Abstract
Purpose
To determine the proportion of lung adenocarcinomas from East Asian never-smokers who harbor known oncogenic driver mutations.
Patients and Methods
In this surgical series, 52 resected lung adenocarcinomas from never-smokers (< 100 cigarettes in a lifetime) at a single institution (Fudan University, Shanghai, China) were analyzed concurrently for mutations in EGFR, KRAS, NRAS, HRAS, HER2, BRAF, ALK, PIK3CA, TP53 and LKB1.
Results
Forty-one tumors harbored EGFR mutations, three harbored EML4-ALK fusions, two harbored HER2 insertions, and one harbored a KRAS mutation. All mutations were mutually exclusive. Thus, 90% (47 of 52; 95% CI, 0.7896 to 0.9625) of lung adenocarcinomas from never-smokers were found to harbor well-known oncogenic mutations in just four genes. No BRAF, NRAS, HRAS, or LKB1 mutations were detected, while 15 had TP53 mutations. Four tumors contained PIK3CA mutations, always together with EGFR mutations.
Conclusion
To our knowledge, this study represents the first comprehensive and concurrent analysis of major recurrent oncogenic mutations found in a large cohort of lung adenocarcinomas from East Asian never-smokers. Since drugs are now available that target mutant EGFR, HER2, and ALK, respectively, this result indicates that prospective mutation testing in these patients should successfully assign a targeted therapy in the majority of cases.
INTRODUCTION
Lung cancer is the leading cause of cancer-related deaths worldwide.1 Most cases are associated with a personal history of direct tobacco smoke exposure. However, approximately 10% to 15% of all lung cancers arise in never-smokers, defined as those who smoked fewer than 100 cigarettes in their lifetime.2 If considered as a separate category, lung cancer in never-smokers would rank among the most common causes of cancer mortality.3
The importance of never-smokers has emerged in lung cancer because of recent clinical observations. During the development of the epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs), gefitinib and erlotinib, retrospective studies showed that never-smoking status, along with adenocarcinoma histology and East Asian ethnicity, were associated with a higher likelihood of responding to treatment.4,5 Subsequently, EGFR kinase domain mutations (ie, deletions in exon 19 and L858R point mutations in exon 21) were found to be enriched in patients with these clinical features and to be highly associated with increased sensitivity to EGFR TKIs.6–8 Furthermore, lung cancers in patients with minimal and/or a remote history of direct tobacco exposure were found to share some molecular features with their never-smoking counterparts. For example, the likelihood of EGFR mutations decreases as the number of pack-years increases.9 Collectively, all of these observations led to a randomized phase III trial open-label study (Iressa Pan Asian Study [IPASS]), in which previously untreated patients in East Asia who had advanced pulmonary adenocarcinomas and who were never- smokers or former light-smokers (those who had stopped smoking at least 15 years previously and had a total of ≤ 10 pack-years of smoking) were randomly assigned to receive gefitinib or conventional chemotherapy with carboplatin plus paclitaxel. Patients with EGFR-mutant tumors experienced longer progression-free survival with gefitinib, and patients without mutations had longer progression-free survival with chemotherapy.10 Similar results have been observed with erlotinib in white never-smokers with lung adenocarcinoma.11
In IPASS, the objective radiographic response rate in all patients treated with gefitinib was 43%. In contrast, for patients with known EGFR mutations, the response rate was 71.2%. A similar high response rate (62.1%) to gefitinib was reported in another recent study, the WJTOG3405 trial, in which only patients with known EGFR tumor mutations were randomly assigned to EGFR TKI or chemotherapy.12 To explore the discrepancy between these results and to better characterize lung adenocarcinomas in never-smokers, we performed a comprehensive analysis of major known driver mutations in 52 East Asian patients from a single institution with resected pulmonary adenocarcinomas and who were never-smokers. Driver mutations occur in genes that encode signaling proteins critical for cellular proliferation and survival. In lung adenocarcinomas, such mutations include EGFR, HER2, KRAS, BRAF, PIK3CA, and EML4-ALK. We also examined tumors for mutations in other genes (ie, the oncogenes, HRAS and NRAS and the tumor suppressor genes TP53 and LKB1). Our results have clear therapeutic implications for never-smokers with lung cancer.
