Abstract
A strong link exists between low aerobic exercise capacity and complex metabolic diseases. To probe this linkage, we utilized rat models of low and high intrinsic aerobic endurance running capacity that differ also in the risk for metabolic syndrome. We investigated in skeletal muscle gene-phenotype relationships that connect aerobic endurance capacity with metabolic disease risk factors. The study compared 12 high capacity runners (HCRs) and 12 low capacity runners (LCRs) from generation 18 of selection that differed by 615% for maximal treadmill endurance running capacity. On average, LCRs were heavier and had increased blood glucose, insulin, and triglycerides compared with HCRs. HCRs were higher for resting metabolic rate, voluntary activity, serum high density lipoproteins, muscle capillarity, and mitochondrial area. Bioinformatic analysis of skeletal muscle gene expression data revealed that many genes up-regulated in HCRs were related to oxidative energy metabolism. Seven mean mRNA expression centroids, including oxidative phosphorylation and fatty acid metabolism, correlated significantly with several exercise capacity and disease risk phenotypes. These expression-phenotype correlations, together with diminished skeletal muscle capillarity and mitochondrial area in LCR rats, support the general hypothesis that an inherited intrinsic aerobic capacity can underlie disease risks.—Kivelä, R., Silvennoinen, M., Lehti, M., Rinnankoski-Tuikka, R., Purhonen, T., Ketola, T., Pullinen, K., Vuento, M., Mutanen, N., Sartor, M. A., Reunanen, H., Koch, L. G., Britton, S. L., Kainulainen, H. Gene expression centroids that link with low intrinsic aerobic exercise capacity and complex disease risk.
Keywords: skeletal muscle, complex metabolic disease, oxygen metabolism, mitochondria, lipid metabolism
There is a strong statistical association between low aerobic capacity and diminished health status. Indeed, low maximal oxygen consumption (Vo2max) and low endurance capacity are stronger predictors of early morbidity and mortality relative to other established risk factors, such as diabetes or hypertension (1, 2). The difference in aerobic endurance capacity between individuals is the sum of inborn capacity and that acquired via lifestyle activity. Despite much study, however, the explicit genetic programs for neither the intrinsic nor acquired components of aerobic capacity have been defined (3).
To facilitate exploration for genes that regulate intrinsic aerobic endurance capacity we started, ∼10 yr ago (4), the development of heterogeneous rat strains artificially selected for low and high inborn exercise capacity. Endurance treadmill running capacity was adopted as the selection criterion because it provides a strong signal corresponding to energy transfer and can be measured objectively on a large scale. This approach to model development is in line with the current shift from focus on manipulating individual genes and pathways to the consideration that biological function resides within highly interconnected modules of molecular networks (5, 6).
As early as generation 7 of selection, we learned that peripheral changes associated with a higher transfer of O2 within skeletal muscle were most responsive to selection and accounted for a large part of the initial divergence in endurance running capacity (7) between the low capacity runner (LCR) and high capacity runner (HCR) rats. By 15 generations of selection, differences for central features of oxygen delivery had also emerged and were accompanied by further differences for O2 transfer capacity in skeletal muscle (8). In a study performed at generation 11, Wisloff et al. (9) found that the LCRs had a higher incidence of cardiovascular and metabolic risk factors consistent with the metabolic syndrome. Here we evaluated rats from generation 18 of selection for phenotypes and gene networks that could explain the differences in exercise capacity and predict disease risk. Gene set enrichment analysis (GSEA), a statistical technique that relies on the principle that genes act as groups in a coordinated manner and not in isolation, was used to identify the subsets of genes that are congruently regulated within a given gene set. Centroids (centroid is the mean expression of the coregulated genes within a subset) are shown to correlate with various systemic metabolic and physiological parameters (10) and may therefore be used to probe for gene expression patterns that are causative of complex metabolic diseases. Our results demonstrate significant differential expression for 239 genes in skeletal muscle that underlie part of the phenotypic differences for low and high intrinsic aerobic endurance capacity. The expression of genes in OXPHOS, TCA cycle, PPAR signaling, and STAT3 pathways was significantly correlated with physical capacity and disease risk variables such as voluntary activity, insulin sensitivity, blood triglycerides, and fatty acids. Oxygen delivery and usage in skeletal muscle were also significantly enhanced in HCRs via increased capillarization and higher mitochondrial content compared with LCRs. Our results reveal mRNA expression signatures that link aerobic endurance capacity to metabolic disease risk factors and suggest that intrinsic aerobic capacity can mediate a difference between health and disease.
MATERIALS AND METHODS
Animal strains
Artificial selective breeding, starting with a founder population of 186 genetically heterogeneous rats (N:NIH stock), was used to produce rat strains differing in inherent aerobic capacity. The procedure is described in detail previously (4). Briefly, endurance running capacity was assessed on a treadmill and the total distance run during the test was used as a measure for maximal aerobic exercise capacity. Rats with the highest running capacity from each generation were bred to produce the HCR strain, and rats with the lowest capacity were bred with each other to produce the LCR strain. The assumption was that selection for divergence for maximal running capacity would capture many contrasting features between the strains for numerous complex physiological and behavioral elements. A subsample of female rats (12 HCR and 12 LCR) from generation 18 was used in the present study. The animals were housed 2/cage in standard conditions (temperature 22°C, humidity 50±10%, light from 8.00 AM to 8.00 PM), and they had free access to tap water and food pellets (R36; Labfor, Stockholm, Sweden). The experimental procedures were approved by the Animal Care and Use Committees of the University of Jyväskylä and the University of Michigan.
Measurements of voluntary running activity, energy intake, and body weight
The timeline of all measurements is presented in Fig. 1. To analyze the voluntary activity of the rats, they were housed individually for 3 wk in a cage with a running wheel, which was connected to a computerized recording system. Energy intake was measured during the 3 wk in a cage with a running wheel and during 6 wk in a normal cage. After the 3 wk period in cages with running wheels, the rats spent ≥6 wk in normal cage conditions before further measures to avoid the possible effects of running on the basal level analyses.
Figure 1.
Course of the experiments.
