | SSTR2 |
| FGF10 | MAFK | TACSTD1 |
| FN1 | Mage3 | TNNC2 |
| GATA3 | MAGEA3 | TOB1 |
| GJA1 | MMP2 | TSG101 |
| GNRH1 | NPPB | VDRIP |
| GRB2 | OAS1 | WNT2 |
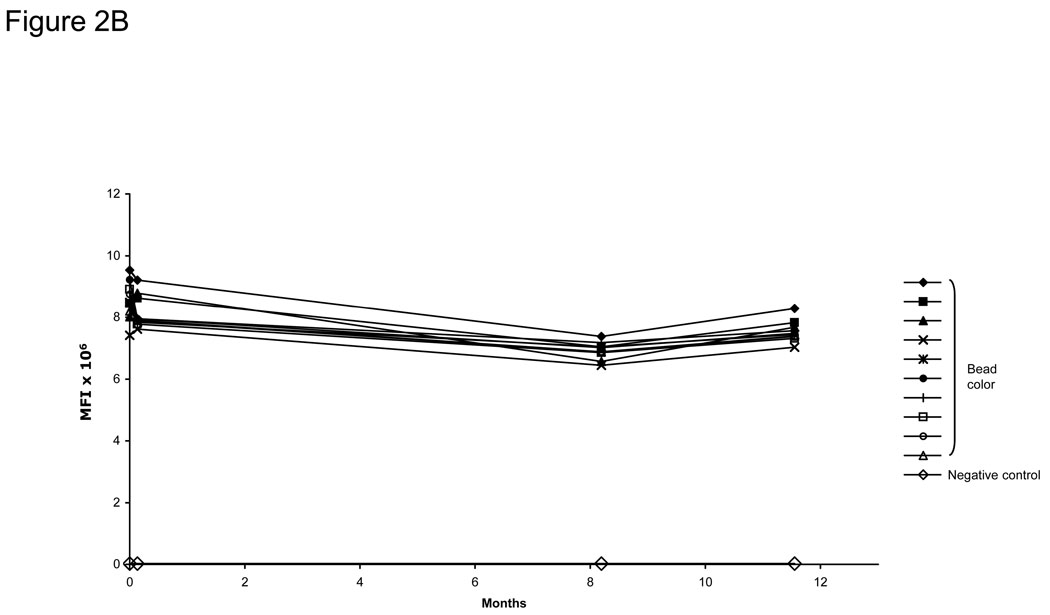
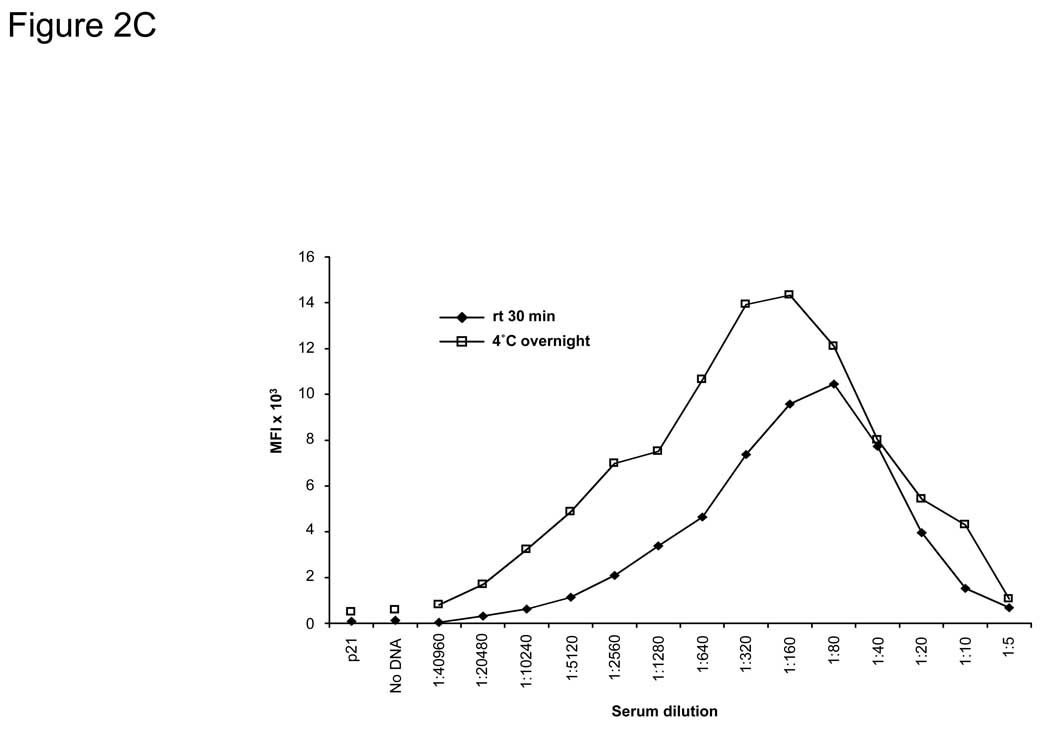
Figure 2. Protein expression, stability, and specificity of the bead ELISA.
A) Specific detection of 72 captured GST-tagged tumor antigens on beads. DNA was prepared using a high-throughput robotic preparation method, expressed in vitro, bound to anti-GST-coupled beads, and detected using an anti-GST MAb. Background expression (10% above the average of non-antigen containing beads) is shown as a dotted line. B) Anti-GST-coupled beads stored over 11 months showed no change in the stability of bound IgG using donkey anti-goat IgG-PE antibody. C) EBNA-1 antibody-positive serum from a healthy donor was tested for anti-EBNA-1 antibodies using the bead ELISA. EBNA-1-GST and p21-GST fusion proteins were expressed by IVTT and bound to anti-GST-coupled Luminex beads. Serum was added in the titrations shown for either 30 minutes at rt or at 4°C overnight. P21-GST and no DNA controls were tested at 1:20 dilution. Incubation overnight increased the sensitivity of the assay 8-fold to >1:20,000, and optimal serum dilution for maximal detection was at 1:80–1:320.