PATIENTS AND METHODS
Specimen Collection
This study was approved by the institutional review board of the Shanghai Cancer Hospital, Fudan University, Shanghai, China. All participants underwent surgery and provided written informed consent. Samples were snap-frozen in liquid nitrogen at the time of resection and stored at −80°C until use. All cases were rereviewed by pathologists for confirmation of tumor histology and tumor content. Patients were considered never-smokers in this study if they reported never smoking any cigarettes in their lifetimes.
Mutational Analyses
All mutational analyses were performed in China. Genomic DNA or RNA was extracted from lung tumors or distant histologically normal lung as per standard protocols (RNeasy Mini Kit, and QiAamp DNA Mini Kit, Qiagen, Hilden, Germany). Total RNA samples were reverse transcribed into single-stranded cDNA using RevertAid First Strand cDNA Synthesis Kit (Fermentas, St Leon-Rot, Germany). Either genomic DNA or cDNA were used for polymerase chain reaction (PCR) amplification and sequencing.
EGFR (exons 18-22), HER2 (exons 18 to 21), KRAS (exons 2 to 3), and BRAF (exons 11 to 15) were PCR amplified using cDNA for further sequencing. All exons of TP53 and LKB1, and exons 9 and 20 of PIK3CA, were sequenced using genomic DNA.
For detection of EML4-ALK fusions, primers were designed to amplify all known fusion variants using cDNA. The forward primers include EML4 E2F (5′-TGATGTTTTGAGGCGTCTTG−3′), EML4 E13F (5′-AGATCGCCTGTCAGCTCTTG−3′), EML4 E18F (5′-TTAGCATTCTTGGGGAATGG−3′), and the reverse primer ALK E20R (5′-TGCCAGCAAAGCAGTAGTTG−3′). Multiplex PCR analysis was done with KOD plus DNA polymerase (Toyobo, Osaka, Japan), with the following program: 94°C 5 minutes; 94°C 30 seconds, 60°C 30 seconds, 68°C 1 minute, 35 cycles; 68°C 10 minutes. PCR products were directly sequenced in forward and reverse directions. All mutations were verified by analysis of an independent PCR isolate.
Statistical Analysis
χ2 and Fisher's exact tests were used to analyze the association of mutations with clinical characteristics, using SPSS 16.0 (SPSS Institute, Chicago, IL). All P values were based on a two-sided hypothesis.
RESULTS
Assembly of Tumor Samples
From January 2008 to June 2009, we collected consecutively a total of 168 resected lung adenocarcinomas at the Shanghai Cancer Hospital, Fudan University, in Shanghai, China. All of the patients (100%) were Chinese. Of these, 52 cases were included in this study based on the following criteria: re-review confirmed a pathologic diagnosis of lung adenocarcinoma; tumor specimen contained a minimum of 70% tumor cells; enough tissue was available for comprehensive analysis; patient was a never-smoker; patient did not receive any neoadjuvant treatment; and corresponding normal tissue was also available for analysis. Eleven cases were from males, and 41 from females. Detailed clinical characteristics are listed in Table 1.
Table 1.
Clinical Characteristics of Never Smokers With Lung Adenocarcinoma (N = 52)
| Characteristic | Total | Sex |
P | |
|---|---|---|---|---|
| Male | Female | |||
| No. of patients | 52 | 11 | 41 | |
| Age, years | 59.04 | 60.73 | 58.59 | .491 |
| SD | 9.04 | 8.09 | 9.32 | |
| Clinical stage | ||||
| I | 24 | 2 | 20 | .205 |
| II | 6 | 3 | 5 | |
| III | 20 | 6 | 14 | |
| IV | 2 | 0 | 2 | |
| Differentiation | ||||
| Well | 13 | 2 | 11 | .113 |
| Moderate | 28 | 4 | 24 | |
| Poor | 11 | 5 | 6 | |
EGFR Mutation Status
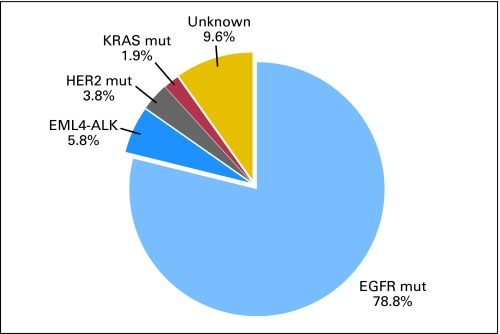
Seventy-eight point eight percent of tumors (41 of 52; 95% CI, 0.6580 to 0.8792) were found to harbor EGFR kinase domain mutations (Fig 1). Among these, 21 were deletions in exon 19 and 18 were L858R missense changes (Table 2). The remaining alterations included an exon 20 insertion and a double mutation involving exon 18 (G719S) and exon 21 (L861Q). We did not detect any T790M mutations, which are found more commonly in patients with acquired resistance to EGFR TKIs13 and rarely in never-smoking patients with lung cancer as a rare germline variant.14 Eighty-two point nine percent of tumors (34 of 41) from female never-smokers harbored an EGFR kinase domain mutation, while 63.6% of tumors (seven of 11) from male never-smokers did (Table 2).
