Summary
Aberrant methylation in the promoter region of cancer-related genes leads to gene transcriptional inactivation and plays an integral role in lung tumorigenesis. Recent studies demonstrated that promoter methylation was detected not only in lung tumors from patients with lung cancer but also in sputum of smokers without the disease, suggesting the potential for aberrant gene promoter methylation in sputum as a predictive marker for lung cancer. In the present study, we investigated promoter methylation of 4 genes frequently detected in lung tumors, including p16, MGMT, RASSF1A and DAPK genes, in sputum samples obtained from 107 individuals, including 34 never-smoking females and 73 mostly smoking males, who had no evidence of lung cancer but who were exposed to smoky coal emission in Xuan Wei County, China, where lung cancer rate is more than 6 times the Chinese national average rate. Forty nine of the individuals showed evidence of chronic bronchitis while the remaining 58 individuals showed no such a symptom. Promoter methylation of p16, MGMT, RASSF1A and DAPK was detected in 51.4% (55/107), 17.8% (19/107), 29.9% (32/107), and 15.9% (17/107) of the sputum samples from these individuals, respectively. There were no differences in promoter methylation frequencies of any of these genes according to smoking status or gender of the subjects or between individuals with chronic bronchitis and those without evidence of such a symptom. Therefore, individuals exposed to smoky coal emissions in this region harbored in their sputum frequent promoter methylation of these genes that have been previously found in lung tumors and implicated in lung cancer development.
Keywords: Smoky coal emissions, Gene promoter methylation, Lung cancer
Introduction
Aberrant promoter methylation of tumor suppressor genes is an important mechanism of gene transcriptional inactivation and has been associated with the development of many kinds of cancers [1], including lung cancer, the most common cause of cancer death worldwide. Although much attention has been paid to understand the molecular and cellular mechanisms of lung cancer, the 5-year overall survival rate for all stages combined is only 15% [2], due primarily to the presence of metastatic tumors in approximately two-third of patients at the time of diagnosis [3]. Detection of lung cancer at earlier stages could potentially increase survival rates by 10–50 folds [4]. Recently, gene promoter methylation has become a target for the development of screening methods for early detection, diagnosis, and treatment of lung cancer [5, 6]. Results from several studies have indeed suggested the potential for gene promoter methylation in sputum as a predictive marker for lung cancer [7–11].
Most studies of gene promoter methylation in sputum involved so far lung cancer patients or smokers from Europe and the United States [7–11]. In Xuan Wei County (XWC), Yunnan Province, China, lung cancer rates for women, who were mostly nonsmokers, and for men, who were mostly smokers, were eight times and four times the Chinese national average rates for women and men, respectively. Several studies demonstrated a strong association between the high lung cancer rate in this region and the use of smoky coal, a low sulfur, medium volatility bituminous coal, for cooking and heating in homes without chimneys [12–14]. These emissions contained a high level of polycyclic aromatic hydrocarbons (PAHs), among which methylated PAHs have higher tumorigenic potency than the parent PAHs [15]. PAHs in XWC smoky coal emission are more carcinogenic than cigarette smoke in a murine skin-tumor assay [16]. Furthermore, these emissions have been associated with high frequencies of p53 and K-ras mutations in lung tumors and in sputum from lung cancer patients from XWC [17, 18]. However, the effects of exposure to smoky coal emissions on epigenetic alterations, specifically gene promoter methylation, in this population remain unclear.
Promoter methylation of the p16, MGMT, RASSF1A, and DAPK genes has been commonly found in lung tumors and implicated in different pathways of lung tumorigenesis, including cell cycle regulation, DNA repair, signal transduction and apoptosis, respectively [19–23]. In the present study, we examined aberrant promoter methylation of these genes in sputum samples obtained from 107 individuals who were exposed to smoky coal emissions in XWC, and who showed no evidence of lung cancer but were at high risk for developing the disease. We analyzed the results in relation to the smoking status, gender, and the presence or absence of symptoms of chronic bronchitis in these individuals.