3.3. Generation of Anti-GST coupled beads
We have had excellent success in protein microarray-based ELISA using anti-GST antisera and anti-FLAG antisera as capture antibodies (Ramachandran et al., 2004; Anderson et al., 2008; Ramachandran et al., 2008b). To adapt this approach to a bead-based array format, we optimized antibody coupling to Luminex SeroMap beads to a ratio of 5 µg antibody to 1 million beads (data not shown). The antibody-coupled beads retain identical sensitivity and linearity of detection and also retain stability over a period of 11 months, with an average decrease in signal of only 11.7% for the 10 analytes (Figure 2B). Similarly, FLAG-tagged proteins can also be captured and specifically detected on the beads (data not shown).
3.4. Adaptation of the bead ELISA to detect antibodies in sera
To optimize the assay for the detection of IgG in human sera, we expressed EBNA-1, a highly immunogenic antigen derived from Epstein-Barr virus (EBV). Over 90% of healthy blood donors have detectable antibodies to EBNA-1 in standard protein ELISA, and the C-terminal portion of EBNA-1 (located after the GA repeat region, amino acid #401 to #641) can be readily expressed using rabbit reticulocyte lysate and detected with healthy donor sera on protein microarrays (Anderson et al., 2008). Using anti-GST-coupled beads, the C-terminal portion of EBNA-1 was expressed with IVTT and captured onto the beads. Protein expression was confirmed with anti-GST antibodies (data not shown). Using serial dilutions of serum from a healthy donor that contains known antibodies to EBNA-1, the optimal serum dilution for detection of EBNA-1 antibodies was determined to be between 1:80–1:320 (Figure 2C). More concentrated sera interfered with detection in this assay, consistent with previous observations of the “hook effect” seen in other immunoassays (Cole et al., 1993; Jassam et al., 2006). Background with no DNA or p21-GST DNA (controls) at 1:20 serum dilution is shown. Incubation of sera at 4°C overnight increased the sensitivity of antibody detection 8-fold to >1:20,000 compared to a 30-minute incubation of sera at rt.
3.5. Reproducibility and stability of detecting autoantibodies
