To the Editor
Although there is only one human genome sequence, different genes are expressed in many different cell types and tissues, as well as in different developmental stages or diseases. The structure of this ‘expression space’ is still largely unknown, as most transcriptomics experiments focus on sampling small regions. We have constructed a global gene expression map by integrating microarray data from 5,372 human samples representing 369 different cell and tissue types, disease states and cell lines. These have been compiled in an online resource (http://www.ebi.ac.uk/gxa/array/U133A) that allows the user to search for a gene of interest and find the conditions in which it is over- or underexpressed, or, conversely, to find which genes are over- or underexpressed in a particular condition. An analysis of the structure of the expression space reveals that it can be described by a small number of distinct expression profile classes and that the first three principal components of this space have biological interpretations. The hematopoietic system, solid tissues and incompletely differentiated cell types are arranged on the first principal axis; cell lines, neoplastic samples and nonneoplastic primary tissue–derived samples are on the second principal axis; and nervous system is separated from the rest of the samples on the third axis. We also show below that most cell lines cluster together rather than with their tissues of origin.
The widely used GNF Gene Expression Atlas1,2 includes a variety of normal tissue and cell types as well as certain disease states. Many more different biological states, such as rare diseases or particular cell subtypes, exist. It is impractical for a single dedicated experiment to generate a comprehensive expression data set covering all biological conditions, partly owing to cost, but also because some conditions are studied only in specialized laboratories. Even so, we can use computational approaches to integrate the wealth of experiments that already have been performed.
Integration of independent microarray studies is challenging, as microarrays do not measure gene expression in any absolute units. Several studies have integrated single-platform3 and cross platform4-6 data from single-channel oligonucleotide arrays yielding consistent results. It has been generally accepted, however, that only data from the same platform can be reliably integrated on a quantitative level7. Integration is also challenging because of the unavoidable complexity of sample descriptions. The Unified Medical Language System has been used to re-annotate free text-based sample descriptions8; however, extracting information from published data sets and representing it suitably for statistical analysis is a time-consuming process that is difficult to automate and requires expert curation9.
We collected over 9,000 raw data files generated on the human gene expression array Affymetrix U133A from the public databases Gene Expression Omnibus10 and ArrayExpress11. After we removed duplicate files and applied strict quality controls (Supplementary Methods), data on 5,372 samples from 206 different studies generated in 163 different laboratories remained. Using text mining and curation, we binned the samples in 369 biological groups, each representing a particular cell or tissue type, disease state or cell line (Supplementary Fig. 1a). Of these, 96 groups contained at least ten biological replicates. We also introduced ‘meta-groups’ such as cell lines, neoplasms, non-neoplastic diseases, and normal, as well as groups by tissue of origin (Supplementary Figs. 1c–e). The raw data were normalized jointly, producing a gene expression matrix of ~22,000 probe sets (mapping to ~14,000 genes) times 5,372 samples (the complete annotated data set is available from the ArrayExpress repository, accession number E-MTAB-62).
To enable exploration of these data, we have implemented an online query interface (http://www.ebi.ac.uk/gxa/array/U133A). After selecting a particular sample binning (e.g., by tissue of origin), the user can find all genes up- or downregulated in a particular sample class (such as liver). Alternatively, choosing a gene of interest will produce box plots showing the gene's expression across the samples within each of the groups. The coloring of each box plot indicates the outcome of a statistical test for over- or underexpression. Probe set–level queries are also permitted.
As these data were generated in different laboratories, and as laboratory effects are known to be strong12, it is important to assess the impact of these effects on the analysis. Most laboratories predominantly work with particular types of samples, which makes the lab effects hard to assess. Even so, 51 of the 96 larger biological groups (with ten replicates or more) contain assays from at least two different laboratories. In total, 100 different laboratories contributed 3,133 samples to these multi-laboratory biological groups. For each of these biological groups, we computed the average similarity between the assays from different laboratories within the same group. We also computed the average similarity between assays from the same laboratory, but representing different biological groups. The comparison of the two similarity distributions showed that the biological effects were significantly (P < 2.2–16) stronger than the laboratory effects (Supplementary Fig. 2). For sample classes to which only one laboratory contributed, we cannot distinguish directly between the laboratory and biological effects. However, we can analyze our data from a biological perspective and compare the results to existing knowledge.
