Abstract
To initiate homologous recombination, sequence similarity between two DNA molecules must be searched for and homology recognized. How the search for and recognition of homology occurs remains unproven. We have examined the influences of DNA topology and the polarity of RecA–single-stranded (ss)DNA filaments on the formation of synaptic complexes promoted by RecA. Using two complementary methods and various ssDNA and duplex DNA molecules as substrates, we demonstrate that topological constraints on a small circular RecA–ssDNA filament prevent it from interwinding with its duplex DNA target at the homologous region. We were unable to detect homologous pairing between a circular RecA–ssDNA filament and its relaxed or supercoiled circular duplex DNA targets. However, the formation of synaptic complexes between an invading linear RecA–ssDNA filament and covalently closed circular duplex DNAs is promoted by supercoiling of the duplex DNA. The results imply that a triplex structure formed by non-Watson–Crick hydrogen bonding is unlikely to be an intermediate in homology searching promoted by RecA. Rather, a model in which RecA-mediated homology searching requires unwinding of the duplex DNA coupled with local strand exchange is the likely mechanism. Furthermore, we show that polarity of the invading RecA–ssDNA does not affect its ability to pair and interwind with its circular target duplex DNA.
INTRODUCTION
The essential steps in homologous recombination are homologous pairing of two DNA chromosomes and exchange of DNA between them. Most of our knowledge about homologous recombination comes from studies of this process in Escherichia coli. The recombination events begin with a linear duplex DNA generated by double-strand breakage (or conjugation) that is either unwound by a helicase or degraded by a nuclease to produce a single-stranded (ss)DNA tail. RecA then polymerizes onto the ssDNA region to form a helical nucleoprotein filament, the RecA–ssDNA filament then searches for its homologous duplex DNA target. Once homology has been found, subsequent exchange of the individual DNA strands leads to heteroduplex DNA products (reviewed in 1–8).
RecA, a 38 kDa polypeptide, performs a critical role in homologous recombination in E.coli. RecA binds to ssDNA to form a nucleoprotein filament that has been studied by 3D image reconstruction and by X-ray crystallography (9–11). The nucleoprotein filaments have a pitch of 95 Å with six RecA monomers per helical turn (9,10). A eukaryotic protein homologous to RecA, ScRAD51, has been shown to be essential in recombination and DNA damage repair in Saccharomyces cerevisiae (12). The human counterpart is termed human Rad51 (hRad51). Electron microscopy shows that both the yeast and human proteins form nucleoprotein filaments similar to those formed by RecA and in vitro experiments show that both proteins promote strand exchange (12–14). Thus, evidence suggests that the overall mechanism of recombination involving these strand exchange proteins has been conserved.
The E.coli RecA system provides a valuable paradigm for the study of the mechanisms of homology searching and strand exchange. ssDNA coated with RecA searching for its homologous sequence buried in a non-RecA-coated duplex DNA initiates the homologous recombination process. Many biochemical methods (electron microscopy, filter binding, chemical probing and gel electrophoresis) have been used to study the intermediates and products of the strand exchange reaction promoted by RecA (reviewed in 1–8). However, it is still not clear how the RecA–ssDNA filament searches for and recognizes its homologous sequence in a duplex DNA to form a synaptic complex. There are two extreme models for initial recognition of homology. In the first model homologous DNA within the target duplex is locally unwound so that ssDNA in the RecA nucleoprotein filament can test for homology by forming Watson–Crick base pairs with the unwound segments (15–20). Homology searching would therefore involve multiple random encounters of the invading ssDNA–RecA filament with the duplex, followed by local melting of the duplex DNA and pairing with the invading ssDNA at each encounter site. In principle a second model is possible, with an intermediate in which the bases within the duplex target and ssDNA are held in registry; this alignment could be produced by non-Watson–Crick hydrogen bonds between the ssDNAs and double-stranded (ds)DNAs within the RecA filament (17,19,21). Thus, recognition of homology would occur via these non-Watson–Crick hydrogen bonds and the two strands of the target duplex DNA would remain paired by conventional Watson–Crick hydrogen bonds. This searching method might avoid the kinetic barriers associated with repeatedly breaking Watson–Crick hydrogen bonds in non-homologous duplex DNA regions before the homologous target is reached (22–25).
In this paper we perform experiments aimed at deducing the influences of topology and the polarity of the RecA–ssDNA in formation of the synaptic complex (homologously paired joint molecule promoted by RecA), which might shed light on distinguishing between the two models described above. We have used a topological method and a restriction enzyme protection method with various linear or circular ssDNAs that were designed to limit the topological freedom to interwind with their target duplex DNA substrates in a RecA-mediated homologous pairing reaction. We demonstrate that, unlike its linear counterpart, a RecA-coated 520mer circular ssDNA can neither unwind nor form a stable joint molecule with a target circular 3 kb duplex DNA having a 120 bp region of homology with the ssDNA. We also show that supercoiling of the target duplex DNA enhances the efficiency of homologous pairing promoted by a linear, but not circular, RecA–ssDNA filament (with 120 nt homologous to its target duplex DNA) and that the polarity of the invading linear RecA–ssDNA has no effect on the efficiency of interwinding. The results suggest that topological freedom plays an important role in formation of stable synaptic complexes and that the RecA-mediated homology searching process is very likely coupled to unwinding of the dsDNA. Therefore, a triplex structure formed at the homologous region by non-Watson–Crick hydrogen bonds is unlikely to be an intermediate during the homologous searching and pairing processes.
MATERIALS AND METHODS
RecA protein
RecA was purified from E.coli strain JC11272 (a generous gift from Prof. A. John Clark of University of California–Berkeley) following a protocol based on spermidine precipitation (26). It was >99% pure as judged by 7.5% SDS–PAGE. Activity of the purified protein was confirmed by performing a standard strand exchange reaction with circular ssDNA and homologous linear duplex DNA as substrates to generate circular duplex DNA and linear ssDNA. The RecA concentration was determined by the Bradford method (27).
Topoisomerase I
Wheatgerm topoisomerase I (Topo I) was a generous gift from Prof. Nicholas R. Cozzarelli of University of California (Berkeley, CA).
DNA substrates
Duplex DNA. Plasmid pBluescript KS+ (3 kb) from Invitrogen was purified in a large-scale preparation according to a standard protocol (27). Linear pBluescript DNA was prepared by digesting the supercoiled plasmid with the restriction endonuclease XhoI. Duplex pBluescript KS+ DNA molecules with different degrees of superhelical densities were generated using wheatgerm Topo I in the presence of 0–4 µg/ml ethidium bromide. The reactions contained (in 1 ml), 20 mM Tris–HCl pH 7.5, 300 mM KCl, 5 mM EDTA, 50 µg/ml bovine serum albumin (BSA), 25 µg/ml DNA, 50 U Topo I and ethidium bromide. After 30 min at 30°C the mixture was extracted with phenol and precipitated with ethanol.