The CVs of the bead-based antibody detection in human sera were determined. Replicate assays of C-terminal EBNA-1-GST antigen and p21-GST antigen were expressed in 7 replicates on 5 different days and captured onto anti-GST beads. These were probed with serum positive for EBNA-1 antibodies. The serum signals for EBNA-1 among the beads processed on the same day showed excellent reproducibility with an intra-assay CV of 3–8%. Serum signals for beads processed on 5 different days had an inter-assay CV of 5%.
3.6. Comparison of the bead ELISA to other ELISA methods
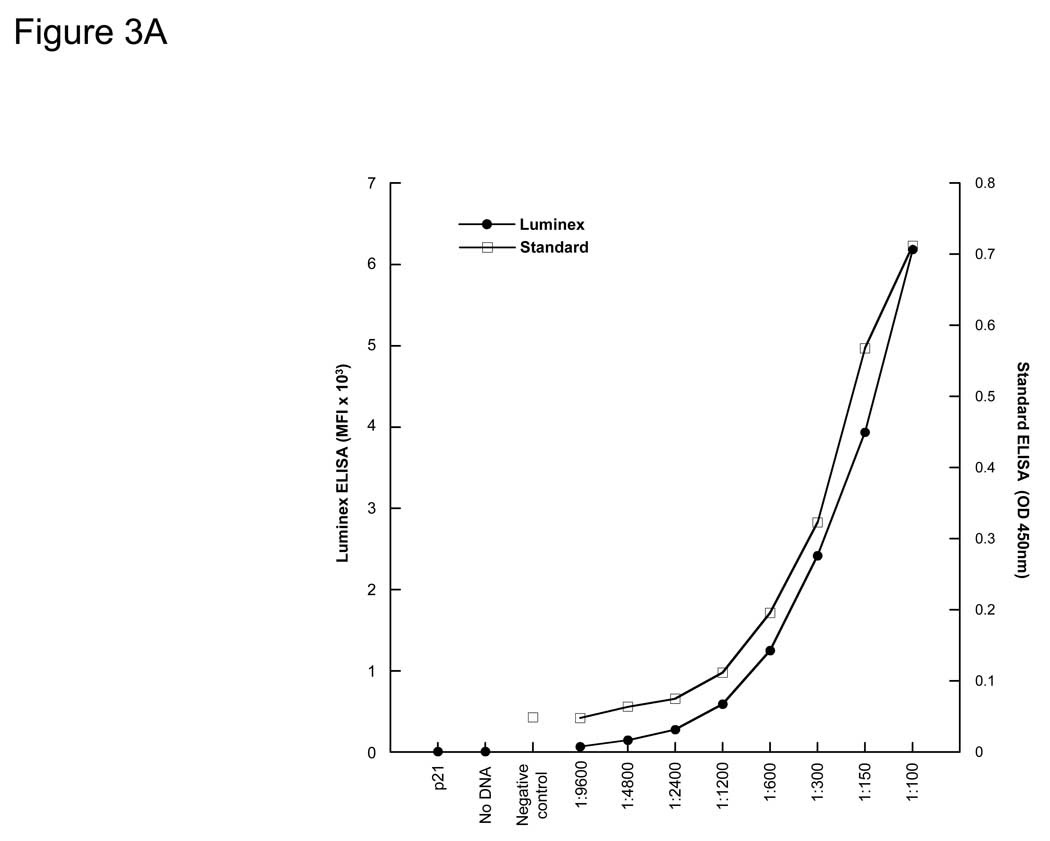
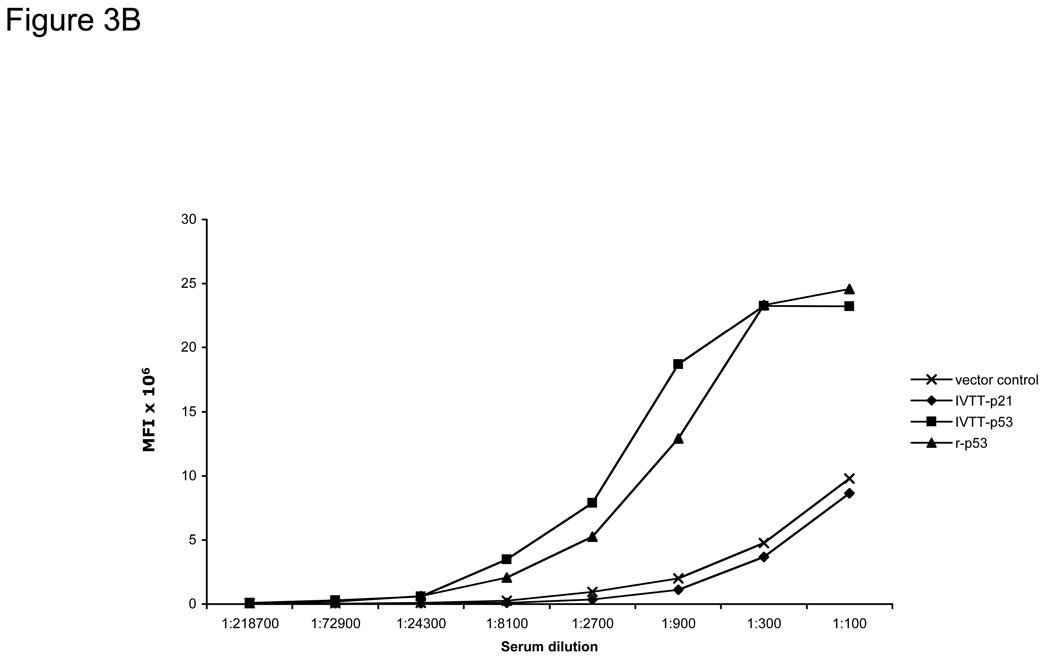
To compare the limits of detection of the bead ELISA with standard recombinant protein ELISA, breast cancer patient sera with known antibodies to p53 were titrated using both assays. The background controls of IVTT with p21-GST DNA or no DNA for the bead ELISA were comparable (Figure 3A). We directly compared the sensitivities of the bead ELISA with purified antigen-coupled bead arrays for the tumor antigen p53, using a p53-positive serum titrated from 1:100 to 1:218700 (Figure 3B). Both methods show comparable sensitivities, with low background of both vector and p21-GST protein.
Figure 3. p53-specific IgG from human sera are specifically detected.
A) Detection of antibodies to p53 by bead ELISA (Luminex) and standard ELISA in a breast cancer patient serum with p53 antibodies. B) Detection of p53 antibodies in the same serum with IVTT-generated p53 is comparable to that with recombinant p53 protein directly captured onto beads.
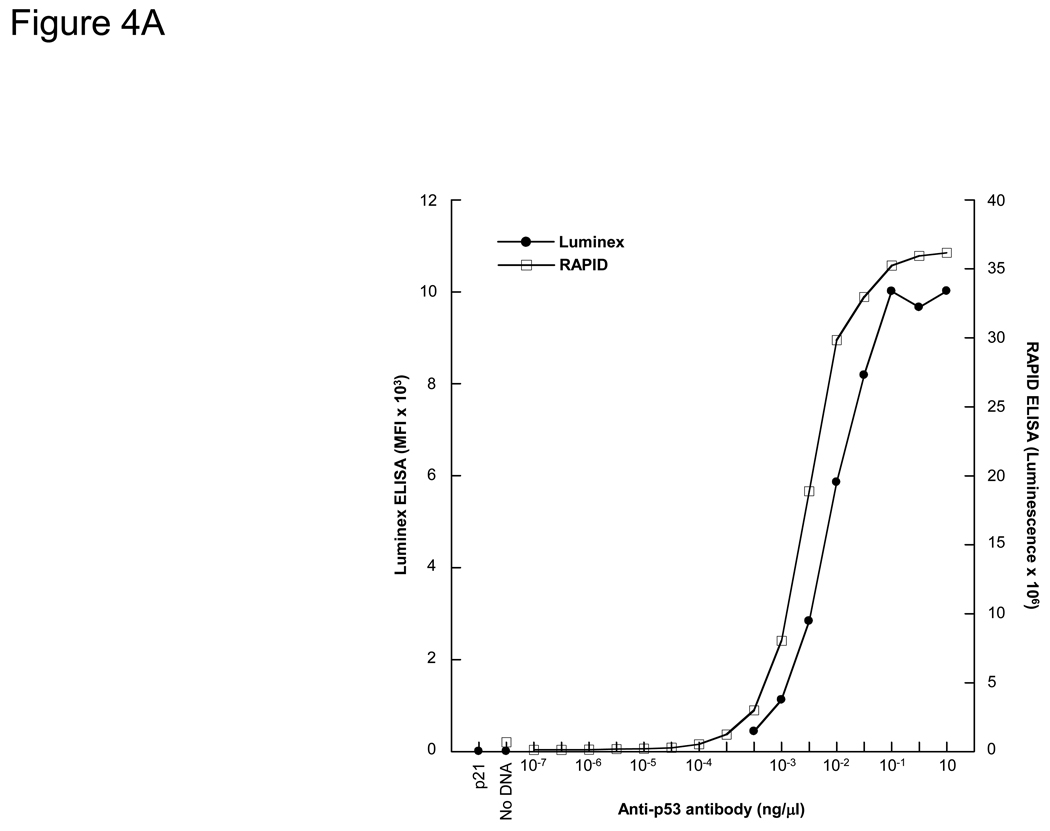
We have previously developed a 96-well format ELISA (RAPID ELISA) using target antigens expressed with IVTT and captured onto anti-GST plates (Ramachandran et al., 2008a). The RAPID ELISA has comparable limits of detection and antibody specificity as standard protein ELISA for the detection of p53 and EBNA-1 antibodies, but cannot be used for multiplexed antibody detection. We determined the relative sensitivities of the bead ELISA with the 96-well plate-based RAPID ELISA (Figure 4A). Both assays showed a >2 log linear range of detection of p53 monoclonal antibodies, and were highly sensitive with detection of the p53 antibody at pM concentration.