Methods
Subject enrollment and sputum collection and processing
All sputum samples were collected from XWC, China. Individuals who donated sputum samples analyzed in this study were part of the subjects involved in a previous study [13]. These individuals showed a minimum of clinical symptoms and chest X-ray analysis at the Xuan Wei Hospital and were found with no evidence of lung cancer. Each individual who provided informed consent to participate in this study also answered a standardized closed questionnaire on demographic information, smoking history, family and personal medical history, as well as information on other variables. For the protection of human subjects, this study was conducted according to recommendations of the World Medical Association Declaration of Helsinki (1989) [24]. The research protocol met the requirements for protection of human subject certification by the US EPA.
The demographic and clinical information of the 107 individuals involved in this study is shown in Table 1. All the male subjects (n = 73) with the exception of one subject are smokers and all the female subjects are nonsmokers. Of these individuals, 49 had symptoms of chronic bronchitis, with excessive bronchial mucus and a chronic cough for three months or more in at least three consecutive years and without any other disease that could account for these symptoms, and 58 had no such symptoms.
Table 1.
Demographic and clinical information of individuals exposed to smoky coal emission in Xuan Wei, China (n=107)
| Variables | Cases (%) |
|---|---|
| Sex | |
| Male | 73 (68.2%) |
| Female | 34 (31.8%) |
| Age (mean ± s.d.) | 57.2 ± 10.8 |
| Smoking status | |
| Smoking | 72 (67.3%) |
| Non-smoking | 35 (32.7%) |
| Bronchitis status | |
| Bronchitis | 49 (45.8%) |
| Non-bronchitis | 58 (54.2%) |
Sputum was collected first-morning on five consecutive mornings. Each subject was instructed to rinse his/her mouth with water to remove extraneous material, to take a deep breath, and cough deeply and expectorate into a plastic cup. Each morning sputum sample was stored in 40 ml of Saccomanno’s fluid (39% ethanol, 3% polyoxyethylene, and 2% isopropanol; Lerner Laboratories, Pittsburgh, PA) to fix and preserve the cells. The sputum samples were stored at 4°C and transported to the U.S. by air. To collect cells, each sputum sample in Saccomanno’s fluid was blended for 8–15 seconds in a blender to break the mucus and free the cells. The sample was then centrifuged at 600-g for 10 minutes. The supernatant was discarded and the cell pellet was resuspended in fresh Saccomanno’s fluid by vortexing to achieve a cell concentration of approximately 106 cells per ml. The cells were subjected to cytological examination using the method described by Saccomanno in order to determine whether the sputum samples were derived from the lower respiratory tracts and also to confirm the presence of bronchial epithelial cells.
DNA extraction and promoter methylation analysis
Genomic DNA was extracted from each sputum sample, using the method combining Proteinase digestion and phenol-chloroform extraction, and recovered by ethanol precipitation and suspended in deionized water.
Each genomic DNA sample was treated with sodium bisulfite (Sigma, Saint Louis, MO) and purified by using a Wizard DNA Clean-Up System (Promega Corporation, Madison, WI), as described previously [25]. Universal methylated human genomic DNA (Chemicon International, Temecula, CA) was treated the same way and was used as a positive control DNA, while water was used as a negative control.
Methylation-Specific Polymerase Chain Reaction (MSP)
Two-step MSP was used for analysis of all 4 genes promoter methylation. The methylation status of the p16 and MGMT were determined by the methods as described before [25]. The methylation status of the RASSF1A and DAPK were determined by using the primers and a protocol modified from that described by Belinsky et al [21]. During the stage-I PCR, the PCR amplification was carried out in a 25-µl reaction mixture containing 10 mM Tris-HCL (pH 8.3), 50 mM KCL, 2 mM MgCl2, 100 µM each dNTP, and 0.2 µM each primer. The reaction was heated at 95°C for 10 min., then amplified for 40 cycles [95°C/30 sec., 64°C (for RASSF1A) or 58°C (for DAPK) /30 sec., and 72°C/30 sec.], followed by a 10-min.-final extension at 72°C. Three microliters of each stage-I PCR product was diluted 10-fold and 1-µl was used for stage-II PCR, using the same reagents and conditions as for stage-I, except that the MgCl2 concentration was reduced to 1 mM and each sample was amplified in duplicated reaction, with one reaction containing primers specific for a methylated C and the other reaction containing primers specific for unmethylated C. Each reaction was heated at 95°C for 10 min., then amplified for 40 cycles each consisting, for the reaction containing methylated primers, of 95°C/30 sec., 68°C (for RASSF1A) or 66°C (for DAPK)/30 sec., and 72°C/30 sec., and, for the reaction containing unmethylated primers, of 95°C /30 sec., 64 °C (for RASSF1A) or 68°C (for DAPK)/30 sec., and 72°C/30 sec., followed by a 10-min.-final extension at 72°C. Five microliters of each stage-II PCR product was separated on an 8% polyacrylamide gel. The gel was stained with ethidium bromide and photographed under UV illumination. Promoter methylation of p16, MGMT and DAPK were further confirmed by using digestion of the resulting PCR products with the restriction enzymes FNU4HI, TaqI, and BstUI, respectively. Promoter methylation of RASSF1A was confirmed by direct sequence.