Respirometry
We measured energy metabolism of weighed individuals in a postabsorptive stage (access to food was restricted 12 h before measurements) at 22°C. The open flow respirometry system consisted of 4 measurement chambers (1728 cm3) and empty baseline chamber that were measured sequentially for 20 min each, for 3.5 to 7 h. The incoming air (3100 ml/min) was controlled (Flow Bar 8; Sable Systems, Las Vegas, NV, USA), subsampled (RM-8; Sable Systems), and dried (Drierite; Hammond Drierite, Xenia, OH, USA) before entering the CO2 analyzer (LI-6252; LI-COR, Lincoln, NE, USA). The flow to analyzer was controlled by the gas analyzer subsampler (Sable Systems). The minimum CO2 production values (acquired by Datacan V software; Sable Systems), when the animals were not moving, were used to resemble minimal energy use requirements.
Intraperitoneal glucose tolerance test
Rats were deprived of food for 12 h, blood glucose was measured, and 2 g/kg of glucose was injected into the peritoneal cavity. Glucose concentration was measured at 0, 30, 60, and 90 min after the injection with HemoCue B-Glucose Analyzer (HemoCue AB, Ängelholm, Sweden). The area under the curve was used to assess in vivo glucose clearance.
Tissue preparation
Rats were killed at 9 mo of age. Soleus, extensor digitorum longus (EDL), and gastrocnemius muscles and the heart were excised, and blood samples were taken. Samples were snap-frozen in liquid nitrogen. In addition, the midbelly of the muscles was transversally oriented, mounted on Tissue-Tek, and snap-frozen in isopentane cooled with liquid nitrogen for histology.
Serum analyses
Insulin was analyzed with an Ultra Sensitive Rat Insulin ELISA kit (Crystal Chem Inc., Downers Grove, IL, USA). Triglycerides, total cholesterol, and HDL-cholesterol were measured using the VITROS DT60 II chemistry system (Ortho-Clinical Diagnostics, Rochester, NY, USA). The LDL-cholesterol concentration was estimated using Friedewald's equation (10). Free fatty acids were determined by an enzymatic colorimetric method of the Wako nonesterified fatty acid (NEFA) C test kit (Wako Chemicals GmbH, Neuss, Germany), scaled down to a microplate format. The homeostatic model assessment (HOMA) index was calculated as the product of the insulin level (μU/ml) and the glucose level (mM).
Immunohistochemistry and ATPase staining
Serial cross-sections (8 μm) were cut from soleus, extensor digitorum longus, and gastrocnemius muscles in a cryomicrotome (n=12 LCR+12 HCR for all muscles). Capillaries were visualized by staining with Isolectin-GS-IB4 containing fluorescent Alexa Fluor 488 label (Molecular Probes, Eugene, OR, USA; 1:200 dilution in 1% BSA/PBS). This was combined with antidystrophin staining (Novocastra, Newcastle on Tyne, UK; 1:500 dilution in 1% BSA/PBS) with Alexa Fluor 555 secondary antibody (Molecular Probes; 1:300 dilution in 1% BSA/PBS) to visualize muscle fibers. For light microscopy, soleus and EDL muscle sections were stained with dystrophin antibody (1:500 dilution) using DAB as a substrate for fiber size analysis. ATPase staining was used to analyze type I and type II fiber type proportions (12). Sections were visualized with Olympus BX-50 fluorescent microscope, and capillary density (cap/mm2), capillary-to-fiber ratio, fiber size, and fiber type distribution were analyzed with AnalySIS (Olympus, Jena, Germany) and TEMA (Scanbeam, Hapsund, Denmark) software in a blinded manner.
Myosin heavy chain (MHC) composition
The MHC isoform composition of the soleus, EDL, and gastrocnemius muscles was determined by SDS-PAGE according to a previously described method with slight modifications (13). Samples were loaded into the SDS-PAGE gel system consisting of stacking gel with 3% acrylamide and separating gel with 6% acrylamide and 30% glycerol. The gels were run at a constant voltage of 70 for 42 h at 4°C. In the Coomassie blue stained gels, 3 distinct protein bands could be separated and identified as MCH I, IIb, and IIa+x according to their migration characteristics in the gel. The relative proportion of each MHC isoform was determined by densitometric analysis using Quantity One software (Bio-Rad, Hercules, CA, USA).
Electron microscopic analysis of mitochondrial content
Pieces of soleus muscles were fixed with 3% glutaraldehyde in phosphate buffer for 2–3 h. After postfixation with 1% osmiumtetroxide for 1 h, the specimens were dehydrated and embedded in LX-112 (Ladd, Williston, VT, USA). Thin sections were cut and double stained with uranyl acetate and lead citrate and examined with a Jeol JEM-1200 electron microscope (Jeol Ltd., Akishima, Japan). The best section of each block was chosen, and 10–15 micrographs were taken from different cells at ×1500–2500 primary magnification so that sarcolemmal areas were included. The amount of subsarcolemmal mitochondria was expressed as mitochondrial area (μm2) and related to the length of sarcolemma (1000 μm) under which it was measured. Images from 10 LCR and 8 HCR rats were analyzed. Intermyofibrillar mitochondria were calculated as a percentage of the total muscle fiber area, and micrographs from 5 rats from both groups were analyzed.
RNA extraction
Total RNA was isolated from soleus, EDL, and gastrocnemius muscles with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. The quality of RNA was confirmed by spectrophotometry and agarose gel electrophoresis.
Illumina microarray
RNA samples from gastrocnemius muscle of all 24 rats were analyzed with genome-wide Illumina Sentrix RatRef-12 BeadChip (BD-27-303; Illumina Inc., San Diego, CA, USA) that contained 22,523 probes. The microarray hybridizations were performed by the Finnish DNA Microarray Center at Turku Center for Biotechnology according to the manufacturer's instructions. Data analyses were performed with an R software environment for statistical computing, including BioConductor development software. The data were normalized with quantiles normalization from an affy package of BioConductor (14). The quality of the data was assessed by calculating Pearson correlations and clustering. Statistically significant differences in individual genes between HCR and LCR rats were tested using t statistics and Benjamin-Hochberg algorithm controlling false discovery rate (FDR). Adjusted values of P < 0.05 were considered statistically significant.