Fig 1.
Oncogenic driver mutations in East Asian never-smokers with lung adenocarcinomas. In tumors from 52 patients, 78.8% (41 of 52) harbored EGFR kinase domain mutations, 5.8% (three of 52) harbored EML4-ALK fusions, 3.8% (two of 52) harbored HER2 mutations, and 1.9% (one of 52) harbored KRAS mutations. Only 9.6% (five of 52) of tumors did not harbor any of these known oncogenic driver mutations.
Table 2.
Association of EGFR Kinase Mutations and Clinical Characteristics in Lung Adenocarcinomas From Never-Smokers
| Characteristic |
EGFR Mutation |
||
|---|---|---|---|
| Total | Male | Female | |
| Patient No. | 41 | 7 | 34 |
| Age, years | 59.10 | 60.14 | 58.88 |
| SD | 9.58 | 10.19 | 9.58 |
| Clinical stage | |||
| I | 22 | 2 | 20 |
| II | 2 | 1 | 1 |
| III | 15 | 4 | 11 |
| IV | 2 | 0 | 2 |
| Differentiation | |||
| Well | 11 | 2 | 9 |
| Moderate | 23 | 2 | 21 |
| Poor | 7 | 3 | 4 |
| Mutation type | |||
| L858R | 18 | 4 | 14 |
| Exon 19 deletion | 21 | 3 | 18 |
| Other EGFR mutation | 2 | 0 | 2 |
Spectrum of Mutations in HER2, KRAS, BRAF,and EML4-ALK
Five point eight percent (three of 52; 95% CI, 0.0138 to 0.1625) of samples were found to harbor EML4-ALK fusions (Fig 1). The three variants included E13;A20, E20;A20, and E6a/b;A20.15 Three point eight percent (two of 52; 95% CI, 0.0032 to 0.1372) of samples had HER2 kinase domain mutations (Fig 1). One sample (1.9%; 95% CI, < 0.0001 to 0.1107) had a KRAS G12V mutation (Fig 1). No mutations were found in BRAF (95% CI, 0.0000 to 0.0822).
Strikingly, only 9.6% (five of 52; 95% CI, 0.0375 to 0.2104) of samples did not harbor any mutations in EGFR, HER2, KRAS, BRAF, or EML4-ALK (Fig 1). All five of these cases were from females. Among the 11 male never-smokers, seven had an EGFR mutation, one had a KRAS mutation, one had a HER2 insertion, and two had an EML4-ALK fusion. Among the 90.4% of tumors with known mutations, tumors with an alteration in one gene did not harbor a mutation in any of the other genes.
Other Mutations
Four samples (7.7%; 95% CI, 0.0253 to 0.1868) harbored PIK3CA mutations, two occurring in exon 9, and two in exon 20 (Table 3). All the PIK3CA mutations coexisted with EGFR mutations (Table 3). Three were in females. We also searched for mutations in the RAS-related genes, HRAS and NRAS, and in the tumor suppressor genes, LKB1 and TP53. No mutations were found in HRAS, NRAS, or LKB1. Fifteen samples had a TP53 mutation.
Table 3.
Association of PIK3CA Mutations With EGFR Kinase Domain Mutations in Lung Adenocarcinomas From Never-Smokers
| No. | Sex | Age (years) | Stage | PIK3CA Mutation | EGFR Mutation |
|---|---|---|---|---|---|
| 168 | Female | 43 | IIIA | E545K | E746-A750del |
| 202 | Female | 51 | IA | H1047R | L747-S752del P753S |
| 338 | Male | 56 | IIIA | H1047R | L858R |
| 411 | Female | 57 | IA | E545K | L747-S752del P753S |
DISCUSSION
During the past decade, a wealth of data from genomic,16 expression,17 mutational,18 and proteomic profiling studies19 have led to the identification of multiple molecularly distinct subsets of lung cancer. Based on such discoveries, one of the most promising treatment strategies now involves the subdivision of non–small-cell lung cancer histologies into clinically relevant molecular subsets, using a classification schema based on specific driver mutations. Major recurrent mutations in lung adenocarcinoma have been found to occur in EGFR, KRAS, HER2, BRAF, ALK, and PIK3CA.