Statistical analysis
Chi-square test was used in univariate analysis. Logistic regression models were used to assess the effect of multiple variables on methylation status.
Results
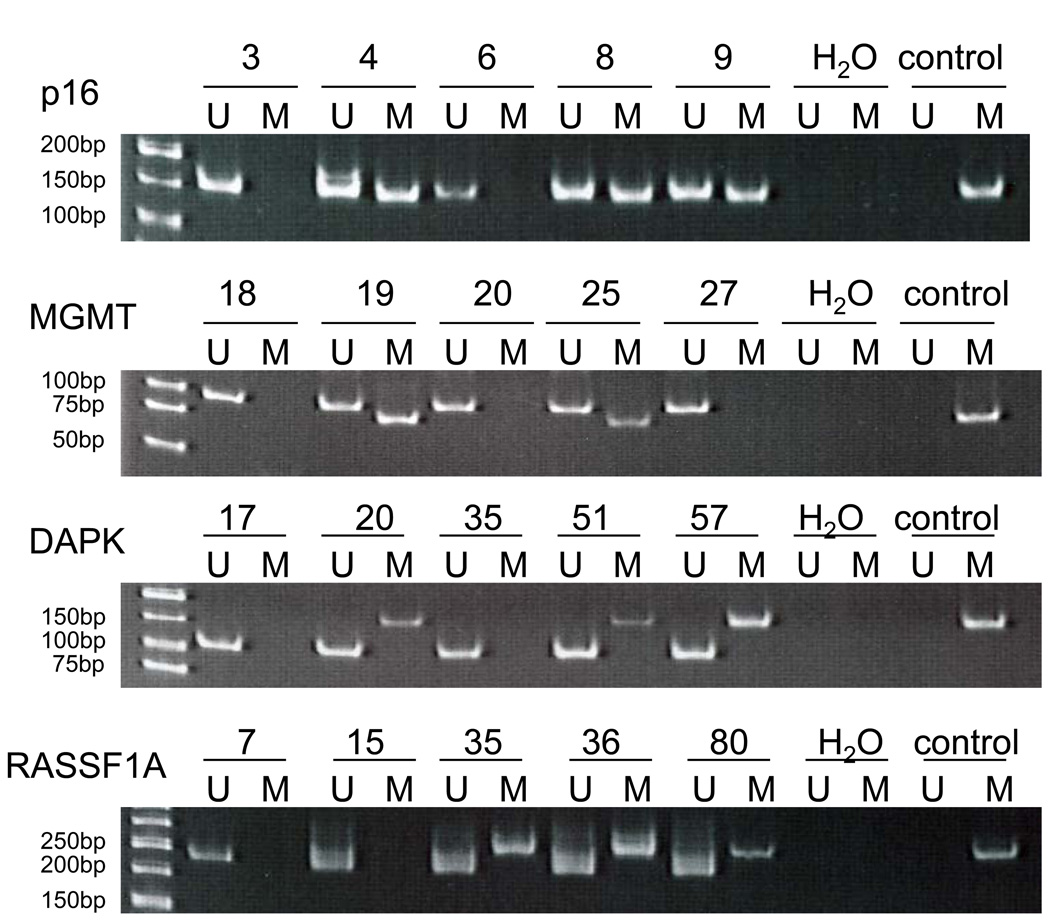
Figure 1 shows a representative example of MSP analysis of sputum DNA. Of the 107 individuals involved in this study, p16, MGMT, RASSF1A and DAPK promoter methylation was detected in 51.4% (55/107), 17.8% (19/107), 29.9% (32/107), 15.9% (17/107), respectively. Seventy-three (68.2%) of the 107 individuals showed promoter methylation of at least one of the genes, including 3 (2.8%), 8 (7.5%), 21 (19.6%), and 41 (38.3%) individuals showing the alteration in all 4 genes, 3 genes, 2, genes, and 1 gene, respectively.