ssDNA. The following oligonucleotides were purchased from The Great American Gene Co. and subsequently purified from 15% polyacrylamide denaturing gels containing 48% (w/v) urea. H-60, 5′-ccgta agatg ctttt ctgtg actgg tgagt actca accaa gtcat tctga gaata gtgta-3′. This molecule was 60 nt long and was completely homologous to pBluescript KS+ in the region that flanks the unique ScaI site (underlined) of the plasmid. H-120, 5′-cactg cataa ttctc ttact gtcat gccat ccgta agatg ctttt ctgtg actgg tgagt actca accaa gtcat tctga gaata gtgta tgcgg cgacc gagtt gctct tgccc ggcgt-3′. This 120mer molecule was produced through ligation of two 60mers, LL and LR, with a linker L-30. H-120 was also completely homologous to the region flanking the unique ScaI site on pBluescript KS+. The sequences of LL, LR and L-30 are: LL, 5′-cactg cataa ttctc ttact gtcat gccat ccgta agatg ctttt ctgtg actgg tgagt-3′; LR, 5′-actca accaa gtcat tctga gaata gtgta tgcgg cgacc gagtt gctct tgccc ggcgt-3′; L-30, 5′-tgact tggtt gagta ctcac cagtc acaga-3′. Aliquots of 5 µg of purified LL and L-30 were added to 5 µg 5′-kinased LR in a volume of 60 µl with 1× TAEMg (40 mM Tris–acetate, 2 mM EDTA, 10 mM Mg acetate). The sample was then heated to 95°C for 3 min and cooled slowly to room temperature. LL and LR anneal with L-30 during the cooling process. Subsequently, 8 µl of 10× ligase buffer and 5 µl of 400 U/ml T4 DNA ligase (New England Biolabs) were added to the sample. The ligation reaction was allowed to proceed overnight at room temperature. The 120mer ligation product, H-120, was then purified by 15% PAGE with 48% urea at 60°C. The bands were visualized by staining with ethidium bromide. The band corresponding to 120mer was cut out and eluted from the gel fragment. Het-124, a 124mer oligonucleotide that is completely heterologous to pBluescript KS+ DNA, is composed of two oligos, 80-A and 44-B. These two fragments were ligated together using a 20 bp linker, L-20B. 44-B was first phosphorylated by T4 polynucleotide kinase at the 5′-end. Then, 80-A and L-20B were added to the sample. Annealing and ligation reactions were carried out as described above. The sequences of 80-A, 44-B and L-20B are: 80A, 5′-cttgc tcata ctgca ttcgg ccagc ctgac atcac cgtgt acgcc caaac ctttc aactt agatg gtaga aggag ggcag-3′; 44B, 5′-ccagg acgct gcata gtatt gcgcc gattt gtcaa tcacc caag-3′; L-20B, 5′-agcgt cctgg ctgcc ctcct-3′. C-520, a 520mer circular ssDNA with only 120 of the 520 nt (the homologous sequence is the same as H-120) homologous to pBluescript KS+. A 412 bp fragment from φX174 was amplified by PCR using φX-primer 1 and φX-primer 2. A 131 bp fragment from pBluescript KS+ was amplified by PCR using 120-primer 1 and 120-primer 2. The two PCR fragments were then phosphorylated and ligated. Using the above ligation product as a template and φX-primer 1 and 120-primer 2 as primers a 543 bp fragment was amplified. The 543 bp fragment was then digested with XhoI and PstI to produce a 520 bp fragment, which contains the 400 bp fragment 163–562 of φX174 at the 5′-end and the 120 bp fragment 2467–2526 of pBluescript KS+ at the 3′-end. This 520 bp fragment was amplified by PCR using φXP1-30bp and 120P2-30bp as primers. The single-stranded 520mer was amplified by one-primer PCR using φXP1-30bp as the primer and the 520 bp dsDNA, described above, as template. The amplified single-stranded 520mer was annealed to a 60mer oligonucleotide (Cir-60), which was complementary to 30 bp of the 5′-end and 30 bp of the 3′-end of the 520mer. The annealed product was treated with Klenow fragment, then phosphorylated and ligated. The ligation product was then subjected to 6% denaturing acrylamide gel electrophoresis; the resulting 520mer circular ssDNA was purified accordingly. φX-primer 1 (151–172 of φX174), 5′-tccttgcgcagctcgagaagct-3′; φX-primer 2 (complementary to 562–541 of φX174), 5′-caagcctcaacgcagcgacgag-3′; 120-primer 1 (2467–2488 of pBluescript KS+), 5′-cactgcataattctcttactgt-3′; 120-primer 2 (partially complementary to 2579–2586 of pBluescript KS+, with the non-homologous region designed to introduce a PstI site at the 3′-end), 5′-gtatgctgcagacgccgggcaa-3′; φXP1-30bp (5′-end of 520 bp chimeric DNA), 5′-tccttgcgcagctcgagaagctcttacttt-3′; 120P2-30bp (3′-end of 520 bp chimeric DNA), 5′-ttattctgcagacgccgggcaagagcaact-3′; Cir-60 (60mers designed as a linker for circularization of single-stranded 520mers), 5′-aaagt aagag cttct cgagc tgcgc aagga ttatt ctgca gacgc cgggc aagag caact-3′.
RecA-promoted synaptic complex formation
The standard synaptic complex reaction with a 5:1 molar ratio of RecA–ssDNA filament to duplex DNA was formed as follows. An aliquot of 0.053 µg ssDNA (80 nt, 2 pmol) was incubated with 2 µg RecA protein (one RecA per 3 nt, 53 pmol) in a total volume of 70 µl in RecA reaction buffer (∼28 nM ssDNA), containing 33 mM HEPES pH 7.2, 15 mM Mg(OAc)2, 2 mM DTT, 1.2 mM ADP, 0.3 mM ATPγS, 0.1 mg/ml BSA. Where indicated, ATPγS was replaced with ATP regeneration buffer containing 33 mM Tris–acetate pH 7.5, 12 mM Mg(OAc)2, 2 mM DTT, 1.2 mM ATP, 8 mM phosphocreatine, 10 U/ml creatine phosphokinase, 100 µg/ml BSA. After incubation for 30 min at 37°C to form the presynaptic filament, 0.8 µg duplex DNA (pBluescript, 3 kb, 0.4 pmol) in 80 µl of the same buffer was then added to the reaction mixture to initiate the homology searching reaction, i.e. formation of synaptic complexes (∼13 nM ssDNA and ∼2.6 nM duplex DNA).
Linking number change assay
Synaptic complexes were formed in 45 µl of ATPγS or ATP regenerating buffer as described above. After 30 min incubation the samples were treated with 30 U wheatgerm Topo I. After incubation for another 5 min at 37°C the reaction was stopped by phenol deproteinization. Samples were then desalted by gel filtration through Chromaspin-10 columns (Clontech). The topoisomers were separated by electrophoresis at 2.5 V/cm in a 1.5% agarose gel in 40 mM Tris–acetate, 2 mM EDTA buffer plus 2 mg/ml chloroquine for 24 h at room temperature with buffer circulation. The gel was then subjected to Southern blotting, probed with 32P-labeled probes generated by random primer extension methods with single-stranded pBluescript KS+ as the template. The autoradiographs were analyzed using the Molecular Analyst Computer Imaging System (Bio-Rad) and the centers of the distributions for individual sets of topoisomers were determined.
Restriction enzyme digestion assay
The synaptic complex formation reaction described above was performed at 37°C. After 0, 10, 20, 30, 50, 70 and 90 min 15 µl aliquots were removed. The samples were then treated with 5 µl of the restriction enzyme ScaI (25 U) in 4× ScaI buffer (New England Biolabs) for 8 min at 37°C. After 8 min the reaction was stopped by addition of 10 µl of 2.5× stop solution, which contained 35 mM EDTA pH 8.0, 3.5% SDS, 38% (w/v) glycerol, 0.05% bromophenol blue and 0.05% xylene cyanol. The digested samples were analyzed by 1% agarose gel electrophoresis in 90 mM Tris–borate, 2 mM EDTA pH 8.3 buffer. Electrophoresis was performed at 6 V/cm for 16 h at 4°C. The gels were then stained using ethidium bromide and analyzed quantitatively using the Molecular Analyst Computer Imaging System (Bio-Rad).
