Abstract
Many genes have been described and characterized that have alternative polyadenylation signals at the 3′-end of their pre-mRNAs. Many of these same messages also contain destabilization motifs responsible for rapid degradation of the mRNA. Polyadenylation site selection can thus determine the stability of an mRNA. Fully modified 2′-O-methoxy ethyl/phosphorothioate oligonucleotides that hybridize to the 3′-most polyadenylation site or signal of E-selectin were able to inhibit polyadenylation at this site and redirect it to one of two upstream cryptic sites. The shorter transcripts produced after antisense treatment have fewer destabilization sequences, increased mRNA stability and altered protein expression. This study demonstrates that antisense oligonucleotides can be successfully employed to redirect polyadenylation. This is the first demonstration of the use of oligonucleotides to increase, rather than decrease, abundance of a message.
INTRODUCTION
Eukaryotic genes often have long 3′-UTRs that contain more than one polyadenylation site (1). Choice of polyadenylation site can be critical to mRNA stability if destabilization signals are present in the longer, but not the shorter, form of the message (2). The site used may also influence translation efficiency or localization of an mRNA in a tissue- or disease-specific manner (3–5). At least two sequence elements determine the precise site of 3′-end cleavage and polyadenylation of pre-mRNAs: a highly conserved AAUAAA signal located 10–30 bases 5′ of the cleavage site and a variable GU-rich element 20–40 bases 3′ of the site. A battery of proteins is needed to recognize a polyadenylation signal and cleave the RNA (6,7). These proteins include the 3–4 subunit cleavage and polyadenylation specificity factor (CPSF) which binds the AAUAAA signal, and the 3 subunit cleavage stimulatory factor (CstF) which interacts with the GU-rich element. Two multi-subunit cleavage factor proteins (CFI and CFII) are also required for efficient 3′-end processing. Addition of poly(A) to the newly formed 3′-end is catalyzed by poly(A) polymerase, a single polypeptide. This activity requires RNA bound CPSF and yet another polypeptide called poly(A) binding protein II to help make the full ∼200 nt poly(A) tail.
E-selectin is a cell adhesion molecule transiently expressed on the surface of endothelial cells (8). E-selectin is an example of an mRNA with multiple polyadenylation sites and is one of a number of genes containing AUUUA destabilizing elements. These elements have been shown to confer instability to the mRNA in many systems (2,9,10). It has previously been demonstrated that persistent E-selectin expression in chronically inflamed skin results from the use of an alternative upstream polyadenylation site that yields a shorter mRNA transcript missing several of the destabilizing elements present in the full-length message (11). This shorter transcript has been shown to be more stable than the full-length transcript in endothelial cells, resulting in prolonged expression of E-selectin on the cell surface.
The intent of this study was to determine if antisense oligonucleotides could be used to promote specific regulation of polyadenylation site usage. Since messages containing fewer destabilization elements should have increased stability, targeting certain polyadenylation sites with antisense oligonucleotides should enhance gene expression. The best-characterized class of antisense oligonucleotides are phosphorothioate oligodeoxynucleotides (12) that exert their activity primarily through an RNase H-mediated mechanism (13–16). For this study RNase H activity was not desired, as our goal was to control polyadenylation site selection, not degrade the message. Many nucleotide modifications have been designed that do not support RNase H activity (17–19), including the 2′-O-methoxy ethyl/phosphorothioate (MOE) antisense oligonucleotides used here. In addition to being incapable of supporting RNase H cleavage, oligonucleotides with the MOE modification have increased affinity for target RNA and increased nuclease stability relative to unmodified phosphorothioate oligodeoxynucleotides (20).
MOE oligonucleotides complementary to E-selectin polyadenylation sites and signals were tested for the ability to force usage of alternate polyadenylation signals and sites on the E-selectin message. Oligonucleotides directed to either the 3′-most polyadenylation site or signal of E-selectin were able to inhibit polyadenylation and redirect it to one of two upstream cryptic sites. These alternative transcripts, containing fewer destabilization sequences, had increased mRNA stability and altered protein expression. We believe that this is the first demonstration of the use of oligonucleotides to increase, rather than decrease, abundance of a message.