Microarray data analysis
Functional enrichment testing was performed using 4 different methods. First, the clustering of differentially expressed genes into functional groups and their significance of overrepresentation among the groups were estimated with the DAVID functional annotation tool (http://david.abcc.ncifcrf.gov/home.jsp; refs. 15, 16). Genes were clustered according to enrichment of Gene Ontology (GO) terms within differentially expressed genes (P<0.05). The statistical significance of clusters was estimated by modified Fisher exact P value. As an alternative to DAVID, we also used LRpath (17) to identify functional groups with significantly higher levels of differential expression. As opposed to DAVID, LRpath allows the data to remain on a continuous scale and, in doing so, allows identification of both functional groups that contain many genes with low differential expression and functional groups that contain few genes with very high differential expression. LRpath uses logistic regression to calculate P values for GO terms and KEGG pathways, and Benjamini-Hochberg algorithm is used to control the FDR. In addition, Reactome Skypainter (http://www.reactome.org/) was used to determine which biological events are statistically overrepresented in our differentially expressed gene set. For a given list of genes, Skypainter can identify common events (functions and pathways) for these genes. Further, GSEA (www.broad.mit.edu/gsea/; ref. 18) was applied for functional clustering of all genes without a priori filtering of the expression data. In the analysis, predetermined curated and motif gene sets of MSigDB database (www.broad.mit.edu/gsea/msigdb/) were utilized to compare HCR and LCR phenotypes.
To compare pheno- and genotypes, the most significant gene clusters obtained from the GSEA analysis were further correlated to physiological and biochemical parameters. For this purpose, leading-edge subsets (i.e., genes producing the GSEA enrichment score) were determined. Common leading-edge genes of several clusters were determined using the leading-edge analysis feature of GSEA (18). The mean centroid of each leading-edge subset was computed by normalizing the expression levels of all subset genes to a mean of 0. Correlation analysis was performed as described later in Statistical Analysis.
Real-time quantitative PCR (qPCR)
Microarray data were verified and supplemented with qPCR measurements for selected genes involved in energy metabolism. In addition to gastrocnemius muscle, which was studied with microarray, we analyzed slow soleus and fast EDL muscles with qPCR. One microgram of total RNA was reverse-transcribed to cDNA using random primers. The ABI Prism 7300 Sequence Detection System was used to perform TaqMan probe-based real-time PCR reactions (Applied Biosystems, Foster City, CA, USA). The primer and probe sets for Lpl (Rn00561482_m1), Cpt1a (Rn00580702_m1), Cpt1b (Rn00566242_m1), Cpt2 (Rn00563995_m1), Hadhb (Rn00592435_m1), Aco1 (Rn00569045_m1), Aco2 (Rn00577876_m1), Ucp2 (Rn01754856_m1), and Ucp3 (Rn00565874_m1) were designed and synthesized by Applied Biosystems. Expression data were normalized to the levels of 2 housekeeping gene mRNAs: GAPDH (Rn99999916_s1) and 18S (Rn01428915_g1). Both housekeeping genes showed no difference between the groups when related to the amount of total RNA or compared with each other and, thus, gave similar results (18S normalized results are presented in Fig. 5). All samples were analyzed in triplicate. Relative quantification was calculated by 2−ΔΔCt using the comparative Ct method as outlined in the Applied Biosystems User Bulletin 2. Amplification efficiencies of primer pairs were checked to be close to each other with dilution series.
Figure 5.
Real-time PCR results from the 3 studied muscles showed that several genes related to lipid oxidation were up-regulated in HCR rats compared with LCR rats: Lpl (A); Cpt1a; (B) Cpt1b (C); Cpt2 (D); Hadhb (E); Ucp2 (F); Ucp3 (G); Aco1 (H); Aco2 (I). *P < 0.05; **P < 0.01; ***P < 0.001.
Statistical analysis
All data are presented as means ± sd. Statistical analyses for all variables except the microarray data (described above) were carried out using SPSS for Windows 13.0 statistical software (SPSS Inc., Chicago, IL, USA). All data were analyzed for normality. Depending on the variable, a 2-tailed Student's t test, 1-way analysis of variance, or analysis of covariance was used to analyze differences between the groups. The significance level was set at P < 0.05. For phenotype microarray correlations, we used Pearson's correlation coefficient to test all gene-phenotype pairs. A histogram of P values was created for each physiological variable to visualize the relative overall level of correlation with gene expression levels. FDR adjustment of P values was performed for genes correlated with different phenotypes.
From respirometry data, we had to exclude 2 individuals due to activity during all measurement cycles. These 2 individuals were also clear outliers (2 sd above the mean). The commonly found effect of mass on respiratory values was accounted for in the analysis of covariance (19). In this analysis, we utilized a hierarchical method (20) in which we explored the effects of genetic background, age, and body mass on metabolic traits. Before analysis, covariates were standardized to a mean of 0, and before this, body mass was ln-transformed. Analysis was initiated from the full model, containing all main effects and second-order interactions between covariates and factor. Subsequently, starting from the 2-way interactions, each nonsignificant interaction was removed at the time.
Linear univariate analysis of variance of physiological/biochemical phenotype-defining parameters with expression data was performed using SPSS. Phenotype-defining parameters were used as dependent variables, rat strain as a fixed factor, and mean centroids of the most significant gene subsets (derived from GSEA analyses) as the independent explanatory covariates.
RESULTS
Physiological characteristics of HCR and LCR rats
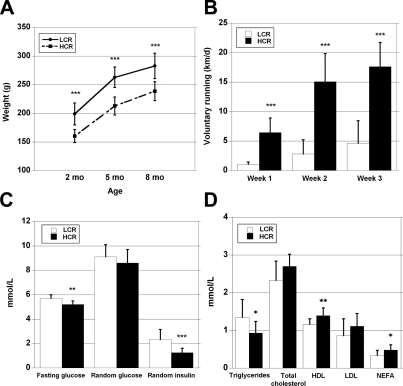
Maximal exercise capacity was determined at 11 wk of age using a standard incremental treadmill running test (4) in 12 LCR and 12 HCR rats born at 18 generations of selection (all females). The mean distance run at exhaustion was 305 ± 16 m in LCRs vs. 1883 ± 45 m in HCRs, which correspond to maximal running times of 20.8 ± 0.8 min and 69.8 ± 1.0 min, respectively. In previous work (9), we showed that selective breeding for running capacity produced a correlated change in body weight such that LCR rats became heavier and the HCR rats lighter. For this subgroup, LCR females were 38 g (24%) heavier (P<0.001) than the HCRs at 2 mo of age and this weight differential was retained at 5 and 8 mo of age (Fig. 2A). Relative weights (see Supplemental Table S1) of the heart and skeletal muscles (soleus, EDL, and quadriceps femoris) normalized to body weight were greater in HCRs (P<0.05).