Figure 1.
Detection of p16, MGMT, RASSF1A and DAPK promoter methylation by MSP. M indicates the presence of methylated p16, MGMT, RASSF1A or DAPK. U indicates the presence of unmethylated p16, MGMT, RASSF1A or MGMT. All samples were performed twice and representative data are shown.
As shown in Table 2, there were no differences in promoter methylation frequencies between the smoking group and nonsmoking group for either the p16 (50.0% vs. 54.3%, p = 0.677), MGMT (18.1% vs. 17.1%, p = 0.908), RASSF1A (31.9% vs. 25.7%, p = 0.509), or DAPK (18.3% vs. 11.4%, p = 0.364) gene, or between the group of individuals with chronic bronchitis and the group of those without this symptom (51.0% vs. 51.7% for p16, p = 0.938; 14.3% vs. 20.7% for MGMT, p = 0.377; 30.6% vs. 29.3% for RASSF1A, p = 0.899; 14.3% vs. 17.2% for DAPK, p = 0.710).
Table 2.
Promoter methylation of p16, MGMT, RASSF1A and DAPK according to gender, smoking and bronchitis status in Xuan Wei, China
| P16 | MGMT | RASSF1A | DAPK | |
|---|---|---|---|---|
| Gender | ||||
| Male (73) | 49.3% (36/73) | 17.8% (13/73) | 31.5% (23/73) | 18.1% (13/72) |
| Female (34) | 55.9% (19/34) | 17.6% (6/34) | 26.5% (9/34) | 11.8% (4/34) |
| Bronchitis status | ||||
| Bronchitis (49) | 51.0% (25/49) | 14.3% (7/49) | 30.6% (15/49) | 14.6% (7/48) |
| Non-bronchitis (58) | 51.7% (30/58) | 20.7% (12/58) | 29.3% (17/58) | 17.2% (10/58) |
| Smoking status | ||||
| Smoking M (72) | 50.0% (36/72) | 18.1% (13/72) | 31.9% (23/72) | 18.3% (13/71) |
| Nonsmoking (35) | 54.3% (19/35) | 17.1% (6/35) | 25.7% (9/35) | 11.4 % (4/35) |
Multivariate logistic regression models were employed to control for potential confounding effects of variables such as gender, age, smoking status and bronchitis. As shown in Table 3, only age showed significant effect on DAPK methylation status (odds ratio (OR) = 1.072; 95% confidence interval (CI) = 1.008 – 1.141, p = 0.027, table 3), while these variables did not have any effect on the promoter methylation status of the other 3 genes.
Table 3.