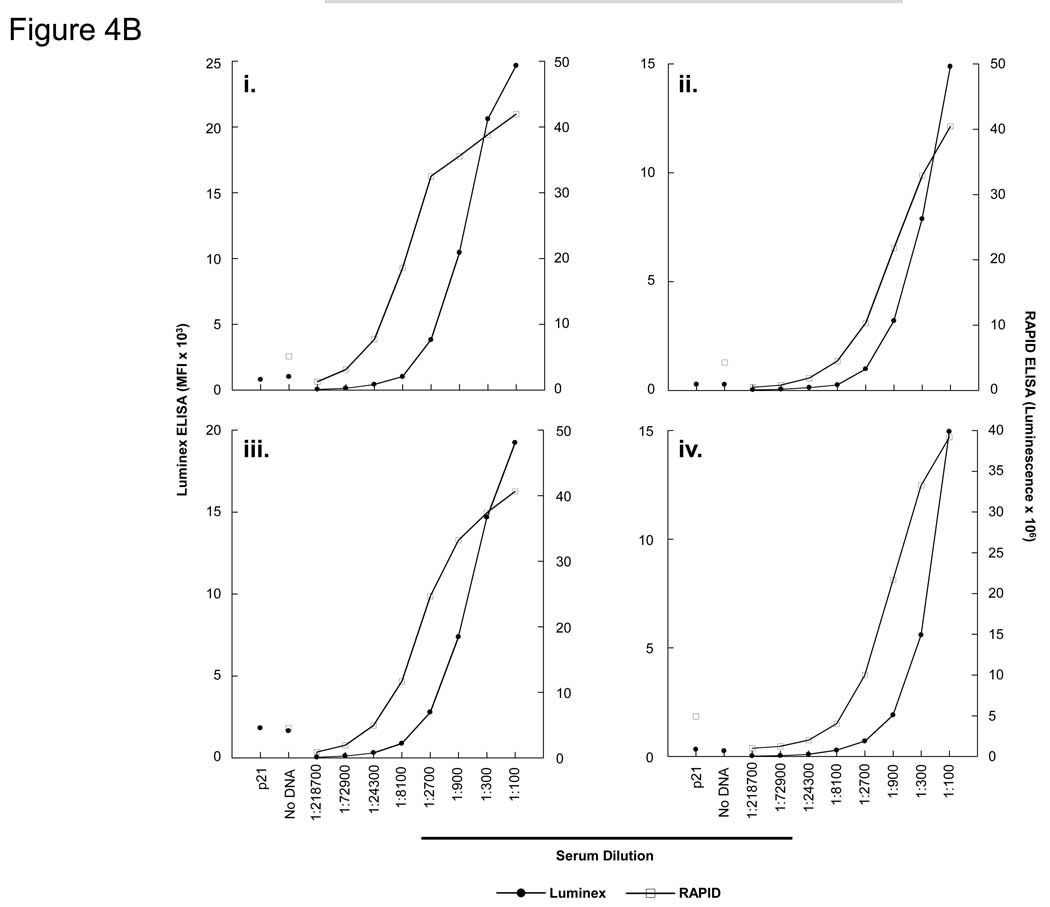
Figure 4. Detection of p53 antibodies by bead ELISA (Luminex) and RAPID (96-well format) ELISA.
A) Quantification of p53 antigen detection by bead ELISA and RAPID ELISA. P53-GST fusion protein was expressed by bead ELISA and RAPID ELISA, and detected with titrations of an anti-p53 MAb. B) i–iv. Detection of anti-p53 antibodies by bead ELISA and RAPID ELISA from 4 breast cancer patient sera containing p53 autoantibodies.
Using four breast cancer patient sera that contain p53 autoantibodies (Anderson et al., 2008), a direct comparison of Luminex ELISA with 96-well format RAPID ELISA was performed (Figure 4B). Both p21-GST fusion protein and no DNA controls were performed at a 1:100 dilution, and the background for each serum is shown. Overall, the RAPID ELISA was 3-fold more sensitive than the bead ELISA, but both assays were highly sensitive (to 1:2700 serum dilution for three sera) for the detection of p53-specific autoantibodies.