To our knowledge, this study represents the first comprehensive and concurrent analysis of these known recurrent driver mutations in a large cohort of lung adenocarcinomas from never-smokers. Previous studies have examined lung adenocarcinomas from never-smokers for multiple mutations, but did not include ALK fusions, which had just been reported.18,20 Other studies on ALK fusions have examined tumors for EGFR and KRAS mutations, but not other driver mutations.21,22 Many studies have examined lung adenocarcinomas from never-smokers and smokers, but this study focused only on lung cancers from patients who never smoked cigarettes.
The main finding of this study is that the majority of lung adenocarcinomas from East Asian never-smokers can be defined molecularly by targetable oncogenic mutant kinases. Eighty-eight percent of the 52 samples examined in this study were found to harbor well-known oncogenic alterations in EGFR, HER2, or ALK. Conversely, only approximately 10% of tumors did not have an identifiable mutation, including in BRAF, HRAS, or NRAS. Erlotinib/gefitinib are already available to target mutant EGFR; a recent clinical trial showed that the EGFR/HER2 inhibitor, BIBW2992, induced partial responses in three of three patients with HER2 mutant lung tumors;23 and the ALK inhibitor, crizotinib (PF-02341066), has demonstrated remarkable efficacy against ALK fusion–positive lung cancers.24 Thus, our molecular data in conjunction with the emerging clinical data indicates that prospective genotyping of lung adenocarcinomas from never-smokers for these genetic alterations could lead to rationally chosen targeted therapy in the overwhelming majority of cases.
PIK3CA mutations have been reported to be less common in lung adenocarcinomas than squamous cell carcinomas.25–27 Interestingly, we found PIK3CA mutations in 7.7% (four of 52) of lung adenocarcinomas from never-smokers, which is higher than previous reports.25–27 PIK3CA alterations were also more commonly found in younger patients. Since the total number of cases was low, in future studies, we plan to clarify in additional samples if these observations remain constant.
Otherwise, our data confirm and extend a number of other published observations. For example, we found only 1 KRAS and no LKB1 mutations; these are rare in never-smokers, especially from East Asia.28,29 The TP53 mutation rate in this group was 28.8%, similar to previous studies.30 We also found a high proportion of tumors to harbor EGFR mutations, the majority of which included deletions in exon 19 and the L858R point mutation. Among EGFR wild-type tumors (n = 11), three (28%) harbored EML4-ALK fusions. Consistent with this, others recently reported that 33% of adenocarcinomas negative for EGFR mutations were positive for ALK fusions.21
Our results can help explain the radiographic response rates observed with gefitinib in the recent IPASS trial. In all never/light smokers in that study, the collective response rate to the EGFR TKI was only 43%. In our study of only never-smokers, 41 of 52 patients had EGFR kinase domain mutations. However, among the 41 mutated cases, one harbored an exon 20 insertion mutation, which has been associated with drug resistance.31 Another four tumors had concurrent PIK3CA mutations, which have been associated with resistance to EGFR TKIs at least in vitro.32 Thus, only 36 (69%) of the 52 tumors would be predicted to be sensitive to TKI treatment. Since EGFR mutant tumors in general display a 70% response rate to EGFR TKIs, 70% of 69% would result in a 48% true response rate, which is very close to the response rate seen in IPASS.
We are now actively trying to identify the driver mutations in the five tumors with no detectable gene alterations. It is possible that they harbor ALK fusions that do not involve EML4; for example, recently, lung tumors have been found to harbor ALK alterations including KIF5B-ALK.33 However, we thus far have no evidence of other fusion events involving ALK in these tumors. Using Affymetrix HuEX-1.0 exon array analysis (Affymetrix, Santa Clara, CA), these samples did not display disparate levels of expression between exons at the 5′ and 3′ ends of ALK (data not shown), which is suggestive of fusions events.34 By contrast, the three samples with EML4-ALK fusions did. Other candidate drivers include ROS fusion or PDGFRα amplification;19 however, our preliminary analysis of both exon array and single nucleotide polymorphism array data have not found evidence for these types of events. More comprehensive genomic analysis and deep sequencing may need to be performed on these pan–wild-type samples, so that the disease is defined completely at the molecular level.