Logistic Regression Models of p16, MGMT, RASSF1A and DAPK Promoter Methylation
| P16 | MGMT | RASSF1A | DAPK | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P | |
| Sex | 0.646 | (0.133–3.135) | 0.588 | 0.822 | (0.093–7.243) | 0.859 | 0.983 | (0.170–5.688) | 0.985 | 1.130 | (0.085–14.999) | 0.926 |
| Age | 0.999 | (0.963–1.036) | 0.960 | 1.031 | (0.980–1.085) | 0.234 | 1.012 | (0.971–1.054) | 0.583 | 1.072 | (1.008–1.141) | 0.027 |
| Smoking | 1.219 | (0.255–5.835) | 0.804 | 1.230 | (0.140–10.812) | 0.852 | 1.355 | (0.236–7.790) | 0.734 | 1.537 | (0.115–20.454) | 0.745 |
| Bronchitis | 0.954 | (0.432–2.107) | 0.908 | 0.540 | (0.187–1.559) | 0.255 | 0.999 | (0.421–2.369) | 0.998 | 0.588 | (0.193–1.792) | 0.350 |
Discussion
In this study, we demonstrated that gene promoter methylation occurred frequently in DNA extracted from sputum of individuals exposed to smoky coal emissions in XWC, with a frequency varying from 51.4% (55/107) for p16 gene, to 29.9% (32/107) for RASSF1A gene, 17.8% (19/107) for DAPK gene, and 15.9% (17/107) for MGMT gene. Furthermore, 3 individuals (2.8%) showed the co-occurrence of promoter methylation of all four genes, while 8 (7.5%), 21 (19.6%), and 41 (38.3%) other individuals showed this alteration in 3 genes, 2 genes, and 1 gene, in their sputum, respectively. The clinical implication of the co-occurrence of promoter methylation on lung cancer risk remains unclear since these subjects were anonymous and were not followed up further. However, results from previous studies of smokers from Europe and the United States suggested that gene promoter methylation in sputum may provide a useful predictive biomarker for lung cancer. For example, Palmisano et al reported that aberrant methylation of the p16 and MGMT was detected in sputum of all patients with squamous cell lung carcinoma up to 3 years before clinical diagnosis. Moreover, the prevalence of these markers in sputum from cancer-free, high-risk subjects approximates lifetime risk for lung cancer [7]. A study by Kersting et al of p16 gene promoter methylation, and p53 and K-ras mutations in exfoliated cells from 51 NSCLC patients and 25 chronic smokers showed that 8 of the chronic smokers harbored a genetic and/or epigenetic alteration, and 3 of whom were subsequently diagnosed with lung cancer [8]. Two recent studies, both from the Belinsky’s group, showed that the co-occurrence of promoter methylation of three genes and of more than 3 genes in sputum was associated with a 3.6- and 6.5-fold increased risk of lung cancer, respectively [10, 11]. Therefore, individuals with sputum positive for aberrant promoter methylation, especially those showing such an alteration in multiples genes, may be at high risk for lung cancer.
In this study, the gene promoter methylation frequencies found in sputum of the XWC population are higher than those reported previously in sputum from smokers from Europe and the United States [7, 8, 26, 27]. For instance, Destro et al reported that only 4 cases (4%) among 100 heavy smokers (age>60, >20 cigarettes/day last at least 20 years) showed p16 promoter methylation [26]. This discrepancy was unlikely attributable to technical problems because similar detection methods were used in both our present study and these studies. Ethnic differences might play a factor to account for this difference. It has been reported that there is a relationship between gene methylation and geography [28] and the Chinese lung cancer patients harboured higher frequency of gene hypermethylation in their tumor tissues, plasma and sputum [29] and bronchoaleolar lavage than Western cases [30]. Furthermore, in this study, there were no differences in promoter methylation frequencies for any of the genes in sputum between the nonsmokers and the smokers, by using either univariate (table 2) or logistic (table 3) analysis. This result is in disagreement with the significantly higher promoter methylation frequencies observed for p16 and RASSF1A genes in sputum samples from smokers, compared with nonsmokers [31]. Taken together, the higher promoter methylation frequencies found in sputum of the XWC individuals, compared with those found in sputum of the European and American smokers, and the similar promoter methylation frequencies between smokers, mostly men, and nonsmokers, mostly women, in the XWC population, may be due to the exposure of the XWC subjects to smoky coal combustion. These emissions contained 81% of organic matter, of which 43% were PAHs [32] and were previously associated with the detection of a higher level of benzo(a)pyrene-adducted guanine in urine from nonsmokers exposed smoky coal emissions, compared with urine from smokers not exposed to these emissions [33]. Furthermore, exposure to these emissions has been associated with high frequencies of p53 and K-ras mutations in lung tumors from lung cancer patients [17, 18] and in sputum from individuals who showed no evidence of lung cancer [34] in XWC.
Finally, there were no differences in the frequencies of promoter methylation in any of the genes between the groups of individuals diagnosed with chronic bronchitis and the group of those without such a symptom, by using either univariate (table 2) or logistic (table 3) analysis. This result is in line with our previous study of these same sputum samples showing no differences in p53 mutation frequencies between these two groups of individuals [34]. These results suggests that promoter methylation of these genes, like p53 mutations, was associated primarily with exposure to smoky coal emissions.