MATERIALS AND METHODS
Cell culture
Human umbilical vein endothelial cells (HUVEC) were purchased from Clonetics (San Diego, CA) and cultivated in endothelial basal media supplemented with 10% FBS. Cells were treated with oligonucleotides as described previously (21,22). Briefly, cells were incubated with a mixture of Lipofectin reagent and oligonucleotide for 4 h. Cells were incubated from 2 to 24 h then treated with 5 ng/ml TNF-α for 2–3 h to induce expression of E-selectin. Following induction with TNF-α, cells were washed extensively with PBS, fresh complete media was added and cells incubated at 37°C until the time of harvest.
Analysis of RNA
Total RNA was harvested from HUVEC at various times following TNF-α induction using a ToTALLY RNA kit (Ambion) according to the manufacturer’s protocol. For northern blots, RNA was separated on a 1.2% agarose gel containing 1.1% formaldehyde, then transferred to nylon membranes. Blots were hybridized with [32P]dCTP random prime-labeled cDNA probes specific for E-selectin or G3PDH for 2 h in Rapid-hyb solution (Amersham). Blots were washed with 2× SSC containing 0.1% SDS at room temperature, followed by 0.1× SSC containing 0.1% SDS at 60°C. Quantitation of RNA expression was performed using a Molecular Dynamics PhosphorImager and ImageQuant software. Anchored RT–PCR was carried out essentially as described by Chu et al. (11).
Flow cytometry
Following oligonucleotide treatment, cells were detached from the plates with D-PBS (without calcium and magnesium) supplemented with 4 mM EDTA. Cells were transferred to microcentrifuge tubes, pelleted at 1500 g for 1 min and washed in 2% BSA, 0.2% sodium azide in D-PBS at 4°C. E-selectin-phycoerythrin antibody (Ancell) was then added at 1:100 in 0.1 ml of the above buffer. The antibody was incubated with the cells for 30 min at 4°C in the dark. Cells were washed again as above and resuspended in 0.3 ml of PBS buffer with 0.5% formaldehyde. Cells were analyzed for E-selectin expression on a Becton Dickinson FACScan. Results are expressed as a percentage of control expression based upon the mean fluorescence intensity.
RESULTS
E-selectin mRNA includes three polyadenylation signals and a number of destabilizing elements (AUUUA) within the 3′-UTR. All three polyadenylation signals are functional and result in three distinct forms of the transcript (Types I, II and III) (23). The polyadenylation signal for the Type I message is located at position 2823 on the E-selectin message (GenBank accession no. M24736), the Type II at position 2981 and the Type III at position 3816 (Fig. 1). The longest form, Type III, is the predominant form in HUVEC (11). HUVEC were treated with MOE antisense oligonucleotides directed to the Type III signal and site (Table 1) as detailed in Materials and Methods. After ~24 h, cells were treated with TNF-α to induce E-selectin expression. Cells were harvested after 2 h and RNA isolated. E-selectin expression was analyzed by northern blot. Treatment with 106344, an oligonucleotide covering the polyadenylation signal, or 106345, covering the polyadenylation site, resulted in the appearance of a more rapidly migrating RNA species detected by the E-selectin probe (Fig. 2A). We assume that this shorter band represents a combination of Type I and II messages because the Type II message is only 158 bases longer than the Type I and the difference in sizes cannot be resolved on this gel. Quantification of the northern image reveals that with increasing oligonucleotide dose, the longer Type III message decreases while the shorter message increases (Fig. 2B). Although at the highest oligonucleotide doses the Type III message remains in great excess over the Type I/II message, oligonucleotide administration clearly inhibits polyadenylation at the preferred Type III site, enabling utilization of Type I and/or II sites.