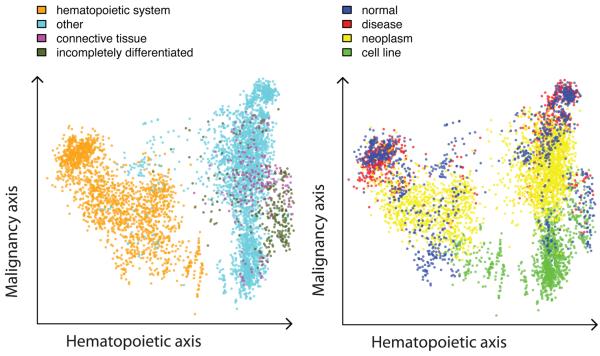
We applied principal component analysis (PCA) to the expression matrix, and produced visualizations in which each sample was represented by a point in the plane formed by two principal axes, and colors were assigned to each point according to the biological class (Fig. 1 and Supplementary Fig. 3a–e). We found that the first three principal components have biological interpretations; we named them the hematopoietic, malignancy and neurological axes. Three groups—hematopoietic system, solid tissues and a mixture of incompletely differentiated cell types and connective tissues—were consecutively arranged on the hematopoietic axis. The malignancy axis differentiates three other groups: cell lines, neoplasms and a mixture of normal tissues and non-neoplastic disease tissues. The neurological axis separates nervous system from other samples. The fourth principal component correlates with an array quality metric RLE (relative log expression). The first three principal components explain ~37% of variability in the data (Supplementary Fig. 3f). Note that the full expression space consists of thousands of dimensions.
Figure 1.
Principal component analysis. Each dot represents one of the 5,372 samples in a multidimensional gene expression space projected on the principal plane formed by the first (hematopoietic) and second (malignancy) principal axes. The dots are colored semitransparently according to the biological group the sample belongs to. (a) The first principal component separates hematopoietic system–derived samples from the rest of the samples, with connective tissues and incompletely differentiated cell–based samples forming a relatively compact group on the right. The cyan dots among the blood samples on the right side represent samples from bronchoalveolar lavage cells (a possible sample contamination with blood) and kidney. The dark green dots at the center include embryonic stem cells. (b) The second principal axis predominantly arranges cell line samples at the bottom, neoplasm samples in the middle and a mixture of nonneoplastic disease and normal samples at the top.
We also used hierarchical clustering to investigate the expression space from a different perspective. We first clustered the 96 larger biological groups (with ≥10 replicates), representing each group by its mean expression profile. Six major clusters emerged: (i) cell lines derived from solid tissues, (ii) incompletely differentiated cell types and connective tissues, (iii) solid normal and neoplastic tissues, (iv) hematopoietic system, (v) brain, and (vi) muscle and heart (Supplementary Fig. 4a). This clustering is robust: we obtained similar results when samples from different laboratories were kept in separate groups (Supplementary Fig. 4b) and by clustering all 369 sample groups (Supplementary Fig. 4c). To see how each of the 273 smaller groups relates to the six original clusters, we computed the pairwise distances between the members of the 96 and 273 groups and applied hierarchical clustering (Supplementary Fig. 4d). The smaller-group clusters correspond well to the six original clusters, although an additional small cluster of liver and small-intestine samples emerged. This analysis is driven by the original clustering; nevertheless, if there were new major expression pattern groups, we would expect to observe them. We conclude that the large-scale structure of our data can be explained by six major sample expression profile groups, corresponding to transcriptional states, and some smaller outliers.
Various observations can be made by examining the sample annotations in more detail. For instance, skeletal and heart muscle cluster together, whereas smooth muscle belongs to the incompletely differentiated cell type cluster, which is dominated by fibroblasts. This cluster includes bonemarrow mesenchymal stem cells, but not the hematopoietic bone-marrow stem cells, which are located in the hematopoietic cluster together with other blood-cell precursors. The embryonic stem cell line (HES2; ref. 13) does not belong to the cluster of incompletely differentiated cell types; its expression profile is similar to those of both fibroblasts and neoplastic cell lines.
Next, we studied which genes are expressed in various biological conditions. We applied hierarchical clustering to gene expression profiles across the 96 larger groups, representing the expression of a gene in each group by its mean. We visualized the 1,000 most variable probe sets mapping to 907 different genes and visually identified 50 gene clusters (Supplementary Fig. 5a). As our data set represents a wide range of biological conditions, we can study the overall variability of gene expression. For the majority of genes, the normalized signal is largely constant across the 5,372 samples; there are only 1,034 probe sets with a standard deviation >2 (Supplementary Fig. 6a,b). The sample clustering obtained using only the 350 most variable probe sets produced similar results to that based on all data and is retained to some extent even when only the 30 most variable probe sets are used (Supplementary Figs. 4e and 5b). Although it is not surprising that only a small number of genes are needed to define six transcriptional states, it is worth noting that the highest expression variance can identify these genes.