Figure 2.
A) LCR rats were heavier from young to adult age (P<0.001); however, the relative difference remained similar (∼20%) throughout the study. B) HCR rats ran ∼4 times the distance in a day compared with LCR rats (P<0.001) during the 3 wk period in cages with running wheels. C) LCR rats had higher unfed (fasting) blood glucose and serum insulin than HCR rats, but no statistical difference was observed in intraperitoneal glucose tolerance test or random glucose. D) LCR rats had higher serum triglyceride concentration, and HCR rats, in turn, had higher HDL cholesterol, and NEFA. *P < 0.05; **P < 0.01; ***P < 0.001.
Daily voluntary running distance was ∼4-fold greater (P<0.001) in HCRs compared with LCRs during 3 wk spent in the cages with running wheels (Fig. 2B). HCRs spent more time in the running wheel than LCRs (2.6-fold; P<0.001), which explained most of the difference in the running distance (r=0.96; P<0.001). HCRs consumed more energy than LCRs when the rats had access to running wheels and after the running period (P<0.05). HCR energy intake also tended to be higher before they were given running wheels (7%; P=0.13).
Unfed blood glucose and random serum insulin levels were higher in LCRs relative to HCRs (Fig. 2C), but no significant difference was found between the strains for glucose tolerance (area under the curve analysis). The HOMA index was significantly higher in LCRs (21.6±8.3 vs. 10.8±3.9; P<0.001). Serum triglycerides were higher and high density lipoprotein and free fatty acids were lower in the LCRs compared with the HCRs (Fig. 2D). Blood hemoglobin (135.8±7.6 g/L in LCRs vs. 135.1±9.7 g/L in HCRs) and hematocrit (47.1±1.5 vs. 47.1±1.7) did not differ between the groups.
Resting metabolic rate higher in HCRs
From analysis of covariance, we found that HCR rats had higher CO2 production at rest than LCR rats (150.3±5.8 vs. 125.9±6.4 ml/h; F1,17=5.848; P<0.05; primary data are presented in Supplemental Table S2). This result was evident when the significant effect of body mass (F1,17=14.755; P<0.001) and HCR/LCR-age interaction effect (F1,17=6.663; P<0.05) on CO2 production were taken into account. There was no evidence for significant age effect on CO2 production (F1,17=0.116; P=0.738) or other interaction effects between covariates and selection lines. Regression analyses (regression slopes are shown as correlations r to facilitate comparisons) revealed that in the LCR line, age tended to decrease CO2 production (r=−0.608; n=11; P=0.060), whereas in HCR line, the effect of age was not significant (r=0.371; n=11; P=0.184). These regressions also included the effects of ln-transformed body mass on CO2 production (LCR line: r=0.867; n=11; P<0.05; and HCR line: r=0.662; n=11 P<0.05). Body mass alone increased the CO2 production (whole data: r=0.481; n=22; P<0.05).
Differences in skeletal muscle structure
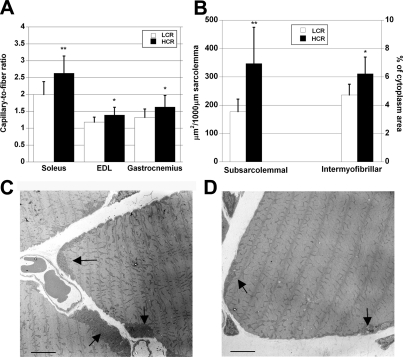
Capillary density (capillaries/mm2) and capillary-to-fiber ratio were significantly greater in soleus (P<0.01) and in mixed gastrocnemius muscle (P<0.05) from HCRs. The capillary-to-fiber ratio in fast EDL muscle (P<0.05) was also greater in the HCRs relative to LCRs (Fig. 3A). No difference between the LCRs and HCRs was observed in the fiber cross-sectional area or fiber type distribution in the analyzed soleus and EDL muscles. The MHC composition analyzed by SDS-PAGE showed a slightly more oxidative phenotype in HCR than in LCR rats for gastrocnemius muscle, with a similar tendency in EDL and soleus (Supplemental Table S3). Subsarcolemmal mitochondrial area, which was analyzed in soleus, was 96% (P<0.01) larger in HCR rats (Fig. 3B), and the largest stocks of mitochondria were localized close to capillaries and around nuclei (Fig. 3C, D). Also, intermyofibrillar mitochondrial area was 32% larger in HCRs (P < 0.05). Only a few intramyocellular lipid droplets were found in either HCR or LCR soleus muscles with EM, and there were no differences between the strains.
Figure 3.
A) Capillary-to-fiber ratio was higher in HCR rats in all studied muscles: slow soleus, fast EDL and mixed gastrocnemius muscles. B) HCR rats had significantly larger subsarcolemmal and intermyofibrillar mitochondrial area compared with LCR rats. C) Large mitochondrial stocks were found under sarcolemma in HCR animals. These stocks were located close to nuclei and blood capillaries (arrows). D) In LCR rat muscle, subsarcolemmal mitochondrial stocks were usually much smaller than in HCR rats (arrows). Scale bars = 5 μm. *P < 0.05; **P < 0.01.
Mitochondrial and lipid metabolism genes up-regulated in HCR muscles
In genome-wide microarray analysis, 239 known or predicted genes were differently expressed between HCR and LCR lines (n=12 HCR+12 LCR; FDR≤0.05). One hundred twenty-six of the genes were up-regulated in HCR vs. LCR, and 113 genes were down-regulated (data not shown). The whole-genome expression patterns were analyzed individually for all 24 rats, which added power and reliability to the results. The expression data were analyzed with 4 different clustering methods (DAVID, Reactome Skypainter, LRPath, GSEA), which all produced notably similar results. The most enriched gene clusters that came up in all analyses were related to mitochondria and lipid metabolism. Functional clustering of the differentially expressed genes according to their GO annotations with DAVID revealed that many of the genes were related to mitochondria, carboxyl acid metabolism, lipid metabolism, oxido-reductase activity, immune response, and protein metabolism (Table 1) In addition, the following events were statistically overrepresented within the set of significantly changed genes by Reactome Skypainter: lipid and lipoprotein metabolism (P<0.02), activated AMPK stimulated fatty acid oxidation in muscle (P<0.004), and metabolism of amino acids (P<0.04).