3.7. Multiplexed detection of antibodies using the bead ELISA
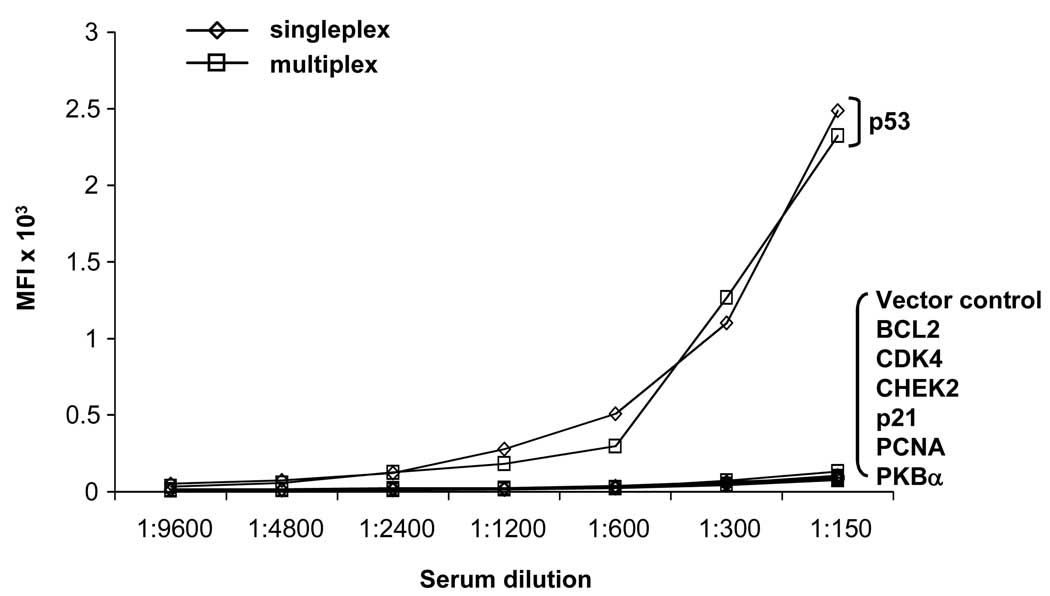
To demonstrate that the bead ELISA can be used as a multiplexed serologic assay without loss of sensitivity, anti-GST antibodies were coupled to 8 different fluorescent beads. P53-GST protein was expressed and bound to one of the 8 beads. In addition, the vector control and 6 other antigens (BCL2, CDK4, CHEK2, p21, PCNA, PKBα) were expressed and bound to the remaining 7 beads. Expression of the antigens was confirmed using an anti-GST MAb (data not shown). The sensitivity of detection of p53 autoantibodies in sera using the p53-GST beads (singleplex) and the p53-GST beads mixed with 7 sets of control and negative antigens (multiplex) was equivalent (Figure 5). No increase in signal was observed in the control beads, demonstrating that there is no diffusion of specific protein antigen onto other beads (data not shown).
Figure 5. Multiplexed detection of antibodies in patient sera.
The p53 autoantibodies were detected by bead ELISA using p53-loaded anti-GST beads either in singleplex or in multiplex with 7 other antigen-loaded beads, including a vector control, BCL2, CDK4, CHEK2, p21, PCNA, and PKB . The p53 autoantibody detection was not diminished by the addition of other antigen-loaded beads.
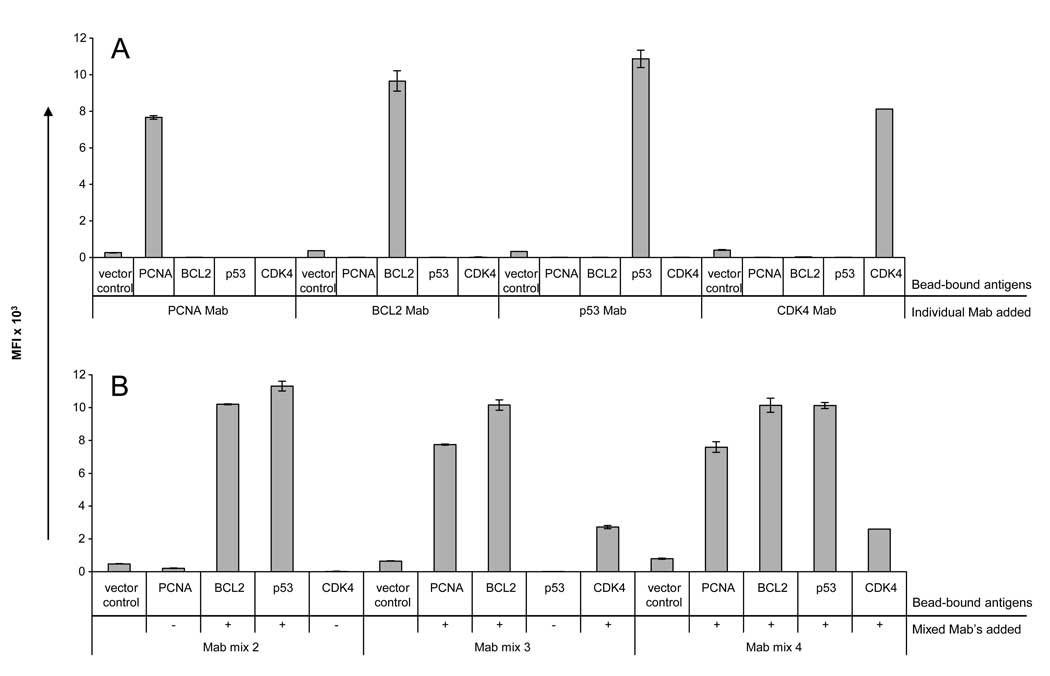
A major limitation of multiplexed bead arrays occurs when interference arises among multiple different antibodies added to detect different analytes. The assay presented here requires only two different antibody reagents: the anti-GST capture antibody and the anti-IgG detection antibody, which are coordinated to avoid interference. To confirm this, four tumor antigens (PCNA, BCL2, CDK4, and p53) were expressed as GST fusion proteins, bound to individual anti-GST-coupled beads, and pooled. Expression of each antigen was confirmed with Mab’s specific for the antigens (Figure 6A and data not shown). To simulate the presence of multiple antibodies in sera, the monoclonal antibodies were diluted at 1:400 and pooled as a mix of 2, 3, or 4 Mab’s (Figure 6B). In all cases, specificity was maintained when both the antigens and the antibodies were multiplexed.
Figure 6. Multiplexed detection of antibodies by bead ELISA.
CDK4, p53, BCL2, and PCNA antigens and a vector control were individually expressed by IVTT as GST fusion proteins, bound to anti-GST beads, and pooled. A) Within the pooled beads, individual antigens could be specifically detected with monoclonal antibodies specific to PCNA, BCL2, p53, and CDK4 at a 1:400 dilution. B) The monoclonal antibodies specific for each antigen were then pooled in different combinations: mix 2 (BCL2 and p53 Mab’s), mix 3 (PCNA, BCL2, and CDK4 Mab’s), and mix 4 (all 4 Mab’s). Each antigen could be specifically detected within the pooled antigens without loss of sensitivity or specificity.