To identify genes differentially expressed in specific biological groups, we performed one-way analysis of variance (Supplementary methods). For instance, we found 243 genes differentially expressed in 567 samples grouped under ‘leukemia’. Many of these are known to be implicated in leukemia (for example, BCR, ETV6, FLT3, HOXA9, MYST3, PRDM2, RUNX1 and TAL1), and we confirmed many others through literature searches. Similarly, 1,217 genes are differentially expressed in all cell lines: the upregulated genes are most overrepresented in gene ontology categories related to M phase, cell division, mitosis, cell cycle and primary metabolic processes, and downregulated genes are most overrepresented in immune and defense response.
Our study demonstrates that analysis of a large microarray data set compiled from many laboratories can reveal the overall structure of gene expression space, which could not be observed in any of the contributing studies individually. A particularly important finding is that solid-tissue cell lines form a distinct group, clustering with each other rather than with their respective tissues of origin (Supplementary Figs. 4a,i). Moreover, they show high similarity to blood cell lines. An exception to this rule is incompletely differentiated cell types, for which cell lines cluster with the primary cells. Note that on the PCA's malignancy axis, neoplasm samples are located between the cell line and the normal and non-neoplastic disease samples, characterizing neoplasm as an intermediate state between normal samples and immortalized cell lines.
When interpreting these results, several limitations concerning the data set must be taken into account. First, there may be gaps in our data; for instance, there are few normal solid-tissue samples besides muscle, heart and brain. More data may reveal other major transcriptional classes. Second, it is possible that the laboratory effects are too strong to achieve resolution beyond the six major classes. Although the PCA shows samples from more specific groups (such as leukemia) located together (Supplementary Fig. 3c), and supervised analysis reveals that genes specific to such sample classes are often known to be involved in the relevant biological conditions, the results of hierarchical clustering did not conclusively reveal finer structures.
To summarize, we have constructed a global map of human gene expression from a large microarray data set. Our analysis reveals six major ‘continents’ on the map. We acknowledge that there may be more continents that we were not able to find owing to incompleteness of the data, and it is to be expected that finer structures exist within the six we found.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Adler, J. Bahler, E. Birney, D. Brazma, I. Dunham, N. Gehlenborg, F. Holstege, S. Kaski, M. Krestyaninova, J. Rung, G. Rustici, T. Schlitt and H. Zang-Bradley for helpful comments and discussion. This research was partly funded by FELICS (021902) and EMERALD (LSHG-CT-2006-037689) grants from the European Commission, a MAGE grant from the National Human Genome Research Institute and National Institutes of Biomedical Imaging and Bioengineering from the National Institute of Health (1 P41 HG003619-01), an ALGODAN CoE grant of the Academy of Finland, and grant no. 40274/06 from Finnish Funding agency for Technology and Innovation (TEKES).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Su AI, et al. Proc. Natl. Acad. Sci. USA. 2002;99:4465–4470. doi: 10.1073/pnas.012025199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Su AI, et al. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day A, Carlson MR, Dong J, O'Connor BD, Nelson SF. Genome Biol. 2007;8:R112. doi: 10.1186/gb-2007-8-6-r112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kilpinen S, et al. Genome Biol. 2008;9:R139. doi: 10.1186/gb-2008-9-9-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapushesky M, et al. Nucleic Acids Res. 2010;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morgan AA, Dudley JT, Deshpande T, Butte AJ. Physiol. Genomics. 2009;40:128–140. doi: 10.1152/physiolgenomics.90403.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi L, et al. Nat. Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Butte AJ, Kohane IS. Nat. Biotechnol. 2006;24:55–62. doi: 10.1038/nbt1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malone J, et al. Bioinformatics. 2010 published online 3 March 2010, doi:10.1093/bioinformatics/btq099. [Google Scholar]
- 10.Barrett T, et al. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parkinson H, et al. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zilliox MJ, Irizarry RA. Nat. Methods. 2007;4:911–913. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hirst CE, et al. Dev. Biol. 2006;293:90–103. doi: 10.1016/j.ydbio.2006.01.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.