TABLE 1.
GO categories that were overrepresented among the significantly differentially expressed genes between HCRs and LCRs by functional clustering with DAVID
| Category | ↑ | ↓ |
|---|---|---|
| Mitochondrion** | 7 | |
| Carboxyl acid metabolism** | 9 | |
| Lipid and lipoprotein metabolism** | 7 | |
| Lipid catabolism** | 3 | |
| Oxido reductase activity** | 7 | |
| Immune response** | 8 | |
| Protein metabolism* | 16 | |
| Attachment of cytoskeletal proteins** | 6 |
Values indicate number of genes. ↑, gene expression higher in HCRs than in LCRs; ↓, gene expression lower in HCRs than in LCRs.
FDR P < 0.05;
FDR P < 0.01.
GSEA (18) and logistic regression based method LRpath (17), both exploiting array expression data without requiring a significance cutoff, further accentuated the stature of mitochondrial oxidative and lipid metabolism processes in intrinsic exercise capacity and the development of metabolic risk disease traits. LRpath analysis against GO and KEGG databases revealed 56 enriched gene sets (only sets with <1000 genes/set included) with FDR < 0.01 (Supplemental Table S4). Most enriched gene sets included biological processes such as oxido-reductase activity; valine, leucine,and isoleucine degradation; fatty acid β -oxidation; carboxylic acid metabolic processes; as well as cellular compartments like mitochondrion and ribosome. Furthermore, LRpath analysis of the human homologues of differentially expressed genes against various gene interaction databases revealed 8 gene sets with FDR < 0.01 (Supplemental Table S5), most of the sets showing protein interactions with mitochondrial energy production. GSEA performed on curated gene sets of cellular processes compiled from several databases (MSigDB) revealed 8 gene sets enriched in HCR rats with value of FDR q ≤ 0.001. These sets included oxidative phosphorylation; fatty acid metabolism; Krebs cycle; valine, leucine, and isoleucine degradation; and PPAR-signaling pathway (Table 2). The 2 most enriched sets in LCR gastrocnemius muscle (FDR q≤0.001) spanned genes expressed in renal cell carcinoma and chronic myeloid leukemia (Supplemental Table S6), both sets including genes of various signaling pathways (e.g., MAPK signaling route). GSEA analysis against the gene sets in Biocarta database yielded β -oxidation pathway as the most enriched set in HCRs (FDR q<0.001) and STAT3 pathway as the most enriched set in LCRs (FDR q=0.033). GSEA motif analysis revealed binding sites for 6 known transcription factors upregulated in HCRs (NR2F1, SF1, ER-α, Tel2, Errα, and Gabpa or NRF2) and for 16 factors upregulated in LCRs (PAX3, FOXO4, Nkx6–1, POU6F1, MEF2D, GATA2, MEF2A, ATF3, HOXA4, RORα2, CRX, FOXA1, PTF1A, NRF1, GCM1, and GATA1) with nominal value of P < 0.05.
TABLE 2.
GSEA data
| Gene set name | Size (n) | ES | NES | NOM P | FDR q | FWER P |
|---|---|---|---|---|---|---|
| Overrepresented at HCR | ||||||
| HSA00190_oxidative_ phosphorylation | 78 | 0.4963 | 4.8228 | 0.000 | 0.000 | 0.000 |
| Oxidative_ phosphorylation | 46 | 0.4619 | 3.4915 | 0.000 | 0.000 | 0.000 |
| HSA00071_fatty_acid_ metabolism | 36 | 0.5165 | 3.4853 | 0.000 | 0.000 | 0.000 |
| HSA00020_citrate_cycle | 22 | 0.5801 | 3.1583 | 0.000 | 0.000 | 0.000 |
| Valine_leucine_and_isoleucine_degradation | 29 | 0.4819 | 3.0034 | 0.000 | 0.000 | 0.000 |
| Krebs_tca_cycle | 24 | 0.5318 | 2.9096 | 0.000 | 0.000 | 0.000 |
| HSA00280_valine_leucine_and_isoleucine_ degradation | 35 | 0.4221 | 2.8212 | 0.000 | 0.000 | 0.000 |
| HSA03320_ppar_signaling_pathway | 55 | 0.3290 | 2.6523 | 0.000 | 0.001 | 0.006 |
| Overrepresented at LCR | ||||||
| HSA05211_renal_cell_ carcinoma | 58 | −0.3222 | −2.9330 | 0.000 | 0.001 | 0.0010 |
| HSA05220_chronic_ myeloid_leukemia | 65 | −0.2910 | −2.9188 | 0.000 | 0.001 | 0.0010 |
GSEA performed on curated gene sets of cellular processes compiled from several databases (MSigDB) revealed 8 gene sets enriched in HCR rats and 2 gene sets enriched in LCR rats with FDR q ≤ 0.001. Size, number of genes in the gene set; ES, enrichment score; NES, normalized enrichment score; NOM P, nominal P value; FDR q, false discovery rate q value; FWER P, familywise error rate P value.
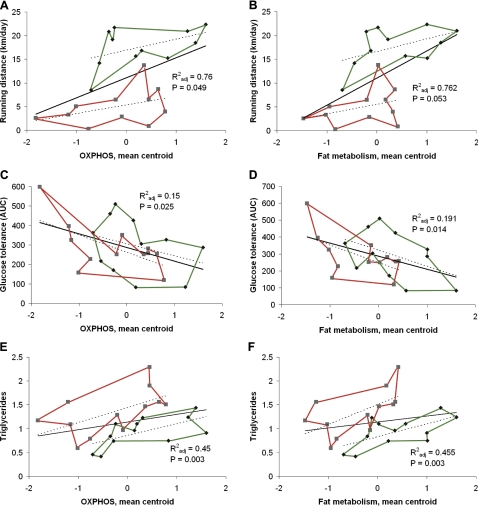
As an unbiased approach for linking phenotypes with gene signatures, we estimated the correlation between individual expression values for each gene probe (22,523 probes) and physiological, biochemical, and disease risk variables for each rat. Voluntary running time and distance, treadmill running performance, energy uptake during voluntary running period, body weight, and relative heart weight, as well as serum insulin and NEFA concentrations were significantly related to skeletal muscle gene expression patterns (Fig. 4 and Supplemental Fig. S1). Voluntary wheel running time correlated significantly with 687/22,523 probe expression levels (FDR q<0.10). Further clustering of these significantly correlated genes revealed enrichment of similar metabolic processes as found from the functional clustering of significantly differentially expressed genes, such as fatty acid metabolic processes and oxidoreductase activity.