RESULTS
Topological constraint of a circular RecA–ssDNA filament prevents it from interwinding with its homologous circular target duplex DNA
In most of the published results regarding in vitro strand exchange reactions promoted by RecA or eukaryotic Rad51 two types of substrates have been used: (i) linear ssDNA with linear duplex DNA or (ii) large circular ssDNA (3–8 kb) with homologous linear duplex DNA (reviewed in 1–8). Topologically, as long as one of the substrates is linear (either ssDNA or duplex DNA) the ssDNA would have the topological freedom to interwind, at least locally, with its complementary strand of the target duplex DNA in the homologous region. One of the possible models for homology searching is that the step that leads to formation of a final heteroduplex DNA product involves interwinding between the invading RecA–ssDNA filament and its complementary strand in the target duplex DNA. Thus, in this model, if one can limit the topological freedom of the two substrates to interwind, the homologous pairing would be blocked or would be much less efficient. On the other hand, if the homology search involved formation of a triplex intermediate where the target duplex DNA remained base paired, homologous pairing should still be detected even if the topological freedom of the two molecules to interwind had been restricted. This would imply the existence of an intermediate complex in which the two partners are aligned but not interwound (triplex model).
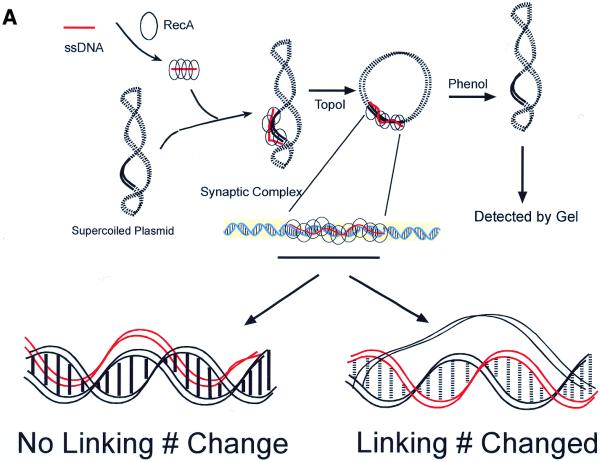
We have used a topological assay (28–30) to determine unwinding of the target duplex DNA in a synaptic complex (Fig. 1A). This method allowed us to probe the topological effect that synaptic complex formation (promoted by RecA) exerts on the target duplex DNA. A RecA–ssDNA filament was used to search for and pair with its homologous sequence (120 bp) on a duplex plasmid DNA (pBluescript, 3 kb). The synaptic complex was then treated with Topo I to relax the target plasmid. After dissociating the RecA protein with phenol the duplex plasmid DNA would become negatively supercoiled if the incoming RecA–ssDNA filament had wound itself around the complementary strand of the target double helix and thus unwound the homologous region of the duplex (Fig. 1A, lower right). This occurs because before removing RecA the homologous region of the target duplex DNA is still trapped by the RecA–ssDNA filament. The degree of supercoiling reflects the extent of unwinding induced by the RecA–ssDNA filament. The degree of unwinding (change in linking number) is then determined by running the final products in agarose gels with chloroquine, which induces positive supercoiling of the plasmid. Thus, the introduction of negative supercoiling by the joint complex will decrease the final degree of positive supercoiling induced by chloroquine. These plasmids will run slower than plasmids in the corresponding control reaction in which ssDNA of the same length without the homologous sequence is used.
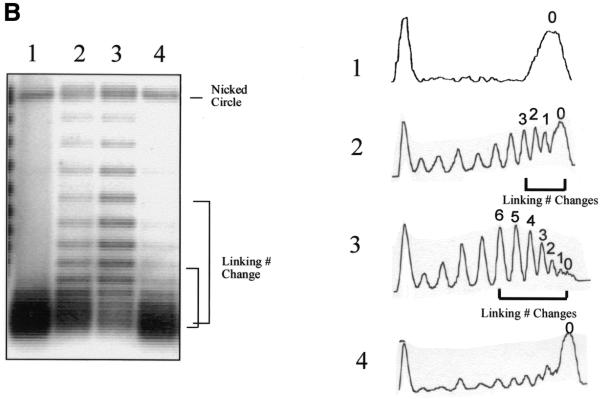
Figure 1.
Unwinding of a target duplex DNA by a RecA–ssDNA filament. (A) A linear (or circular in the experiments shown in Fig. 2) RecA–ssDNA filament was used to search for its homologous sequence (120 bp) on a circular duplex plasmid DNA (pBluescript, 3 kb). The synaptic complex was then treated with Topo I to relax the target plasmid. If the invading RecA–ssDNA filament had interwound with its complementary strand in the duplex (as shown lower right), after dissociating the protein with phenol the duplex plasmid DNA would become negatively supercoiled. The degree of supercoiling reflects the extent of unwinding induced by the RecA–ssDNA filament. The change in linking number could then be determined by running the final products in agarose gels with chloroquine. If the invading RecA–ssDNA filament just wound itself around the duplex, then no linking number change should be detected (as shown lower left). Solid lines represent homologous regions. Red represents the invading ssDNA. (B) (Left) An agarose gel containing 2 mg/ml chloroquine; (right) densitometric scans of each lane of the gel. Lane 3 demonstrates that a linear 120mer RecA–ssDNA filament can cause a target supercoiled DNA (pBluescript, 3 kb) with 120 bp of homology to unwind on average about six helical turns. Lane 1 is the same experiment as in lane 3, but no RecA has been used. With a 60mer homologous RecA–ssDNA as the searching entity, the linking number change is on average ∼3 (lane 2). When a RecA-coated heterologous 124mer ssDNA is used, no linking number change can be detected (lane 4).
On the other hand, if the incoming RecA–ssDNA filament just wound itself around the target double helix, i.e. the target duplex DNA stayed base paired, and no unwinding of the duplex DNA has occurred, then one should not detect any linking number change in the duplex DNA (Fig. 1A, lower left).
Using this topological assay (Fig. 1A; 28–30) we found that a linear 120mer RecA–ssDNA filament (H-120) can cause a target supercoiled DNA (pBluescript, 3 kb) with 120 bp of homology to unwind on average by about six helical turns (Fig. 1B, lane 3). Note that even though six is a special number relative to the extent of homology (120 bp, 12 turns), the results indicated that four, five or seven and eight are similar to six in their intensities. Therefore, we would not emphasize the significance of this average linking number change at this point. These changes require the presence of RecA (lane 1). The degree of unwinding is proportional to the length of homology. Thus, in the same experiment using 60mer RecA–ssDNA (H-60) as the invading strand the average unwinding (linking number change) is ∼3 (lane 2). When a RecA-coated heterologous 124mer (Het-124) is used no linking number change can be detected (lane 4). Thus, the observed unwinding of the target duplex DNA is RecA- and homology-dependent. The experiments shown in Figure 1B establish the feasibility of the topological assay to probe unwinding of target duplex DNA upon homologous pairing with RecA–ssDNA.
Next, in a RecA-promoted homologous pairing reaction we used a 520mer circular RecA–ssDNA (C-520) as the invading strand and covalently closed circular duplex DNA (pBluescript, 3 kb) as the target. The topological constraints of both circular DNA molecules should prevent them from interwinding with each other (31).
As shown in Figure 2, when a RecA-coated circular ssDNA (C-520, 520 nt long with 120 nt homologous to the 3 kb long supercoiled target plasmid) is used as the invading strand there is no detectable unwinding of the target duplex DNA (lane 5). However, with the linear form of the same 520mer ssDNA (the 3′-end with 120 nt homologous to the target duplex) as the invading strand there is unwinding of the target duplex DNA, similar to that observed with the 120mer homologous ssDNA (lanes 2 and 3). As expected, without RecA protein or with heterologous ssDNA as the invading strand there was no detectable unwinding of the target duplex DNA (lanes 1 and 4). These results suggest that the topological constraints of the circular ssDNA prevent it from interwinding with its circular target duplex DNA. Thus, no unwinding of the target duplex DNA was observed, unlike with the linear version of the same ssDNA as the invading ssDNA (compare lanes 3 and 5).