Acknowledgment
We thank Wenjing Zhang, Xiaolei Ye, and Zuoyun Wang for technical support.
Footnotes
Supported by Grant No. 2010CB912102 from the National Basic Research Program of China; Grants No. 30623003, 30740084, and 30871284 from the National Natural Science Foundation of China; Grants No. 2008KIP101 and KSCX1-YW-22 from the Chinese Academy of Sciences; and Grants No. 08PJ14105 and 09JC1416300 from the Science and Technology Commission of Shanghai Municipality. H.J. is a scholar of the Hundred Talents Program of the Chinese Academy of Sciences; W.P. received funding from the National Cancer Institute (Grant No. R01 CA121210), the Specialized Program of Research Excellence in Lung Cancer (Grant No. CA90949), and the Vanderbilt-Ingram Cancer Center Core (Grant No. CA68485).
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
Although all authors completed the disclosure declaration, the following author(s) indicated a financial or other interest that is relevant to the subject matter under consideration in this article. Certain relationships marked with a “U” are those for which no compensation was received; those relationships marked with a “C” were compensated. For a detailed description of the disclosure categories, or for more information about ASCO's conflict of interest policy, please refer to the Author Disclosure Declaration and the Disclosures of Potential Conflicts of Interest section in Information for Contributors.
Employment or Leadership Position: None Consultant or Advisory Role: William Pao, MolecularMD (C), AstraZeneca (C), Symphony Evolution (C), Bristol-Myers Squibb (C) Stock Ownership: None Honoraria: None Research Funding: William Pao, Xcovery Expert Testimony: None Other Remuneration: William Pao, EGFR T790M patent licensed to MolecularMD
AUTHOR CONTRIBUTIONS
Conception and design: William Pao, Haiquan Chen, Hongbin Ji
Financial support: Hongbin Ji
Administrative support: Haiquan Chen
Provision of study materials or patients: Yihua Sun, Chenguang Li, Haiquan Chen
Collection and assembly of data: Yihua Sun, Yan Ren, Rong Fang, Bin Gao, Xiangkun Han, Hongbin Ji
Data analysis and interpretation: Yihua Sun, Yan Ren, Zhaoyuan Fang, Weidong Tian, William Pao, Haiquan Chen, Hongbin Ji
Manuscript writing: Yihua Sun, Yan Ren, Zhaoyuan Fang, Chenguang Li, Rong Fang, Bin Gao, Xiangkun Han, Weidong Tian, William Pao, Haiquan Chen, Hongbin Ji
Final approval of manuscript: Yihua Sun, Yan Ren, Zhaoyuan Fang, Chenguang Li, Rong Fang, Bin Gao, Xiangkun Han, Weidong Tian, William Pao, Haiquan Chen, Hongbin Ji
REFERENCES
- 1.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–249. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Samet JM. Adverse effects of smoke exposure on the upper airway. Tob Control. 2004;13(suppl 1):i57–60. doi: 10.1136/tc.2003.005454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Samet JM, Avila-Tang E, Boffetta P, et al. Lung cancer in never smokers: Clinical epidemiology and environmental risk factors. Clin Cancer Res. 2009;15:5626–5645. doi: 10.1158/1078-0432.CCR-09-0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fukuoka M, Yano S, Giaccone G, et al. Multi-institutional randomized phase II trial of gefitinib for previously treated patients with advanced non-small-cell lung cancer (The IDEAL 1 trial) J Clin Oncol. 2003;21:2237–2246. doi: 10.1200/JCO.2003.10.038. [DOI] [PubMed] [Google Scholar]
- 5.Miller VA, Kris MG, Shah N, et al. Bronchioloalveolar pathologic subtype and smoking history predict sensitivity to gefitinib in advanced non-small-cell lung cancer. J Clin Oncol. 2004;22:1103–1109. doi: 10.1200/JCO.2004.08.158. [DOI] [PubMed] [Google Scholar]
- 6.Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Paez JG, Janne PA, Lee JC, et al. EGFR mutations in lung cancer: Correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 8.Pao W, Miller VA. Epidermal growth factor receptor mutations, small-molecule kinase inhibitors, and non-small-cell lung cancer: Current knowledge and future directions. J Clin Oncol. 2005;23:2556–2568. doi: 10.1200/JCO.2005.07.799. [DOI] [PubMed] [Google Scholar]
- 9.Pham D, Kris MG, Riely GJ, et al. Use of cigarette-smoking history to estimate the likelihood of mutations in epidermal growth factor receptor gene exons 19 and 21 in lung adenocarcinomas. J Clin Oncol. 2006;24:1700–1704. doi: 10.1200/JCO.2005.04.3224. [DOI] [PubMed] [Google Scholar]