Figure 4.
Expression of oxidative phosphorylation (A, C, E) and lipid metabolism genes (B, D, F) predict aerobic capacity (A, B; running distance), insulin sensitivity (C, D; glucose tolerance), and serum triglyceride concentration (E, F). Diamonds indicate HCR values; green lines indicate outer values. Squares indicate LCR values; red lines indicate outer values. Explanatory power and significance are shown.
Univariate analysis of variance showed strong positive relationships between parameters related to running performance and serum triglycerides and mRNA expression centroids of mitochondrial oxidative phosphorylation and fat metabolism. The centroids of mitochondrial oxidative phosphorylation and fat metabolism correlated negatively with glucose tolerance (Fig. 4). Supplemental Fig. S2 shows additional correlations of measures with expression centroids. NEFA correlated positively with 5 centroids, and soleus weight correlated with 3 centroids. Supplemental Table S6 lists the 141 genes that comprise the 7 centroids that ranged in size from 8 genes (STAT3 signaling pathway) to 38 (renal cell carcinoma).
The genes involved in lipid metabolism were more closely examined from the microarray data, and 9 were chosen for further evaluation with real-time qPCR in 3 different muscles: soleus, EDL, and gastrocnemius. Even though the magnitude of the expression changes in individual genes was not large, 7 of the 9 genes showed significantly higher expression in the HCR rats for ≥1 of the 3 muscles tested (Fig. 5)
DISCUSSION
Development of LCR and HCR rats by 2-way artificial selection produced a useful model system for study of aerobic endurance capacity and its correlated traits. The major hypothesis is that alleles at multiple interacting loci that affect intrinsic aerobic capacity have been enriched or fixed differentially between the LCRs and HCRs (4). It is critical that the models are maintained as genetically heterogeneous lines by using a rotational mating paradigm that minimizes inbreeding. Compared with inbred strains, in which essentially all loci have been taken to fixation, outbred selected lines maintain genetic complexity, allowing combinations of allelic variants at multiple interacting loci to be enriched by selection pressure. As a result, the LCR-HCR model system is better suited to discover epistatic interactions, modifier genes, and synergistic actions (21). Importantly, the concurrent breeding of the LCRs and HCRs at every generation allows the lines to serve as reciprocal controls for unknown environmental changes. A limitation of the models is that founder rats contained a finite population of genes such that not all combinations of genes that underlie low and high endurance capacity phenotypes will be represented.
Our data link the segregation of low intrinsic aerobic running capacity with impaired oxidative metabolism of skeletal muscle and increased disease risk factors. Seven mean mRNA centroids comprised of 141 leading-edge genes correlated with 9 endurance exercise capacity and disease risk phenotypes. These results support the hypothesis that small expression differences between LCRs and HCRs for pathways of oxidative energy metabolism (fat metabolism, branched-chain amino acid metabolism, Krebs cycle, and oxidative phosphorylation including electron transport chain) and pivotal regulatory pathways (PPAR and MAPK signaling) account at least partially for the differences in exercise capacity and disease risk phenotypes (Supplemental Table S6). Transcription factor analysis of the mRNA expression data further supports this paradigm. For example, binding sites of the transcriptional factors Errα and Gabpa that were overrepresented in HCRs are partners with PGC-1α, a key regulator of oxidative metabolism and possible modulator of the transcriptional program in diabetic muscle (22). Another enriched site in HCRs binds ERα whose down-regulation is related to insulin resistance and glucose intolerance (23).
Interestingly, the MAPK-signaling pathway is not only implicated in the development of insulin resistance and related diseases but also in the exercise-induced regulation of mitochondrial biogenesis and muscle glucose uptake (24, 25). Our data support the view that MAPK signaling (included in the RCCa-centroid) in skeletal muscle is, along with PPARsignaling, an essential factor and descriptor of intrinsic aerobic capacity. The proteasome-centroid did not correlate with the measured physiological parameters. It is known, however, that saturated fatty acids induce the expression of IL-6 in human myotubes through proteasome-dependent activation leading to phosphorylation of STAT3 (26). In addition to STAT3, IL-6 activates both AMPK and MAPK pathways (27). It is worth noting that these pathways are similar to those that influence disease risks factors and acute exercise-induced metabolic homeostasis but unlike those that seem permissive for determining skeletal muscle adaptations from aerobic exercise training (28).
At 2 mo of age, the LCR rats were significantly heavier than the HCRs and remained ∼20–25% heavier throughout the study. As the relative muscle weights were higher in HCR rats, it is likely that the greater body weight in LCR rats came from adipose tissue. Male LCR rats indeed have been shown to have greater percentage fat mass than HCRs. The difference in body composition between the strains may have affected some of the variables, but it is an integral part of these phenotypes in the selection continuum, as is for the case in humans. In addition, LCR rats had higher blood glucose, serum insulin, and triglyceride concentrations and lower HDL cholesterol, which are all risk factors of the metabolic syndrome. In contrast, HCR rats had significantly higher serum free fatty acid concentratio,n which may reflect enhanced lipolysis from adipose tissue and increased usage of free fatty acids for energy (29). Consistent with this result, it has been demonstrated that HCRs have enhanced capacity for hepatic and skeletal muscle oxidation of lipids relative to LCRs (30, 31).
We confirmed previous reports (32, 33) that lower resting metabolism and lower voluntary activity (and not a higher energy intake) are associated with the higher body weight of the LCRs. These lower expenditures can account for the higher body weight, lower heart weight, and higher body fat in the LCRs relative to HCRs. Of these metabolic disease risk factors, body weight and relative heart weight as well as blood lipids were significantly correlated in GSEA analysis with OXPHOS, TCA cycle, and PPAR centroids. In addition, glucose tolerance was also highly correlated with these centroids. Resting metabolic rate, however, did not correlate significantly with individual genes or gene clusters in either GSEA or whole-genome correlation analysis.