Figure 2.
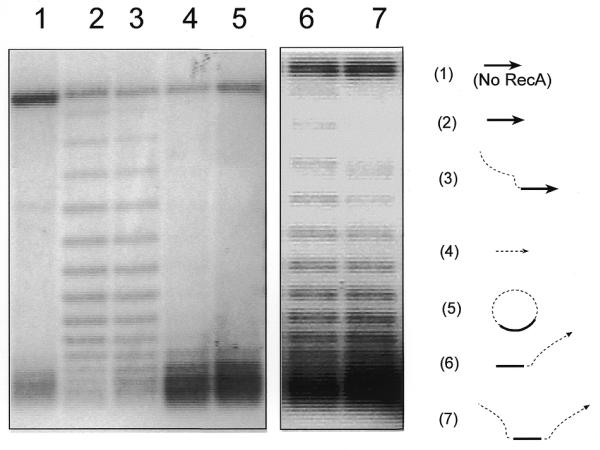
A circular RecA–ssDNA filament cannot unwind its supercoiled target DNA. Using the assay described in Figure 1A, shown here is an agarose gel with 2 mg/ml chloroquine. The diagrams to the right of the gel indicate the ssDNA substrates used in each reaction; the target duplex DNA used in all reactions was the 3 kb supercoiled pBluescript plasmid. The solid line indicates the homologous region (120 nt) and the dashed line indicates the heterologous region (400 nt). Lane 5 shows a reaction in which circular RecA–ssDNA filament (C-520, 520 nt long with 120 nt homologous to the target duplex DNA) was used as the searching entity. Lane 3 shows the same reaction but linear 520mer ssDNA (with the 3′-end 120 nt homologous to the target duplex) was used as the searching entity. Lane 2 is the reaction in which a 120mer homologous ssDNA (H-120) was used. Lanes 1 and 4 are control reactions either without RecA protein or with heterologous 124mer ssDNA (Het-124). Lanes 6 and 7 show two reactions with linear 520mer ssDNA with 120 nt homologous to the target duplex located at the 5′-end (lane 6) or at the center (lane 7).
It is important to note that unwinding of the target duplex DNA detected by this method could potentially arise either from interwinding of the invading ssDNA with its complementary strand in the target duplex DNA or just unwinding of the target duplex DNA itself. In the second case the homologous ssDNA and the duplex DNA are aligned but strand exchange does not occur (triplex model). For example, the target duplex DNA could be unwound from ∼10 bp/turn to ∼18 bp/turn to match the pitch of 95 Å with six RecA monomers per helical turn of the invading RecA–ssDNA nucleoprotein filament (9–11), but without interwinding with the invading ssDNA. However, if the change in linking number had arisen from a change in helical pitch of the duplex DNA itself by pairing side-by-side with the invading RecA–ssDNA we should observe the same linking number change when either linear or circular RecA–ssDNA was used as the invading strand. Thus, the implications from the results shown in Figure 2 are that the observed linking number change for the duplex DNA when homologous linear RecA–ssDNA was used as the invading strand is due to interwinding of the complementary strands of the two DNA molecules (Fig. 1A, lower right).
Furthermore, with the 120 nt homologous region located at the 5′-end or at the center of the linear 520mer RecA–ssDNA we also detected similar linking number changes as that with the homologous 120mer ssDNA (compare lanes 6 and 7 to lane 2). However, the linear ssDNA in which the homologous region is located at its center (lane 7) caused less unwinding of the target duplex substrate. This indicates relatively more topological constraints for the two molecules to interwind because on this linear DNA there are heterologous 200 nt sequences flanking both ends of the homologous region (see Fig. 2, lane 7).
These results also suggest that even though the strand exchange reaction has been shown to have polarity (32–37) (5′→3′ relative to the invading ssDNA), the polarity of the invading RecA–ssDNA apparently does not affect the efficiency of interwinding with the target duplex DNA to form a homologous pairing synaptic complex (compare lanes 3, 6 and 7 in Fig. 2).
Interwinding between a linear RecA–ssDNA filament and its target duplex DNA is affected by the superhelicity of the target duplex DNA
DNA supercoiling has been shown to play many important physiological roles in the cell (38). Because the linking number is constant for each topologically closed DNA domain, writhe must change in an equal and compensating fashion for all changes in twist of the double helix (39). Hence, the relaxation of supercoiled dsDNA tends to promote separation of the two single strands of duplex DNA. Thus, many biological processes that require separation of the single strands cannot proceed without the duplex DNA substrate being negatively supercoiled (31,38,39).
The results shown in Figure 2 imply that the RecA–ssDNA filament might search for its homologous sequence by a mechanism in which the invading ssDNA interwinds with its complementary strand in the duplex target DNA to check for homology. If this is true, a negatively supercoiled duplex DNA should be a better substrate for a homology search.
Therefore, instead of constraining the topological freedom of the invading ssDNA, we attempted to investigate the influence of the extent of supercoiling of the target dsDNA on the process of synaptic complex formation. For the target dsDNA substrates, covalently closed circular duplex DNA (pBluescript, 3 kb) with various degrees of supercoiling (highly supercoiled to relaxed) were constructed using Topo I to relax circular plasmid DNA in the presence of different concentrations of ethidium bromide (∼0–4 µg/ml) (40). After removing the ethidium bromide, plasmid DNAs with different degrees of supercoiling were formed (Fig. 3A; for details see Materials and Methods).
Figure 3.
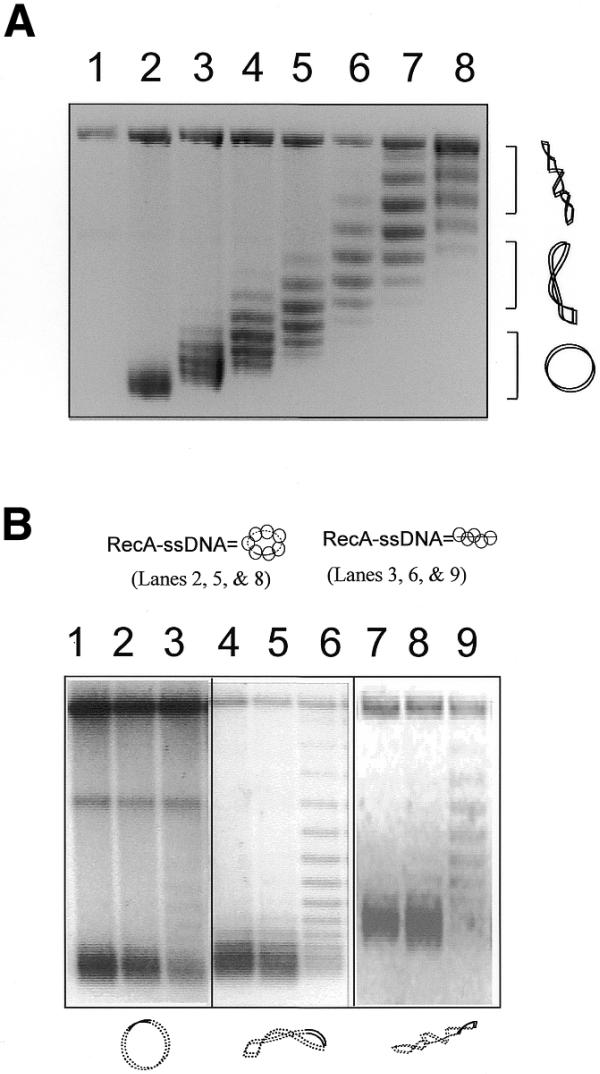
Unwinding of target DNAs with different degrees of supercoiling by a linear RecA–ssDNA filament. (A) An agarose gel with chloroquine (2 mg/ml) shows the covalently closed circular duplex DNA (pBluescript, 3 kb with 120 bp homologous to the ssDNA used) with high to zero supercoiling that was used in the reactions shown in (B). In the chloroquine gel the relaxed duplex DNA will become positively supercoiled. Therefore, it runs faster than the negatively supercoiled DNA that has become relaxed under these conditions. Lane 1 is a nicked circle as the marker for relaxed plasmid. Lanes 2–8 are the covalently closed plasmids (pBluescript, 3 kb) with increasing negative superhelicity. (B) An agarose gel with chloroquine (2 mg/ml) showing unwinding of the circular target DNA caused by either linear or circular homologous RecA–ssDNA. The diagrams at the top and bottom of the gel indicate the RecA–ssDNA and the duplex target DNA used in the reactions; solid lines represent homology between the two molecules. Lanes 1–3 are reactions where relaxed duplex was used as the target. Lanes 4–6 are reactions where medium supercoiled duplexes (superhelical density ∼–0.016) were used as target. Lanes 7–9 are reactions where highly supercoiled duplex (superhelical density ∼–0.033) were used as the target. Lane 3, 6 and 9 show reactions in which a homologous linear 120mer RecA–ssDNA filament (H-120) was used as the searching molecule. Lanes 1, 4 and 7 are reactions in which 124mer heterologous RecA–ssDNA filament (Het-124) was used as the searching molecule. Lanes 2, 5 and 8 show a reaction in which circular RecA–ssDNA filament (C-520, 520 nt long with 120 nt homologous to the target duplex DNA) was used as the searching molecule.