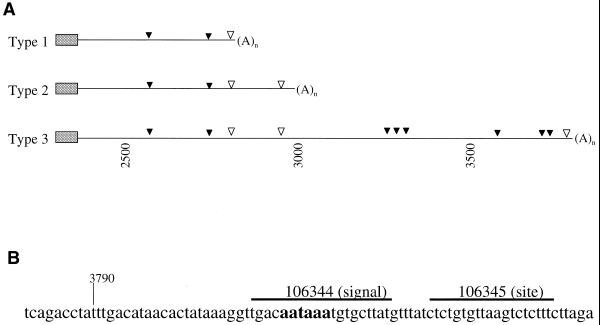
Figure 1.
Schematic illustration of human E-selectin 3′-UTR. (A) Closed triangles, AUUUA mRNA destabilizing elements; open triangles, position of the polyadenylation signal (AAUAAA). (B) The position of antisense oligonucleotides near the Type III polyadenylation site and signal are shown.
Table 1. Oligonucleotides used in the study.
| ISIS no. |
Sequence |
Target |
| 106344 | CATAAGCACATTTATTGTCA | Type III poly(A) signal |
| 106345 | AGAAAGAGACTTAACACAGA | Type III poly(A) site |
| 106346 | CATCAGAACTTATATAGTCA | 106344 mismatch control |
| 106347 | AGACAGTGAATCAACTCAGA | 106345 mismatch control |
| 135568 | GCTTTTATTAGTTCAAAACGTTTGG | Type II poly(A) signal |
| 135570 | CAGAACTTTATTCTGGTTAACATCATG | Type I poly(A) signal |
Figure 2.
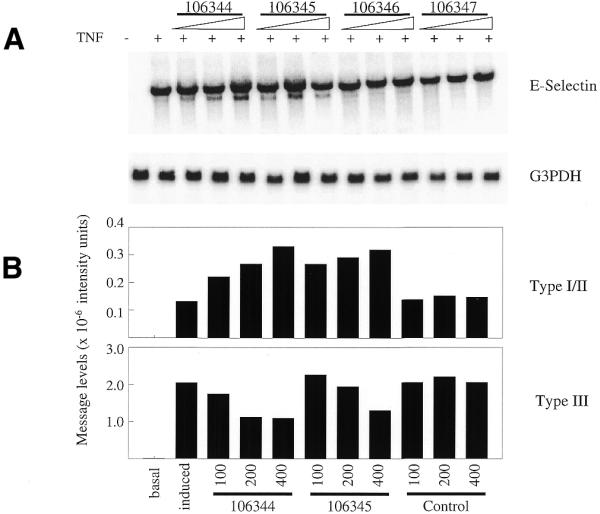
Northern blot analysis of antisense-treated HUVEC. (A) HUVEC were oligonucleotide-treated with antisense oligonucleotides directed to the Type III E-selectin poly(A) signal and site or mismatch controls at doses of 50, 100 or 200 nM. E-selectin expression was then stimulated by the addition of TNF. Total RNA was isolated and analyzed by northern blot using an E-selectin probe. The blot was then stripped and probed for G3PDH RNA expression. (B) Following PhosphorImager scanning, intensity of E-selectin expression was accessed using ImageQuant software. Signals were normalized to corresponding G3PDH expression. Corrected image intensity levels of both the long (Type III) and short (Types I and II) messages are plotted.
Type I and II transcripts lack six of the mRNA destabilizing elements found in the Type III message (Fig. 1). In order to determine if targeting the Type III polyadenylation signal affects RNA stability, cells were treated with 106344 at 200 nM to induce production of the Type I/II message. After overnight incubation the cells were stimulated for 2 h with TNF-α, then washed and fresh media was added without TNF. Cells were harvested at various time-points following the removal of the TNF and total RNA isolated for northern analysis. Approximately one-half of the Type III message remains 2.5 h following TNF removal (Fig. 3). The results are identical in cells not treated with oligonucleotide (data not shown). The stability of the Type III message is in sharp contrast to that of the Type I/II message. More than half of the shorter messages remains 5 h after TNF removal.