Higher capillarization and larger mitochondrial content in HCR muscles are likely directly causative of both better exercise capacity and reduced disease risk. A dense capillary network enables efficient oxygen delivery to muscle fiber mitochondria, which then can use abundantly available oxygen to provide energy via β-oxidation and the TCA cycle. These structural changes were accompanied with higher expression of mitochondrial, lipid metabolism, and oxidoreductase activity genes in HCRs. These data provide evidence that high intrinsic exercise capacity evolves from both structural components and the regulation of oxygen metabolism pathways. We found no difference in fiber cross-sectional area or type I and II fiber distribution in any of the studied muscles. Analysis of MHC composition, however, showed that HCR rats had more 2a/x MHC compared with LCRs, in which the MHC 2b isoform was more pronounced. This increase in oxidative fibers in HCRs relative to LCRs was very modest compared with the differences in exercise capacity, capillarization, and mitochondrial density.
We previously performed a genome scan on an F2 segregating population derived from crossing Copenhagen (COP; low running capacity) with Dark Agouti (DA; high running capacity) inbred strains. Linkage analysis revealed a significant quantitative trait loci for aerobic running capacity on chromosome 16 that contained several molecular targets relevant to lipid metabolism, but no causative genes were defined (35). Genome scan approaches using inbred strains provide the benefits of experimental and computational simplicity but are problematic because they can only identify genes that have large effects, low epistasis, and high penetrance (18). Because a small variation in the expression of multiple pathways appears to underlie common disease (6), complex animal model systems and approaches as employed here can perhaps provide more realistic information. The exact formula by which mRNA expression differences for intrinsic aerobic capacity transduce into divergence for health risks is unknown. The pathways represented by the 141 genes that comprise the 7 mRNA centroids that correlated significantly with exercise capacity and disease risk phenotypes may provide clues for development of diagnostics and therapeutic agents.
Supplementary Material
Acknowledgments
The study was financially supported by LIKES Foundation for Promotion of Sport and Health Sciences, the Ministry of Education of Finland, and the Academy of Finland. L.G.K. and S.L.B. were supported by grant RR17718 from the National Center for Research Resources of the National Institutes of Health (NIH), U.S. Department of Health and Human Services. M.A.S. was supported by NIH grant 1R01DK077200. The authors acknowledge the expert care of the LCR/HCR rat colony provided by Lori Gilligan and Nathan Kanner. The authors thank Raija Vassinen, Virpi Miettinen, Veijo Kinnunen, Leena Salminen, and Sira Torvinen for help in the cutting and analysis of EM samples.
The authors thank Paavo Niutanen for help with electron micrographs. Microarray experiments were conducted at the Finnish DNA Microarray Center at Turku Center for Biotechnology. The microarray data accession number in GEO is GSE17190.
REFERENCES
- 1. Kokkinos P., Myers J., Kokkinos J. P., Pittaras A., Narayan P., Manolis A., Karasik P., Greenberg M., Papademetriou V., Singh S. (2008) Exercise capacity and mortality in black and white men. Circulation 117, 614– 622 [DOI] [PubMed] [Google Scholar]
- 2. Newman A. B., Simonsick E. M., Naydeck B. L., Boudreau R. M., Kritchevsky S. B., Nevitt M. C., Pahor M., Satterfield S., Brach J. S., Studenski S. A., Harris T. B. (2006) Association of long-distance corridor walk performance with mortality, cardiovascular disease, mobility limitation, and disability. JAMA 295, 2018– 2026 [DOI] [PubMed] [Google Scholar]
- 3. Bray M. S., Hagberg J. M., Perusse L., Rankinen T., Roth S. M., Wolfarth B., Bouchard C. (2009) The human gene map for performance and health-related fitness phenotypes: The 2006–2007 update. Med. Sci. Sports Exerc. 41, 35– 73 [DOI] [PubMed] [Google Scholar]
- 4. Koch L. G., Britton S. L. (2001) Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol. Genomics 5, 45– 52 [DOI] [PubMed] [Google Scholar]
- 5. Jeong H., Tombor B., Albert R., Oltvai Z. N., Barabasi A. L. (2000) The large-scale organization of metabolic networks. Nature 407, 651– 654 [DOI] [PubMed] [Google Scholar]
- 6. Zhu J., Zhang B., Smith E. N., Drees B., Brem R. B., Kruglyak L., Bumgarner R. E., Schadt E. E. (2008) Integrating large-scale functional genomic data to dissect the complexity of yeast regulatory networks. Nat. Genet. 40, 854– 861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Howlett R. A., Gonzalez N. C., Wagner H. E., Fu Z., Britton S. L., Koch L. G., Wagner P. D. (2003) Selected contribution: Skeletal muscle capillarity and enzyme activity in rats selectively bred for running endurance. J. Appl. Physiol. 94, 1682– 1688 [DOI] [PubMed] [Google Scholar]
- 8. Howlett R. A., Kirkton S. D., Gonzalez N. C., Wagner H. E., Britton S. L., Koch L. G., Wagner P. D. (2009) Peripheral oxygen transport and utilization in rats following continued selective breeding for endurance running capacity. J. Appl. Physiol. 106, 1819– 1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wisloff U., Najjar S. M., Ellingsen O., Haram P. M., Swoap S., Al-Share Q., Fernstrom M., Rezaei K., Lee S. J., Koch L. G., Britton S. L. (2005) Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science 307, 418– 420 [DOI] [PubMed] [Google Scholar]
- 10. Mootha V. K., Lindgren C. M., Eriksson K. F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstrale M., Laurila E., Houstis N., Daly M. J., Patterson N., Mesirov J. P., Golub T. R., Tamayo P., Spiegelman B., Lander E. S., Hirschhorn J. N., Altshuler D., Groop L. C. (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267– 273 [DOI] [PubMed] [Google Scholar]
- 11. Friedewald W. T., Levy R. I., Fredrickson D. S. (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin. Chem. 18, 499– 502 [PubMed] [Google Scholar]