Taken together, these results suggest that chemicals in the smoky coal emissions, particularly the high concentration of PAHs and other chemicals [35], may play a primary role in the formation of genetic and epigenetic alterations found in sputum from individuals without evidence of lung cancer in XWC.
Conclusion
In this study, we investigated gene promoter methylation in sputum of 107 individuals who had no evidence of lung cancer but were at high risk for developing lung cancer because of their history of long-term exposure to smoky coal emissions in XWC. We found that promoter methylation of these genes was relatively frequent in this population. This alteration was not associated with the smoking status, gender, or chronic bronchitis diagnosis of the subjects, indicating a dominant role of chemicals present in smoky coal emissions in the formation of promoter methylation in sputum of these individuals. Weaknesses of this study include the relatively small number of subjects and the lack of individuals not exposed to smoky coal emissions who might serve as a control group and that fact that these individuals were not followed up. Nevertheless, the results of this study suggest that detection of epigenetic alterations, such as aberrant promoter methylation of these genes or additional genes, in sputum from a larger number of followed up individuals in XWC who had been exposed to smoky coal emissions may provide a useful means for early detection of lung cancer.
Acknowledgement
This research has been reviewed by US EPA and approved for publication. Approval does not signify that the contents reflect views of the Agency or endorse the trade names mentioned.
Reference
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Cancer facts and figures. 2004 http://www.cancer.org/
- 3.Ellis JR, Gleeson FV. Lung cancer screening. Br J Radiol. 2001;74:478–485. doi: 10.1259/bjr.74.882.740478. [DOI] [PubMed] [Google Scholar]
- 4.Wingo PA, Ries LA, Giovino GA, Miller DS, Rosenberg HM, Shopland DR, Thun MJ, Edwards BK. Annual report to the nation on the status of cancer, 1973–1996, with a special section on lung cancer and tobacco smoking. J Natl Cancer Inst. 1999;91:675–690. doi: 10.1093/jnci/91.8.675. [DOI] [PubMed] [Google Scholar]
- 5.Belinsky SA. Gene-promoter hypermethylation as a biomarker in lung cancer. Nat Rev Cancer. 2004;4:707–717. doi: 10.1038/nrc1432. [DOI] [PubMed] [Google Scholar]
- 6.Belinsky SA. Silencing of genes by promoter hypermethylation: key event in rodent and human lung cancer. Carcinogenesis. 2005;26:1481–1487. doi: 10.1093/carcin/bgi020. [DOI] [PubMed] [Google Scholar]
- 7.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 8.Kersting M, Friedl C, Kraus A, Behn M, Pankow W, Schuermann M. Differential frequencies of p16(INK4a) promoter hypermethylation, p53 mutation, and K-ras mutation in exfoliative material mark the development of lung cancer in symptomatic chronic smokers. J Clin Oncol. 2000;18:3221–3229. doi: 10.1200/JCO.2000.18.18.3221. [DOI] [PubMed] [Google Scholar]
- 9.Honorio S, Agathangglelou A, Schuermann M, Pankow W, Viacava P, Maher ER, Latif F. Detection of RASSF1A aberrant promoter hypermethylation in sputum from chronic smokers and ductal carcinoma in situ from breast cancer patients. Oncogene. 2003;22:147–150. doi: 10.1038/sj.onc.1206057. [DOI] [PubMed] [Google Scholar]
- 10.Belinsky SA, Klinge DM, Dekker JD, Smith MW, Bocklage TJ, Gilliland FD, Crowell RE, Karp DD, Stidley CA, Picchi MA. Gene promoter methylation in plasma and sputum increases with lung cancer risk. Clin Cancer Res. 2005;11:6505–6511. doi: 10.1158/1078-0432.CCR-05-0625. [DOI] [PubMed] [Google Scholar]