Figure 3B shows that a 120mer RecA–ssDNA filament (H-120) caused a highly supercoiled target duplex (with 120 bp homology to the ssDNA and a superhelical density of ∼–0.033) to unwind by about 6 turns (lane 9). However, when a totally relaxed target duplex was used, unwinding was much less extensive (lane 3). An intermediately supercoiled target duplex (superhelical density ∼–0.016) was unwound less extensively compared to the highly supercoiled duplex (∼3–4 turns; lane 6) yet was more extensively supercoiled when compared to the relaxed substrate (compare lane 6 to lane 3). When a 124mer heterologous RecA–ssDNA filament (Het-124) was used in the reaction there was little detectable unwinding of any of the three target duplexes used (lanes 1, 4 and 7). Consistent with previous results (Fig. 2), when circular RecA–ssDNA (C-520) was used as the search entity no obvious change in linking number could be detected for any of the target duplexes with different degrees of supercoiling (lanes 2, 5 and 8).
Relaxation of negative supercoiling promotes separation of the two strands of the circular duplex DNA (31,39) and the results shown in Figure 3 demonstrate that a high degree of negative supercoiling promotes unwinding of the circular duplex DNA by the invading RecA-coated linear ssDNA. Therefore, the results shown in Figure 3 further support the concept that the mechanism for homologous pairing promoted by a RecA–ssDNA filament involves local interwinding of the invading ssDNA with its target duplex DNA.
Homologous pairing between the circular RecA–ssDNA filament and a circular duplex DNA target is undetectable with a restriction enzyme protection assay
Although unlikely, the topological results (Figs 1–3) presented so far cannot absolutely rule out the possibility that when the circular RecA–ssDNA pairs with the duplex DNA through a triplex structure it causes the duplex DNA to extend from ∼10 to ∼18 bp/turn. To address this concern we used a complementary restriction enzyme protection method to characterize the homologous pairing promoted by RecA. In this assay a RecA–ssDNA filament was used to search for its homologous sequence (120 bp) located on a plasmid DNA (pBluescript, linear, covalently closed relaxed or supercoiled circle) that contains a ScaI restriction site in the center of the homologous region. Once the RecA–ssDNA filament finds its homologous target it forms a joint molecule with the duplex DNA that inhibits digestion of the duplex DNA. As ScaI cannot cleave a triple strand, the inability of ScaI to cleave the duplex DNA provides a direct measurement for homologous pairing of the two molecules or synaptic complex formation (17,41). Agarose gel electrophoresis allowed us to separate the unprotected (digested) product from the protected substrate (undigested) (Fig. 4A).
Figure 4.
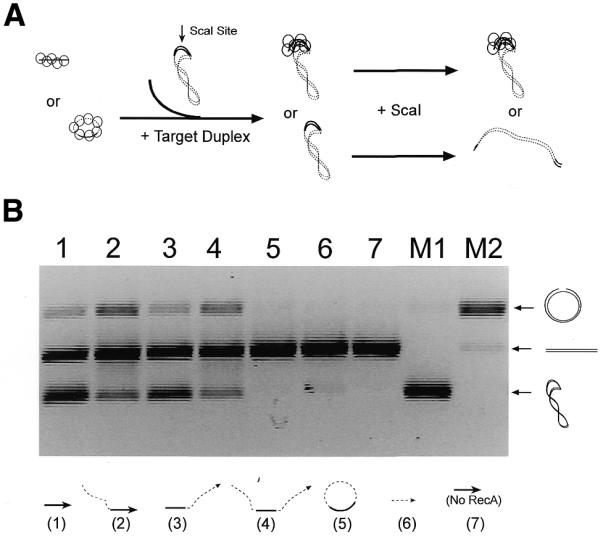
Restriction enzyme protection assay for homologous pairing. (A) In this assay a RecA–ssDNA filament (linear or circular) is used to search for its homologous sequence located on a plasmid DNA (pBluescript, linear, supercoiled or relaxed circular). Once the RecA–ssDNA filament finds its homologous target, it forms a synaptic complex with the duplex DNA that inhibits digestion of the duplex DNA by a restriction enzyme (ScaI) that recognizes the center of the homologous region. The increasing inability of ScaI to cleave the duplex DNA provides a direct measurement of synaptic complex formation. Agarose gel electrophoresis allows us to separate the unprotected (digested) product from the protected substrate. Here, only a linear RecA–ssDNA filament and supercoiled duplex target are shown as an example. (B) This agarose gel shows homologous pairing, assayed by the restriction enzyme protection method, between various RecA–ssDNA filaments (indicated at the bottom of the gel) and supercoiled duplex DNA as target. The solid lines represent the 120 bp of homology between the two molecules. The dashed lines represent 400 nt heterologous sequences in the ssDNA (except for the substrate used in lane 6, which is a 124mer heterologous ssDNA). Lanes M1 and M2 are markers for supercoiled, linear and nicked forms of the duplex substrates.
Using this assay we found that homologous pairing between a circular RecA–ssDNA filament (C-520, 520mer with 120 nt of homology) and a supercoiled target duplex DNA was not detectable (Fig. 4B, lane 5). However, when various 520mer linear RecA–ssDNAs were used, regardless of the positions of the 120mer homologous region, homologous pairing was obvious, as indicated by protection of the supercoiled duplex substrates from complete digestion by the restriction enzyme (Fig. 4B, lanes 1–4; the nicked circles at the top of the gel are incomplete digestion products). When a heterologous linear RecA–ssDNA was used in the reactions, or in the absence of RecA, no protection of the supercoiled duplex DNA could be detected (Fig. 4B, lanes 6 and 7).