- 12. Brooke M. H., Kaiser K. K. (1970) Three “myosin adenosine triphosphatase” systems: The nature of their pH lability and sulfhydryl dependence. J. Histochem. Cytochem. 18, 670– 672 [DOI] [PubMed] [Google Scholar]
- 13. Andersen J. L., Aagaard P. (2000) Myosin heavy chain IIX overshoot in human skeletal muscle. Muscle Nerve 23, 1095– 1104 [DOI] [PubMed] [Google Scholar]
- 14. Gautier L., Cope L., Bolstad B. M., Irizarry R. A. (2004) Affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307– 315 [DOI] [PubMed] [Google Scholar]
- 15. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4, P3 [PubMed] [Google Scholar]
- 16. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44– 57 [DOI] [PubMed] [Google Scholar]
- 17. Sartor M. A., Leikauf G. D., Medvedovic M. (2009) LRpath: A logistic regression approach for identifying enriched biological groups in gene expression data. Bioinformatics 25, 211– 217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545– 15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Packard G. C., Boardman T. J. (1999) The use of percentages and size specific indices to normalize physiological data for variation in body size: Wasted time, wasted effort? Comp. Biochem. Phys. A 122, 37– 44 [Google Scholar]
- 20. Hendrix L. J., Carter M. W., Scott D. T. (1982) Covariance analyses with heterogeneity of slopes in fixed models. Biometrics 38, 641– 650 [PubMed] [Google Scholar]
- 21. Carlborg O., Jacobsson L., Ahgren P., Siegel P., Andersson L. (2006) Epistasis and the release of genetic variation during long-term selection. Nat. Genet. 38, 418– 420 [DOI] [PubMed] [Google Scholar]
- 22. Mootha V. K., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., Willy P. J., Schulman I. G., Heyman R. A., Lander E. S., Spiegelman B. M. (2004) Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. U. S. A. 101, 6570– 6575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Riant E., Waget A., Cogo H., Arnal J. F., Burcelin R., Gourdy P. (2009) Estrogens protect against high-fat diet-induced insulin resistance and glucose intolerance in mice. Endocrinology 150, 2109– 2117 [DOI] [PubMed] [Google Scholar]
- 24. Liu Z., Cao W. (2009) P38 mitogen-activated protein kinase: A critical node linking insulin resistance and cardiovascular diseases in type 2 diabetes mellitus. Endocr. Metab. Immune. Disord. Drug Targets. 9, 38– 46 [DOI] [PubMed] [Google Scholar]
- 25. Wright D. C. (2007) Mechanisms of calcium-induced mitochondrial biogenesis and GLUT4 synthesis. Appl. Physiol. Nutr. Metab. 32, 840– 845 [DOI] [PubMed] [Google Scholar]
- 26. Weigert C., Brodbeck K., Staiger H., Kausch C., Machicao F., Haring H. U., Schleicher E. D. (2004) Palmitate, but not unsaturated fatty acids, induces the expression of interleukin-6 in human myotubes through proteasome-dependent activation of nuclear factor-kappaB. J. Biol. Chem. 279, 23942– 23952 [DOI] [PubMed] [Google Scholar]
- 27. Glund S., Deshmukh A., Long Y. C., Moller T., Koistinen H. A., Caidahl K., Zierath J. R., Krook A. (2007) Interleukin-6 directly increases glucose metabolism in resting human skeletal muscle. Diabetes 56, 1630– 1637 [DOI] [PubMed] [Google Scholar]
- 28. Timmons J. A., Knudsen S., Rankinen T., Koch L. G., Sarzynski M. A., Jensen T., Keller P., Scheele C., Vollaard N. B., Nielsen S., Akerstrom T., Macdougald O. A., Jansson E., Greenhaff P. L., Tarnopolsky M. A., van Loon L. J., Pedersen B. K., Sundberg C. J., Wahlestedt C., Britton S. L., Bouchard C. (2010) Using molecular classification to predict gains in maximal aerobic capacity following endurance exercise training in humans. J. Appl. Physiol. 108, 1487– 1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lessard S. J., Rivas D. A., Chen Z. P., van Denderen B. J., Watt M. J., Koch L. G., Britton S. L., Kemp B. E., Hawley J. A. (2009) Impaired skeletal muscle β-adrenergic activation and lipolysis are associated with whole-body insulin resistance in rats bred for low intrinsic exercise capacity. Endocrinology 150, 4883– 4891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Thyfault J. P., Rector R. S., Uptergrove G. M., Borengasser S. J., Morris E. M., Wei Y., Laye M. J., Burant C. F., Qi N. R., Ridenhour S. E., Koch L. G., Britton S. L., Ibdah J. A. (2009) Rats selectively bred for low aerobic capacity have reduced hepatic mitochondrial oxidative capacity and susceptibility to hepatic steatosis and injury. J. Physiol. 587, 1805– 1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Naples S. P., Borengasser S. J., Rector R. S., Uptergrove G. M., Morris E. M., Mikus C. R., Koch L. G., Britton S. L., Ibdah J. A., Thyfault J. P. (2010) Skeletal muscle mitochondrial and metabolic responses to a high-fat diet in female rats bred for high and low aerobic capacity. Appl. Physiol. Nutr. Metab. 35, 151– 162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Novak C. M., Escande C., Gerber S. M., Chini E. N., Zhang M., Britton S. L., Koch L. G., Levine J. A. (2009) Endurance capacity, not body size, determines physical activity levels: Role of skeletal muscle PEPCK. PloS One 4, e5869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Novak C. M., Escande C., Burghardt P. R., Zhang M., Barbosa M. T., Chini E. N., Britton S. L., Koch L. G., Akil H., Levine J. A. (2010) Spontaneous activity, economy of activity, and resistance to diet-induced obesity in rats bred for high intrinsic aerobic capacity. [E-pub ahead of print] Horm. Behav. doi: 10.1016/j.yhbeh.2010.03.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ways J. A., Cicila G. T., Garrett M. R., Koch L. G. (2002) A genome scan for loci associated with aerobic running capacity in rats. Genomics 80, 13– 20 [DOI] [PubMed] [Google Scholar]
- 35. Ways J. A., Smith B. M., Barbato J. C., Ramdath R. S., Pettee K. M., DeRaedt S. J., Allison D. C., Koch L. G., Lee S. J., Cicila G. T. (2007) Congenic strains confirm aerobic running capacity quantitative trait loci on rat chromosome 16 and identify possible intermediate phenotypes. Physiol. Genomics 29, 91– 97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.