3.8. Multiplexed detection of viral IgG with human sera
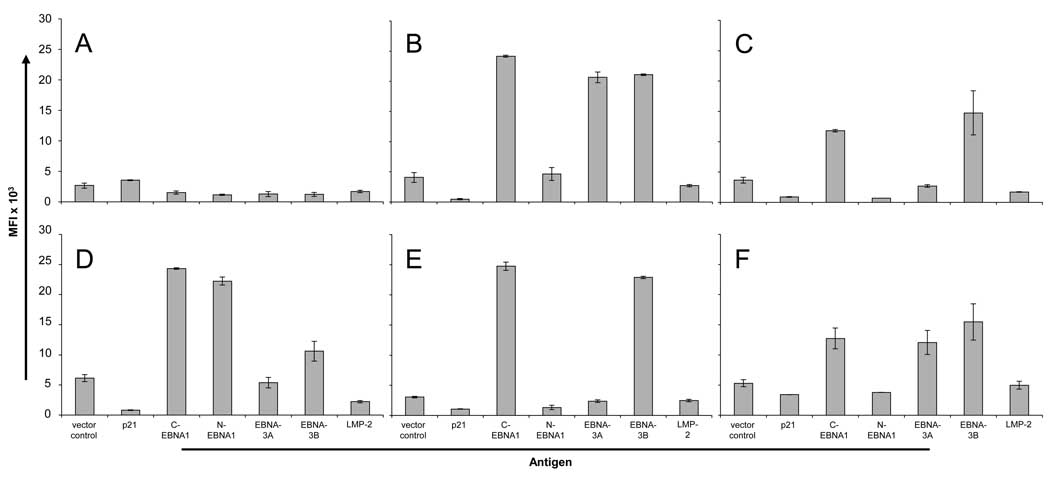
To demonstrate that the bead ELISA can detect multiple IgG antibodies within human sera, we developed a custom assay for the detection of antigens derived from the highly immunogenic Epstein-Barr Virus (EBV). Serologic ELISA assays to EBV target antibody responses to the VCA and EBNA-1 antigens have been adapted by direct protein coupling to Luminex beads (Klutts et al., 2004). Although more is known about the serologic responses to VCA, EBNA-1, and EA (Altuglu et al., 2007; Lunemann et al., 2007), little is known about responses to other latent antigens, such as EBNA-3A, EBNA-3B, and LMP-2. We expressed these antigens along with EBNA-1 by IVTT. To optimize expression of EBNA-1, the cDNA was divided into the N-terminal (N-EBNA1) and C-terminal (C-EBNA1) fragments by removal of the central GA repeat region. Control antigen p21 was also expressed as C-terminal GST fusion proteins. Protein expression was confirmed by GST detection (data not shown). All six antigens, with vector control, were expressed and combined using the bead ELISA. Six healthy donor sera were then tested for antibodies to the EBV antigens (Figure 7). Positive antibodies were defined as >2 S.D.’s over the median of the vector control. One serum (Figure 7A) was negative for all antibodies, consistent with the observed frequencies of EBNA-1 IgG in blood donor sera of 93% (Anderson et al., 2008; Schwenk et al., 2008). C-EBNA1 was the dominant antigen, detected in 5/6 sera (median MFI 18455, range 1531–24745). EBNA-3B antibodies were detected in all C-EBNA1 positive sera (5/6) (median MFI 15120, range 1229–22885). EBNA-3A antibodies were detected in 2/6 sera (median MFI 4044, range 1281.5–20661), and only in sera that were positive for both C-EBNA1 and EBNA-3B alone. LMP-2 antibodies were not detected in this cohort. In contrast to C-EBNA1, N-EBNA1 antibodies were detected in only 1/6 sera (median MFI 2564, range 707–22246).
Figure 7.
4. Discussion
The development of serologic screening of protein microarrays, phage display, and reverse-phase protein arrays has led to the discovery of thousands of novel tumor and infectious antigens. As a discovery platform, these technologies hold the promise of understanding the antibody immunome. We have previously optimized and used protein microarrays (NAPPA ELISA) for the detection of antibodies to a wide variety of tumor antigens, infectious antigens, and autoimmune antigens in patient sera as well as in animal sera (Anderson and LaBaer, 2005; Anderson et al., 2008). By adapting and standardizing the source DNA preparation and protein expression to high-throughput proteomic production, 98% of genes show readily detectable protein. Of 1000 human genes, this system results in success rates of protein expression of kinases (98%), transcription factors (96%), membrane proteins (93%) and large >100 kDa proteins (88%) (Ramachandran et al., 2008b). Using rabbit reticulocyte lysate for in vitro coupled transcription-translation protein expression, specific protein-protein interactions can be maintained, suggesting that protein folding can be preserved (Ramachandran et al., 2008b).
Although proteins produced by in vitro transcription/translation display only a limited number of post translational modifications, this had not limited the ability to detect antibodies to a wide variety of known immunogenic antigens, including antigens to the pathogens F. tularensis and P. aeruginosa, the tumor antigens BCL2 and p53, and the GAD65 antigen in insulin-dependent diabetes mellitus (Ramachandran et al., 2008a). Scientifically, these are highly flexible assays that allow for rapid screening of any target antigen expressed from cDNA. However, protein microarray production requires printing technologies that are not readily available in immunology laboratories. These arrays are excellent discovery tools, but serologic assays for rapid validation of novel antigens that can screen hundreds of sera on select proteins have not been developed.
Here, we developed a programmable bead ELISA for the multiplexed serologic detection of target antigens. The bead platform has been used for simultaneous and specific detection of up to 100 analytes on pooled beads for monospecific antibody proteomics, without tailing, agglutination, and only minor scattering (Schwenk et al., 2008). By coupling antigens or peptides on the beads, they have been adapted for detection of antibody responses to a wide variety of infectious antigens. Binding of bacterially-expressed HPV-derived GST-fusion proteins onto glutathione-tagged beads (Waterboer et al., 2005) demonstrates that bead arrays can be used for multiplexed detection of antibodies in sera, and has been used to detected seropositivity to 34 different HPV L1 proteins (Clifford et al., 2007; Michael et al., 2008). A direct comparison of S. aureus-derived His-fusion proteins directly coupled to beads demonstrated that direct coupling of antigen yielded a higher signal-to-noise ratio for serologic analysis than linking through the penta-His antibody (Verkaik et al., 2008). However, traditional methods that use purified proteins require months of protein preparation and optimization prior to ELISA assay development. This is a labor-intensive process that limits the rapid serologic analysis of novel antigens.