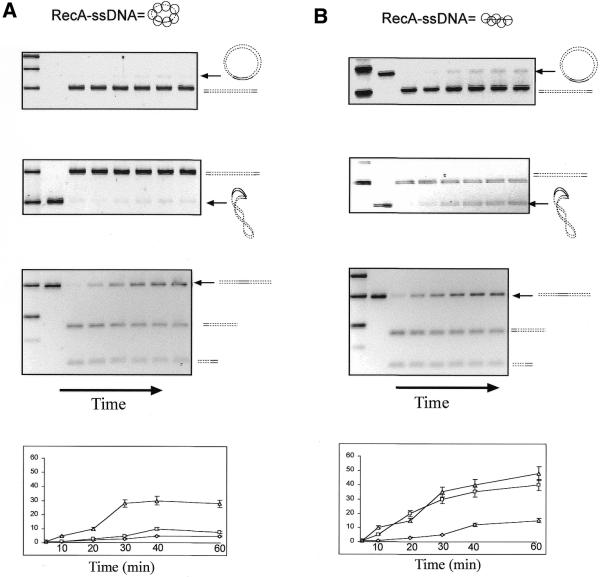
Next, the kinetics of the homologous pairing reaction (detected by restriction enzyme protection assay) were investigated using circular RecA–ssDNA as the invading strand and either supercoiled or relaxed (covalently closed) circular duplex DNA as the target. In both cases protection from restriction enzyme digestion of the circular duplexes was barely detectable (<10%; Fig. 5A, top two panels). However, when linear RecA–ssDNA was used as the invading strand, homologous pairing was demonstrated in both cases (Fig. 5B, top two panels). It is also evident that when linear RecA–ssDNA was used as the invading strand with supercoiled duplex DNA used as the target the amount of joint molecule formation was much more than if relaxed circular duplex DNA was used as the target (40 and 15%, respectively; see quantification in the bottom panels). The latter point also confirms the topological assay results, which showed that supercoiled DNA promotes homologous pairing (Fig. 3).
Figure 5.
Restriction enzyme protection assay for homologous pairing between linear or circular RecA–ssDNA with relaxed, supercoiled or linear duplex DNA with 120 bp of homology. (A) (Top) An agarose gel that shows homologous pairing between a 520mer circular RecA–ssDNA (C-520, with 120 nt homologous to the duplex) filament and relaxed circular duplex target. (Upper middle) A reaction in which supercoiled duplex DNA was used as the target; (lower middle) the same reaction with linear duplex DNA as the target. (Bottom) Quantification of the three reactions (diamonds represent the reaction with relaxed duplex target; squares represent the reaction with supercoiled duplex target; triangles represent the reaction with linear duplex target). The reaction times were 5, 10, 20, 30, 40 and 60 min after formation of synaptic complexes and prior to 8 min of restriction digestion (see Materials and Methods for details). (B) Here, instead of 520mer circular RecA–ssDNA a 120mer homologous linear RecA–ssDNA (H-120) was used as the searching molecule. Top to bottom the panels show the formation of synaptic complexes with all three types of target duplex DNA as in (A). Solid lines indicate homology between the two molecules. Arrows to the right of each gel indicate the initial substrate used in that reaction.
It is possible that circular RecA–ssDNA could not form a stable synaptic complex with its target duplex DNA, as detected by the restriction enzyme assay, because RecA cannot form a stable filament on the relatively small circular ssDNA. To address this concern, as shown in the third panel of Figure 5A, we demonstrated that even though it could not protect the homologous restriction site on a circular target DNA, circular ssDNA–RecA could search for and protect the homologous site on a linear duplex DNA target.
Note that with this restriction protection assay one can detect whether the RecA–ssDNA filament had found its homologous target site on the duplex target molecule and had formed a relatively stable synaptic complex. Formation of this synaptic complex thus hinders the ability of the restriction enzyme to digest duplex DNA with the recognition site in the middle of the homologous region. This protection occurs regardless of whether the invading ssDNA is interwound with its complementary strand in the target duplex DNA or is aligned side-by-side with the target duplex to form a triplex structure. Therefore, with this restriction enzyme protection assay the results demonstrate a lack of any significant amount of homologous pairing between a circular RecA–ssDNA and a circular duplex DNA (Fig. 5A, top two panels). Yet homologous pairing is evident between a circular RecA–ssDNA and a linear duplex (Fig. 5A, third panel), which again supports the concept that to form a stable intermediate synaptic complex one of the two DNA molecules needs to be linear to have the topological freedom to interwind with the other. This topological freedom can be in either the invading ssDNA or its target duplex DNA. Thus, it is unlikely that a triplex type of structure, in which the two DNA molecules align but do not interwind, is an intermediate product prior to the occurrence of strand exchange.
DISCUSSION
Several properties of the mechanism by which RecA mediates homologous pairing have been demonstrated. (i) A circular RecA–ssDNA filament cannot pair to nor unwind the homologous region of a circular target duplex DNA. However, a circular RecA–ssDNA filament can find its homologous region and form a stable synaptic complex with a linear duplex target DNA (as detected by the restriction enzyme protection assay). On the other hand, a linear RecA–ssDNA can bind to its homologous region and form a stable complex with a circular duplex target DNA. These results suggest that at least one free end, either on the ssDNA or dsDNA, is needed for homology recognition promoted by RecA. (ii) Negative supercoiling of the circular target duplex DNA facilitates homologous pairing (synaptic complex formation) by an invading linear RecA–ssDNA filament. This supports the concept that the homology searching process is coupled to local strand exchange, because negative supercoiling promotes separation of the two single strands in a duplex. (iii) The polarity of the invading linear RecA–ssDNA, with the homologous region located 5′, 3′ or in the middle of a linear ssDNA, does not affect the efficiency of its interwinding with the supercoiled target duplex DNA. This result implies that in vivo initiation of strand exchange does not have to start with a RecA-coated ssDNA with the homologous region located at its 3′-end, as has been previously suggested (reviewed in 1,42).
From various in vitro biochemical studies of recombination activities mediated by RecA it is well established that there is an initial phase of the reaction where homology recognition occurs, followed by a slower phase in which extensive strand exchange occurs (3,15,16,42). Recent kinetic studies, using a fluorescence resonance energy transfer method with short oligonucleotides as substrates, demonstrate that there are two, or possibly three, distinct steps in the overall strand exchange reaction promoted by RecA (15–17). A minimum reaction mechanism, therefore, appears to require at least one intermediate. The results shown in this paper demonstrate a requirement for unwinding of the duplex DNA and strand exchange with the invading single strand in the region of homology. Therefore, the results are consistent with the model in which homology searching promoted by a RecA–ssDNA filament is coupled to interwinding of the invading ssDNA with its target duplex DNA (Fig. 6A). The unwinding of the target duplex DNA needed for the invading ssDNA to interwind with it could either be induced through contacts with the RecA-coated invading ssDNA or simply be the result of thermal fluctuations in the duplex DNA itself. However, it must be pointed out that from the results presented in this paper we could not rule out the possibility that a triplex might be formed as a transient intermediate during the homology search, but it is quickly disassembled in the absence of pairing freedom to form the more stable interwound helix.
Figure 6.
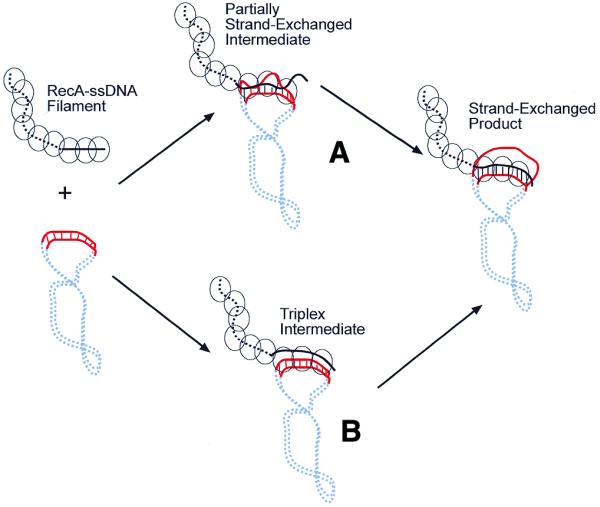
Proposed model for RecA-mediated homology recognition. (A) Homology recognition is likely coupled to local strand exchanges between the two molecules. These locally unwound regions on the target duplex DNA could either be induced through contacts with the RecA–ssDNA filament or simply be the result of thermal fluctuation of the duplex DNA. Thus, the searching RecA–ssDNA filament tests for homology by base pairing with its complementary strand in the target duplex DNA. If homology is found through these short strand exchanges, then branch migration can extend the exchanged region to a greater distance, eventually resulting in full strand exchange. (B) In this model the RecA–ssDNA forms a triplex intermediate with the target duplex DNA prior to strand exchange. The results presented in this paper do not support this model, where in the triplex intermediate the RecA–ssDNA (which has been extended to ∼18 bp/turn) needs to be aligned in register with its target duplex DNA (10 bp/turn) on the homologous region, yet does not unwind the target duplex DNA.