- 11.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, Franklin WA, Herman JG, Baylin SB, Bunn PA, Byers T. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 12.Mumford JL, Li X, Hu F, Lu XB, Chuang JC. Human exposure and dosimetry of polycyclic aromatic hydrocarbons in urine from Xuan Wei, China with high lung cancer mortality associated with exposure to unvented coal smoke. Carcinogenesis. 1995;16:3031–3036. doi: 10.1093/carcin/16.12.3031. [DOI] [PubMed] [Google Scholar]
- 13.Lan Q, He X, Costa L, Rothman N, Hu G, Mumford Indoor coal combustion emissions, GSTM1 and GSTT1 genotypes, and lung cancer risk: a case-control study in Xuan Wei, China. Cancer Epidemiol Biomarkers Prev. 2000;9:605–608. [PubMed] [Google Scholar]
- 14.Lan Q, Chapman RS, Schreinemachers DM, Tian L, He X. Household stove improvement and risk of lung cancer in Xuanwei, China. J Natl Cancer Inst. 2002;94:826–835. doi: 10.1093/jnci/94.11.826. [DOI] [PubMed] [Google Scholar]
- 15.Chuang JC, Cao SR, Xian L, Harris DB, Mumford JL. Chemical characterization of indoor air of homes from communes in Xuan wei, China with high lung cancer mortality rate. Atmos Environ. 1992;26A:2193–2201. [Google Scholar]
- 16.Mumford JL, Helmes CT, Lee XM, Seidenberg J, Nesnow S. Mouse skin tumorigenicity studies of indoor coal and wood combustion emissions from homes of residents in Xuan Wei, China with high lung cancer mortality. Carcinogenesis. 1990;11:397–403. doi: 10.1093/carcin/11.3.397. [DOI] [PubMed] [Google Scholar]
- 17.DeMarini DM, Landi S, Tian D, Hanley NM, Li X, Hu P, Roop BC, Mass MJ, Keohavong P, Gao W, Oliver M, Hainaut P, Mumford JL. Lung tumor KRAS and TP53 mutations in nonsmokers reflect exposure to PAH-rich coal combustion emissions. Cancer Res. 2001;61:6679–6681. [PubMed] [Google Scholar]
- 18.Keohavong P, Lan Q, Gao WM, DeMarini DM, Mass MJ, Li XM, Roop BC, Weifeld J, Tian D, Mumford JD. K-ras mutations in lung carcinomas from nonsmoking women exposed to unvented coal smoke in China. Lung Cancer. 2003;41:21–27. doi: 10.1016/s0169-5002(03)00125-9. [DOI] [PubMed] [Google Scholar]
- 19.Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci USA. 1998;95:11891–11896. doi: 10.1073/pnas.95.20.11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dammann R, Li C, Yoon JH, Chin PL, Bates S, Pfeifer GP. Epigenetic inactivation of a RAS association domain family protein from the lung tumour suppressor locus 3p21.3. Nat Genet. 2000;25:315–319. doi: 10.1038/77083. [DOI] [PubMed] [Google Scholar]
- 21.Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, Crowell RE. Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res. 2002;62:2370–2377. [PubMed] [Google Scholar]
- 22.Pulling LC, Divine KK, Klinge DM, Gilliland FD, Kang T, Schwartz AG, Bocklage TJ, Belinsky SA. Promoter hypermethylation of the O6-methylguanine-DNA methyltransferase gene: more common in lung adenocarcinomas from never-smokers than smokers and associated with tumor progression. Cancer Res. 2003;63:4842–4848. [PubMed] [Google Scholar]
- 23.Russo AL, Thiagalingam A, Pan H, Califano J, Cheng JF, Chinnappan D, Nemani P, Sidransky D, Thiagalingam S. Differential DNA hypermethylation of critical genes mediates the stage-specific tobacco smoke-induced neoplastic progression of lung cancer. Clin Cancer Res. 2005;11:2466–2470. doi: 10.1158/1078-0432.CCR-04-1962. [DOI] [PubMed] [Google Scholar]