We have used in vitro transcription/translation with rabbit reticulocyte lysate to omit potentially immunogenic bacterial products and express the antigens in a mammalian microenvironment. The use of anti-tag antibodies coupled to fluorescent polystyrene beads, rather than glutathione or direct antigen coupling for protein capture, allows for greater flexibility in tag selection, antigen expression, and rapid linking to ORFeome-derived gene sets. The bead ELISA allows for rapid, multiplexed detection of antigen-specific antibodies within a single serum sample, with a system readily adaptable to clinical immunology diagnostics. Depending on the sera, we found a 0 to 3-fold loss of sensitivity of the bead ELISA, but that can be corrected for by using the sera at increased concentration without changing signal-to-noise ratios. We have demonstrated that the bead ELISA is reproducible, stable, can readily detect >70 antigens, and can be multiplexed for serologic detection of antigens, saving time, cost, and sera. With current bead-based technology, there is the potential to rapidly and simultaneously measure up to 100 antibodies per serum sample, although this has not been tested. We did not observe interference of multiplexing in our assay and no diffusion of antigen across the beads. By coupling advances in cDNA ORFs that are publicly available in flexible donor vector systems (http://plasmid.med.harvard.edu/PLASMID/) with multiplexed serologic assays, proteomic-based serologic analysis of the immune response is possible.
ACKNOWLEDGEMENTS
We thank Dr. Niroshan Ramachandran for assistance in setting up the Luminex system. This study was supported by a research grant from the NCI Early Detection Research Network 5U01CA117374-02
Abbreviations
- NAPPA
Nucleic Acid Programmable Protein Array
- RAPID
Rapid Antigenic Protein In situ Display
- IVTT
in vitro transcription/translation
- EBV
Epstein-Barr Virus
- EBNA-1
EBV Nuclear Antigen 1
- CV
coefficient of variation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Altuglu I, Bozkurt H, Samlioglu P, Zeytinoglu A. Evaluation of three different assays for the assessment of Epstein Barr Virus immunological status. New Microbiol. 2007;30:393–398. [PubMed] [Google Scholar]
- Anderson KS, LaBaer J. The sentinel within: exploiting the immune system for cancer biomarkers. J Proteome Res. 2005;4:1123–1133. doi: 10.1021/pr0500814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KS, Ramachandran N, Wong J, Raphael JV, Hainsworth E, Demirkan G, Cramer D, Aronzon D, Hodi FS, Harris L, Logvinenko T, LaBaer J. Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J Proteome Res. 2008;7:1490–1499. doi: 10.1021/pr700804c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagini RE, Sammons DL, Smith JP, MacKenzie BA, Striley CA, Robertson SA, Snawder JE, Quinn CP. Simultaneous measurement of specific serum IgG responses to five select agents. Anal Bioanal Chem. 2005;382:1027–1034. doi: 10.1007/s00216-005-3204-6. [DOI] [PubMed] [Google Scholar]
- Biagini RE, Sammons DL, Smith JP, MacKenzie BA, Striley CA, Semenova V, Steward-Clark E, Stamey K, Freeman AE, Quinn CP, Snawder JE. Comparison of a multiplexed fluorescent covalent microsphere immunoassay and an enzyme-linked immunosorbent assay for measurement of human immunoglobulin G antibodies to anthrax toxins. Clin Diagn Lab Immunol. 2004;11:50–55. doi: 10.1128/CDLI.11.1.50-55.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnicker MJ, Jespersen DJ, Harring JA, Rollins LO, Beito EM. Evaluation of a multiplex flow immunoassay for detection of epstein-barr virus-specific antibodies. Clin Vaccine Immunol. 2008;15:1410–1413. doi: 10.1128/CVI.00082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman C, Murray A, Chakrabarti J, Thorpe A, Woolston C, Sahin U, Barnes A, Robertson J. Autoantibodies in breast cancer: their use as an aid to early diagnosis. Ann Oncol. 2007;18:868–873. doi: 10.1093/annonc/mdm007. [DOI] [PubMed] [Google Scholar]
- Clifford GM, Shin HR, Oh JK, Waterboer T, Ju YH, Vaccarella S, Quint W, Pawlita M, Franceschi S. Serologic response to oncogenic human papillomavirus types in male and female university students in Busan, South Korea. Cancer Epidemiol Biomarkers Prev. 2007;16:1874–1879. doi: 10.1158/1055-9965.EPI-07-0349. [DOI] [PubMed] [Google Scholar]
- Cole T, Johnson D, Eveland B, Nahm M. Cost-effective method for detection of "hook effect" in tumor marker immunometric assays. Clin Chem. 1993;39:695b–696b. [PubMed] [Google Scholar]
- Coronella-Wood JA, Hersh EM. Naturally occurring B-cell responses to breast cancer. Cancer Immunol Immunother. 2003;52:715–738. doi: 10.1007/s00262-003-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias D, Van Doren J, Schlottmann S, Kelly S, Puchalski D, Ruiz W, Boerckel P, Kessler J, Antonello JM, Green T, Brown M, Smith J, Chirmule N, Barr E, Jansen KU, Esser MT. Optimization and validation of a multiplexed luminex assay to quantify antibodies to neutralizing epitopes on human papillomaviruses 6, 11, 16, and 18. Clin Diagn Lab Immunol. 2005;12:959–969. doi: 10.1128/CDLI.12.8.959-969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond JE, Shaw EE, Antonello JM, Green T, Page GJ, Motley CO, Wilson KA, Finnefrock AC, Liang X, Casimiro DR. Design and optimization of a multiplex anti-influenza peptide immunoassay. J Immunol Methods. 2008;334:11–20. doi: 10.1016/j.jim.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Gu AD, Xie YB, Mo HY, Jia WH, Li MY, Li M, Chen LZ, Feng QS, Liu Q, Qian CN, Zeng YX. Antibodies against Epstein-Barr virus gp78 antigen: a novel marker for serological diagnosis of nasopharyngeal carcinoma detected by xMAP technology. J Gen Virol. 2008;89:1152–1158. doi: 10.1099/vir.0.83686-0. [DOI] [PubMed] [Google Scholar]
- Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M. Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc Natl Acad Sci U S A. 2007;104:17494–17499. doi: 10.1073/pnas.0708572104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jassam N, Jones CM, Briscoe T, Horner JH. The hook effect: a need for constant vigilance. Ann Clin Biochem. 2006;43:314–317. doi: 10.1258/000456306777695726. [DOI] [PubMed] [Google Scholar]
- Kellar KL, Iannone MA. Multiplexed microsphere-based flow cytometric assays. Exp Hematol. 2002;30:1227–1237. doi: 10.1016/s0301-472x(02)00922-0. [DOI] [PubMed] [Google Scholar]
- Khan IH, Ravindran R, Yee J, Ziman M, Lewinsohn DM, Gennaro ML, Flynn JL, Goulding CW, DeRiemer K, Lerche NW, Luciw PA. Profiling antibodies to Mycobacterium tuberculosis by multiplex microbead suspension arrays for serodiagnosis of tuberculosis. Clin Vaccine Immunol. 2008;15:433–438. doi: 10.1128/CVI.00354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klutts JS, Liao RS, Dunne WM, Jr, Gronowski AM. Evaluation of a multiplexed bead assay for assessment of Epstein-Barr virus immunologic status. J Clin Microbiol. 2004;42:4996–5000. doi: 10.1128/JCM.42.11.4996-5000.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunemann JD, Kamradt T, Martin R, Munz C. Epstein-Barr Virus: Environmental Trigger of Multiple Sclerosis? J. Virol. 2007;81:6777–6784. doi: 10.1128/JVI.00153-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael KM, Waterboer T, Sehr P, Rother A, Reidel U, Boeing H, Bravo IG, Schlehofer J, Gartner BC, Pawlita M. Seroprevalence of 34 human papillomavirus types in the German general population. PLoS Pathog. 2008;4:e1000091. doi: 10.1371/journal.ppat.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka D, Lachman CE, MacMullen SA, Jansen KU, Smith JF, Chirmule N, Esser MT. Simultaneous quantitation of antibodies to neutralizing epitopes on virus-like particles for human papillomavirus types 6, 11, 16, and 18 by a multiplexed luminex assay. Clin Diagn Lab Immunol. 2003;10:108–115. doi: 10.1128/CDLI.10.1.108-115.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opalka D, Pessi A, Bianchi E, Ciliberto G, Schleif W, McElhaugh M, Danzeisen R, Geleziunas R, Miller M, Eckert DM, Bramhill D, Joyce J, Cook J, Magilton W, Shiver J, Emini E, Esser MT. Analysis of the HIV-1 gp41 specific immune response using a multiplexed antibody detection assay. J Immunol Methods. 2004;287:49–65. doi: 10.1016/j.jim.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Ramachandran N, Anderson KS, Raphael JV, Hainsworth E, Sibani S, Montor WR, Pacek M, Wong J, Eljanne M, Sanda MG, Hu Y, Logvinenko T, LaBaer J. Tracking humoral responses using self assembling protein microarrays. Proteomics - Clinical Applications. 2008a;2:1518–1527. doi: 10.1002/prca.200800034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran N, Hainsworth E, Bhullar B, Eisenstein S, Rosen B, Lau AY, Walter JC, LaBaer J. Self-assembling protein microarrays. Science. 2004;305:86–90. doi: 10.1126/science.1097639. [DOI] [PubMed] [Google Scholar]
- Ramachandran N, Raphael JV, Hainsworth E, Demirkan G, Fuentes MG, Rolfs A, Hu Y, LaBaer J. Next-generation high-density self-assembling functional protein arrays. Nat Methods. 2008b;5:535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson WH, Steinman L, Utz PJ. Protein arrays for autoantibody profiling and fine-specificity mapping. Proteomics. 2003;3:2077–2084. doi: 10.1002/pmic.200300583. [DOI] [PubMed] [Google Scholar]
- Schmetzer O, Moldenhauer G, Riesenberg R, Pires JR, Schlag P, Pezzutto A. Quality of recombinant protein determines the amount of autoreactivity detected against the tumor-associated epithelial cell adhesion molecule antigen: low frequency of antibodies against the natural protein. J Immunol. 2005;174:942–952. doi: 10.4049/jimmunol.174.2.942. [DOI] [PubMed] [Google Scholar]
- Schwenk JM, Gry M, Rimini R, Uhlen M, Nilsson P. Antibody suspension bead arrays within serum proteomics. J Proteome Res. 2008;7:3168–3179. doi: 10.1021/pr700890b. [DOI] [PubMed] [Google Scholar]
- Verkaik N, Brouwer E, Hooijkaas H, van Belkum A, van Wamel W. Comparison of carboxylated and Penta-His microspheres for semi-quantitative measurement of antibody responses to His-tagged proteins. J Immunol Methods. 2008;335:121–125. doi: 10.1016/j.jim.2008.02.022. [DOI] [PubMed] [Google Scholar]
- Waterboer T, Sehr P, Michael KM, Franceschi S, Nieland JD, Joos TO, Templin MF, Pawlita M. Multiplex human papillomavirus serology based on in situ-purified glutathione s-transferase fusion proteins. Clin Chem. 2005;51:1845–1853. doi: 10.1373/clinchem.2005.052381. [DOI] [PubMed] [Google Scholar]