The helical RecA–ssDNA nucleoprotein filament that mediates homologous pairing is assembled in a directional fashion, has a unique asymmetrical structure and has been shown to promote strand exchange in a polarized fashion, 5′→3′ with respect to the ssDNA inside the filament (33–37). However, genetic and biochemical studies in E.coli and S.cerevisiae have suggested that recombination is initiated by invasion of the target duplex DNA by a partial duplex DNA with a single-stranded 3′-tail (43–46). Therefore, paradoxically, the intermediate joint molecules formed by 3′ invasion would be resolved quickly by strand exchange in the 5′→3′ direction. In this paper we have demonstrated that the polarity of the invading linear RecA–ssDNA does not affect the efficiency of its interwinding with the target duplex DNA. In the model shown in Figure 6A the quandary of polarity observed in in vitro biochemical studies may not be an issue in vivo. Once RecA initiates strand exchange over short regions regardless of polarity, subsequent branch migration to produce long heteroduplex regions is likely be a function of other proteins, such as RuvA and RuvB, which specifically recognize Holliday junctions and promote unidirectional branch migration (47–50).
The results described here also agree with earlier reports that analyzed the base pairing status of RecA-promoted joint molecules by chemical probing methods (19–21). Chemical probing results indicated that the original base pairs in the parental duplex are disrupted; one strand is displaced and the other strand appears to be involved in Watson–Crick base pairing with the incoming ssDNA. The state of base pairing revealed by chemical probing thus resembles that of strand-exchanged products and not a canonical DNA triplex involving non-Watson–Crick base pairing.
Contrary to the results described here, there have been observations showing that homologous pairing could occur without any free single-stranded ends in the DNA substrates. These results suggest that homology recognition does not require that the strands of the nascent heteroduplex DNA be truly interwound, as in Watson–Crick duplex DNA (51–55). In most of these studies long circular ssDNA (4–8 kb φX174 or M13) coated with RecA that is homologous to the central region (several kb long) of a linear duplex DNA were used as substrates. Filter binding assays and direct observations by electron microscopy suggested a so-called ‘paranemic joint’ as the intermediate in the strand exchange reaction (51–55). Note that in all these studies the ssDNAs used were usually very long (M13 or φX174) and the duplex DNA used was usually linear. Therefore, as we have shown in this study, a circular RecA–ssDNA can pair to a linear target duplex DNA (with 120 bp of homology; Fig. 5, third panel). Thus, with the topological freedoms of the long flexible ssDNA and its linear duplex DNA partner it is likely that many small local regions of strand exchange have occurred between them to form a joint molecule.
In summary, the results presented in this paper suggest that the homology search might be coupled to the topological freedom of the two molecules to exchange strands, possibly through small regions of duplex unwinding and interwinding with the invading ssDNA. A triplex type of ‘paranemic joint molecule’, in which the two DNA molecules are aligned in register but not interwound, is unlikely to be an intermediate during the homologous pairing process promoted by RecA.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Professors Nick Cozzarelli and Ned Seeman for critical reading of this manuscript. This work was supported by grants from the American Cancer Society (PRG-99-352-01-GMC) and the University of Delaware Research Foundation.
References
- 1.Cox M.M. (2000) Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acid Res. Mol. Biol., 63, 311–366. [DOI] [PubMed] [Google Scholar]
- 2.Kowalczykowski S.C. and Eggleston,A.K. (1994) Homologous pairing and DNA strand exchange proteins. Annu. Rev. Biochem., 63, 991–1043. [DOI] [PubMed] [Google Scholar]
- 3.Rao B.J.,Chiu,S.K., Bazemore,L.R., Reddy,G. and Radding,C.M. (1995) How specific is the first recognition step of homologous recombination? Trends Biochem. Sci., 20, 109–113. [DOI] [PubMed] [Google Scholar]
- 4.West S.C. (1992) Enzymes and molecular mechanisms of genetic recombination. Annu. Rev. Biochem., 61, 603–640. [DOI] [PubMed] [Google Scholar]
- 5.West S.C. (1994) The processing of recombination intermediates: mechanistic insights from studies of bacterial proteins. Cell, 76, 9–15. [DOI] [PubMed] [Google Scholar]
- 6.Camerini-Otero R. and Hsieh,P. (1993) Parallel DNA triplexes, homologous recombination and other homology-dependent DNA interactions. Cell, 73, 217–223. [DOI] [PubMed] [Google Scholar]
- 7.Camerini-Otero R.D. and Hsieh,P. (1995) Homologous recombination proteins in prokaryotes and eukaryotes. Annu. Rev. Genet., 29, 509–552. [DOI] [PubMed] [Google Scholar]
- 8.Eggleston A.K. and West,S.C. (1996) Exchanging partners: recombination in E. coli. Trends Genet., 12, 20–25. [DOI] [PubMed] [Google Scholar]
- 9.Stasiak A. and Egelman,E.H. (1994) Structure and function of RecA–DNA complexes. Experientia, 50, 192–203. [DOI] [PubMed] [Google Scholar]
- 10.Story R.M., Weber,I.T. and Steitz,T.A. (1992) The structure of the E. coli RecA protein monomer and polymer. Nature, 355, 318–325. [DOI] [PubMed] [Google Scholar]
- 11.Egelman E.H. and Stasiak,A. (1993) Electron microscopy of RecA–DNA complexes: two different states, their functional significance and relation to the solved crystal structure. Micron, 24, 309–324. [Google Scholar]
- 12.Ogawa T., Yu,X., Shinohara,A. and Egelman,E.H. (1993) Similarity of the yeast Rad51 filament to the bacterial RecA filament. Science, 259, 1896–1898. [DOI] [PubMed] [Google Scholar]
- 13.Shinohara A., Ogawa,H., Matsuda,Y., Ushino,N., Ikeo,K. and Ogawa,T. (1993) Cloning of human, mouse and fusion yeast recombination genes homologous to RAD51 and recA. Nature Genet., 4, 239–243. [DOI] [PubMed] [Google Scholar]
- 14.Benson F.E., Stasiak,A. and West,S.C. (1994) Purification and characterization of the human Rad51 protein, an analogue of E.coli RecA. EMBO J., 13, 5764–5771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bazemore L.R., Takahashi,M. and Radding,C.M. (1997) Kinetic analysis of pairing and strand exchange catalyzed by RecA: detection by fluorescence energy transfer. J. Biol. Chem., 272, 14672–14682. [DOI] [PubMed] [Google Scholar]
- 16.Gumbs O.H. and Shaner,S. (1998) Three mechanistic steps detected by FRET after presynaptic filament formation in homologous recombination. ATP hydrolysis required for release of oligonucleotides heteroduplex product from recA. Biochemistry, 37, 11692–11706. [DOI] [PubMed] [Google Scholar]