- 24.World Medical Association. Hong Kong: Forty-first World Medical Assembly; Helsinki Declaration. 1989
- 25.Liu Y, Lan Q, Siegfried JM, Luketich JD, Keohavong P. Aberrant Promoter Methylation of p16 and MGMT Genes in Lung Tumors from Smoking and Never-Smoking Lung Cancer Patients. Neoplasia. 2006;8:46–51. doi: 10.1593/neo.05586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Destro A, Bianchi P, Alloisio M, Laghi L, Di Gioia S, Malesci A, Carboni U, Gribaudi G, Bulfamante G, Marchetti A, Bosari S, Infante M, Ravasi G, Roncalli M. K-ras and p16(INK4A)alterations in sputum of NSCLC patients and in heavy asymptomatic chronic smokers. Lung Cancer. 2004;44:23–32. doi: 10.1016/j.lungcan.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Cirincione R, Lintas C, Conte D, Mariani L, Roz L, Vignola AM, Pastorino U, Sozzi G. Methylation profile in tumor and sputum samples of lung cancer patients detected by spiral computed tomography: a nested case-control study. Int J Cancer. 2006;118:1248–1253. doi: 10.1002/ijc.21473. [DOI] [PubMed] [Google Scholar]
- 28.Toyooka S, Maruyama R, Toyooka KO, Mclerran D, Feng Z, Fukuyama Y, Virmani AK, Zochauer-Muller S, Tsukuda K, Sugio K, Shimizu N, Shimizu K, Lee H, Chen CY, Fong KM, Gilcrease M, Roth JA, Minna JD, Gazdar AF. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103:153–160. doi: 10.1002/ijc.10787. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, An Q, Li L, Zhang D, Huang J, Feng X, Cheng S, Gao Y. Hypermethylation of p16INK4a in Chinese lung cancer patients: biological and clinical implications. Carcinogenesis. 2003;24:1897–1901. doi: 10.1093/carcin/bgg169. [DOI] [PubMed] [Google Scholar]
- 30.Chan EC, Lam SY, Tsang KW, Lam B, Ho JC, Fu KH, Lam WK, Kwong YL. Aberrant promoter methylation in Chinese patients with non-small cell lung cancer: patterns in primary tumors and potential diagnostic application in bronchoalevolar lavage. Clin Cancer Res. 2002;8:3741–3746. [PubMed] [Google Scholar]
- 31.Zochbauer-Muller S, Lam S, Toyooka S, Virmani AK, Toyooka KO, Seidl S, Minna JD, Gazdar AF. Aberrant methylation of multiple genes in the upper aerodigestive tract epithelium of heavy smokers. Int J Cancer. 2003;107:612–616. doi: 10.1002/ijc.11458. [DOI] [PubMed] [Google Scholar]
- 32.Granville CA, Hanley NM, Mumford JL, DeMarini DM. Mutation spectra of smoky coal combustion emissions in Salmonella reflect the TP53 and KRAS mutations in lung tumors from smoky coal-exposed individuals. Mutat Res. 2003;525:77–83. doi: 10.1016/s0027-5107(02)00314-7. [DOI] [PubMed] [Google Scholar]
- 33.Casale GP, Singhal M, Bhattacharya S, RamaNathan R, Roberts KP, Barbacci DC, Zhao J, Jankowiak R, Gross ML, Cavalieri EL, Small GJ, Rennard SI, Mumford JL, Shen M. Detection and quantification of depurinated benzo[a]pyrene-adducted DNA bases in the urine of cigarette smokers and women exposed to household coal smoke. Chem Res Toxicol. 2001;14:192–201. doi: 10.1021/tx000012y. [DOI] [PubMed] [Google Scholar]
- 34.Keohavong P, Lan Q, Gao WM, Zheng KC, Mady HH, Melhem MF, Mumford JL. Detection of p53 and K-ras mutations in sputum of individuals exposed to smoky coal emissions in Xuan Wei County, China. Carcinogenesis. 2005;26:303–308. doi: 10.1093/carcin/bgh328. [DOI] [PubMed] [Google Scholar]
- 35.Mumford JL, He XZ, Chapman RS, Cao SR, Harris DB, Li XM, Xian YL, Jiang WZ, Xu CW, Chuang JC, Wilson WE, Cooke M. Lung cancer and indoor air pollution in Xuan Wei, China. Science. 1987;235:217–220. doi: 10.1126/science.3798109. [DOI] [PubMed] [Google Scholar]