- 10.Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin-paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947–957. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 11.Rosell R, Moran T, Queralt C, et al. Screening for epidermal growth factor receptor mutations in lung cancer. N Engl J Med. 2009;361:958–967. doi: 10.1056/NEJMoa0904554. [DOI] [PubMed] [Google Scholar]
- 12.Mitsudomi T, Morita S, Yatabe Y, et al. Gefitinib versus cisplatin plus docetaxel in patients with non-small-cell lung cancer harbouring mutations of the epidermal growth factor receptor (WJTOG3405): An open label, randomised phase 3 trial. Lancet Oncol. 2010;11:121–128. doi: 10.1016/S1470-2045(09)70364-X. [DOI] [PubMed] [Google Scholar]
- 13.Pao W, Miller VA, Politi KA, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girard N, Lou E, Azzoli CG, et al. Analysis of genetic variants in never-smokers with lung cancer facilitated by an Internet-based blood collection protocol: A preliminary report. Clin Cancer Res. 2010;16:755–763. doi: 10.1158/1078-0432.CCR-09-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horn L, Pao W. EML4-ALK: Honing in on a new target in non-small-cell lung cancer. J Clin Oncol. 2009;27:4232–4235. doi: 10.1200/JCO.2009.23.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shedden K, Taylor JM, Enkemann SA, et al. Gene expression-based survival prediction in lung adenocarcinoma: A multi-site, blinded validation study. Nat Med. 2008;14:822–827. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding L, Getz G, Wheeler DA, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rikova K, Guo A, Zeng Q, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 20.Chitale D, Gong Y, Taylor BS, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28:2773–2783. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaw AT, Yeap BY, Mino-Kenudson M, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–4253. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soda M, Choi YL, Enomoto M, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–566. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 23.De Grève J, Teugels E, De Mey J, et al. Clinical activity of BIBW 2992, an irreversible inhibitor of EGFR and HER2 in adenocarcinoma of the lung with mutations in the kinase domain of HER2neu. J Thorac Oncol. 2009;4:S307. abstr A6.7. [Google Scholar]
- 24.Shaw AT, Costa DB, Iafrate AJ, et al. Clinical activity of the oral ALK and MET inhibitor PF-02341066 in non-small lung cancer (NSCLC) with EML4-ALK translocations. Presented at the 13th World Congress on Lung Cancer; July 31-August 4, 2009; San Francisco, CA. [Google Scholar]
- 25.Yamamoto H, Shigematsu H, Nomura M, et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68:6913–6921. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawano O, Sasaki H, Endo K, et al. PIK3CA mutation status in Japanese lung cancer patients. Lung Cancer. 2006;54:209–215. doi: 10.1016/j.lungcan.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 27.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304:554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 28.Wu CC, Hsu HY, Liu HP, et al. Reversed mutation rates of KRAS and EGFR genes in adenocarcinoma of the lung in Taiwan and their implications. Cancer. 2008;113:3199–3208. doi: 10.1002/cncr.23925. [DOI] [PubMed] [Google Scholar]
- 29.Koivunen JP, Kim J, Lee J, et al. Mutations in the LKB1 tumour suppressor are frequently detected in tumours from Caucasian but not Asian lung cancer patients. Br J Cancer. 2008;99:245–252. doi: 10.1038/sj.bjc.6604469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448:807–810. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 31.Wu JY, Wu SG, Yang CH, et al. Lung cancer with epidermal growth factor receptor exon 20 mutations is associated with poor gefitinib treatment response. Clin Cancer Res. 2008;14:4877–4882. doi: 10.1158/1078-0432.CCR-07-5123. [DOI] [PubMed] [Google Scholar]
- 32.Engelman JA, Mukohara T, Zejnullahu K, et al. Allelic dilution obscures detection of a biologically significant resistance mutation in EGFR-amplified lung cancer. J Clin Invest. 2006;116:2695–2706. doi: 10.1172/JCI28656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takeuchi K, Choi YL, Togashi Y, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–3149. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 34.Lin E, Li L, Guan Y, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7:1466–1476. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]