- 17.Yancey-Wrona J.E. and Camerini-Otero,R.D. (1995) The search for DNA homology does not limit stable homologous pairing promoted by RecA protein. Curr. Biol., 5, 1149–1158. [DOI] [PubMed] [Google Scholar]
- 18.Adzuma K. (1998) No sliding during homology search by RecA protein. J. Biol. Chem., 273, 31565–31573. [DOI] [PubMed] [Google Scholar]
- 19.Adzuma K. (1992) Stable synapsis of homologous DNA molecules mediated by the Escherichia coli RecA protein involves local exchange of DNA strands. Genes Dev., 6, 1679–1694. [DOI] [PubMed] [Google Scholar]
- 20.Zhou X. and Adzuma,K. (1997) DNA strand exchange mediated by the Escherichia coli RecA protein initiates in the minor groove of double-stranded DNA. Biochemistry, 36, 4650–4661. [DOI] [PubMed] [Google Scholar]
- 21.Baliga R., Singleton,J.W. and Dervan,P.B. (1995) RecA-oligonucleotide filaments bind in the minor groove of double-stranded DNA. Proc. Natl Acad. Sci. USA, 92, 10393–10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Howard-Flanders P., West,S.C. and Stasiak,A. (1984) Role of RecA protein spiral filaments in genetic-recombination. Nature, 309, 215–219. [DOI] [PubMed] [Google Scholar]
- 23.Lindsley J.E. and Cox,M.M. (1990) Assembly and disassembly of RecA protein filaments occur at opposite filament ends. J. Biol. Chem., 265, 9043–9054. [PubMed] [Google Scholar]
- 24.Haber J.E. (1998) MEIOSIS: searching for a partner. Science, 279, 823–825. [DOI] [PubMed] [Google Scholar]
- 25.Chen J., Kannar,R. and Cozzarelli,N.R. (1994) The Sep1 strand exchange protein from Saccharomyces cerevisiae promotes a paranemic joint between homologous DNA molecules. Genes Dev., 8, 1356–1365. [DOI] [PubMed] [Google Scholar]
- 26.Lavery P.E. and Kowalczykowski,S.C. (1992) Enhancement of recA protein-promoted DNA strand exchange activity by volume-occupying agents. J. Biol. Chem., 267, 9307–9314. [PubMed] [Google Scholar]
- 27.Sambrook J., Fritsch,E. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Rould E., Muniyappa,K. and Radding,C.M. (1992) Unwinding of heterologous DNA by RecA protein during the search for homologous sequences. J. Mol. Biol., 226, 127–139. [DOI] [PubMed] [Google Scholar]
- 29.Kiianitsa K. and Stasiak,A. (1997) Helical repeat of DNA in the region of homologous pairing. Proc. Natl Acad. Sci. USA, 94, 7837–7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Voloshin O.N. and Camerini-Otero,R.D. (1997) The duplex DNA is very underwound in the three-stranded RecA protein-mediated synaptic complex. Genes Cells, 2, 303–314. [DOI] [PubMed]
- 31.Vologodskii A. (1992) Topology and Physics of Circular DNA. CRC Press, Boca Raton, FL.
- 32.Jain S.K., Cox,M.M. and Inman,R.B. (1994) On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. J. Biol. Chem., 269, 20653–20661. [PubMed] [Google Scholar]
- 33.Cox M.M. and Lehman,I.R. (1981) Directionality and polarity in recA protein-promoted branch migration. Proc. Natl Acad. Sci. USA, 78, 6018–6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kahn R., Cunningham,R.P., DasGupta,C. and Radding,C.M. (1981) Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc. Natl Acad. Sci. USA, 78, 4786–4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.West S.C., Cassuto,E. and Howard-Flanders,P. (1981) Heteroduplex formation by recA protein: polarity of strand exchange. Proc. Natl Acad. Sci. USA, 78, 6149–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Register J.C. and Griffith,J. (1985) The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem., 260, 12308–12312. [PubMed] [Google Scholar]
- 37.Stasiak A., Egelman,E.H. and Howard-Flanders,P. (1988) Structure of helical recA–DNA complexes. III. J. Mol. Biol., 202, 659–662. [DOI] [PubMed] [Google Scholar]
- 38.Kannar R. and Cozzarelli,N.R. (1992) Roles of supercoiled DNA structure in DNA transactions. Curr. Opin. Struct. Biol., 2, 369–379. [Google Scholar]
- 39.White J.H. (1989) In Waterman,M.S. (ed.) Mathematical Methods for DNA Sequences. CRC Press, Boca Raton, FL, pp. 225–253.
- 40.Bliska J.B. and Cozzarelli,N.R. (1987) Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J. Mol. Biol., 194, 205–218. [DOI] [PubMed] [Google Scholar]
- 41.Ferrin L.J. and Camerini-Otero,R.D. (1991) Sequence-specific ligation of DNA using RecA protein. Science, 254, 1494–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roca A.I. and Cox,M.M. (1997) RecA protein: structure, function and role in recombinational DNA repair. Prog. Nucleic Acid Res. Mol. Biol., 56, 129–223. [DOI] [PubMed] [Google Scholar]
- 43.Smith G.R. (1987) Mechanism and control of homologous recombination in Escherichia coli. Annu. Rev. Genet., 21, 179–201. [DOI] [PubMed] [Google Scholar]
- 44.Sun H., Treco,D. and Szostak,J. (1991) Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell, 64, 1155–1161. [DOI] [PubMed] [Google Scholar]
- 45.Silberstein Z., Shalit,M. and Cohen,A. (1993) Heteroduplex strand-specificity in restriction-stimulated recombination by the RecE pathway of E. coli. Genetics, 133, 439–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman-Ohana R. and Cohen,A. (1998) Heteroduplex joint formation in Escherichia coli recombination is initiated by pairing of a 3′-ending strand. Proc. Natl Acad. Sci. USA, 95, 6909–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.West S.C. (1997) Processing of recombination intermediates by the Ruv ABC proteins. Annu. Rev. Genet., 31, 213–244. [DOI] [PubMed] [Google Scholar]
- 48.Shinagawa H. and Iwasaki,H. (1996) Processing the Holliday junction in homologous recombination. Trends Biochem. Sci., 21, 107–111. [PubMed] [Google Scholar]
- 49.Ariyoshi M., Vassylyev,D.G., Iwasaki,H., Nakamura,H., Shinagawa,H. and Morikawa,K. (1994) Atomic structure of the RuvC resolvase: a Holliday junction-specific endonuclease from E. coli. Cell, 78, 1063–1072. [DOI] [PubMed] [Google Scholar]
- 50.Iype L.E., Inman,R.B. and Cox,M.M. (1995) Blocked RecA protein-mediated DNA strand exchange reactions are reversed by the RuvA and RuvB proteins J. Biol. Chem., 270, 19473–19480. [DOI] [PubMed] [Google Scholar]
- 51.Wu A.M., Kahn,R., DasGupta,C. and Radding,C.M. (1982) Formation of nascent heteroduplex structures by RecA protein and DNA. Cell, 30, 37–44. [DOI] [PubMed] [Google Scholar]
- 52.Schutte B.C. and Cox,M.M. (1988) Homology-dependent underwinding of duplex DNA in recA protein generated paranemic complexes. Biochemistry, 27, 7886–7894. [DOI] [PubMed] [Google Scholar]
- 53.Conley E.C. and West,S.C. (1989) Underwinding of DNA associated with duplex-duplex pairing by RecA protein. Cell, 56, 987–995. [PubMed] [Google Scholar]
- 54.DasGupta C., Shibata,T., Cunningham,R.P. and Radding,C.M. (1980) The topology of homologous pairing promoted by RecA protein. Cell, 22, 437–446. [DOI] [PubMed] [Google Scholar]
- 55.Cunningham R.P., Wu,A.M., Shibata,T., DasGupta,C. and Radding,C.M. (1981) Homologous pairing and topological linkage of DNA molecules by combined action of E. coli RecA protein and topoisomerase I. Cell, 24, 213–223. [DOI] [PubMed] [Google Scholar]