Figure 3.
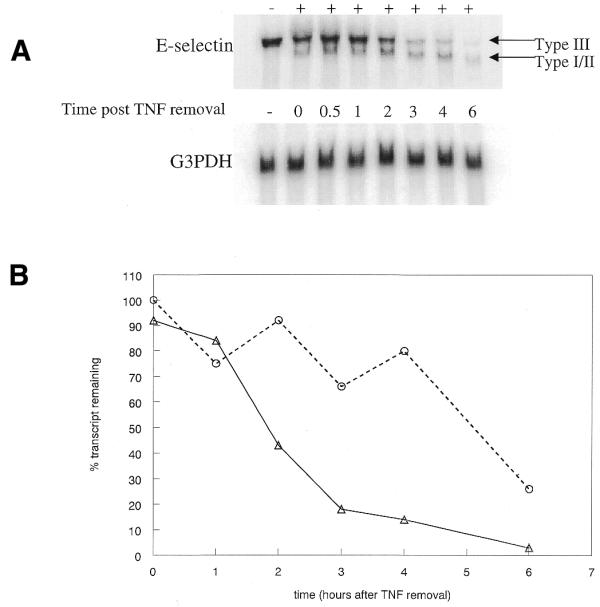
Oligonucleotide-induced Type I/II message has an altered stability. (A) Cells were treated with 200 nM 106344 then E-selectin expression was stimulated by the addition of TNF at 5 ng/ml. After 2 h, TNF was removed and replaced with fresh media. Cells were harvested at various time-points following TNF removal, RNA isolated, then expression of E-selectin and G3PDH analyzed by northern blot. (B) Type III and Type I/II E-selectin RNA levels were normalized to G3PDH expression then plotted relative to the initial E-selectin level following TNF removal. Triangles, Type III; circles, Type I/II.
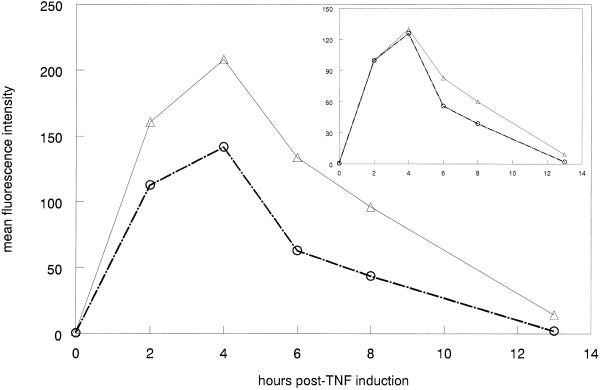
Experiments were also performed to determine if oligonucleotide administration had an effect on protein expression. Cells were treated with 106344 or a control oligonucleotide as above. The treated cells were stimulated with TNF for 2 h then the cells analyzed for E-selectin protein expression by flow cytometry. As previously reported (8), E-selectin is maximally expressed 4 h after addition of TNF for both control and 106344-treated cells (Fig. 4). However, cells treated with 106344 show a sustained expression of E-selectin protein compared to those treated with the control oligonucleotide. The increased duration of protein expression is likely to be due to increased levels of the more stable Type I/II message in antisense-treated cells. Results at the protein level are not as dramatic as at the RNA level due to the fact that the Type III message remains the predominant RNA species (Fig. 2). Any change in protein stability resulting from the presence of the Type I/II message would be partially masked by the more abundant Type III RNA.
Figure 4.
Time course of protein expression in 106433 or mismatch-control-treated HUVEC. HUVEC were treated with 106344 or control at 250 nM for 4 h. E-selectin expression was induced by the addition of TNF at 5 ng/ml, which was removed and replaced with fresh media after 2 h. Cells were harvested 0, 2, 4, 6, 8 or 13 h after initiation of TNF treatment, fixed and stained for E-selectin. Cell surface expression was assessed by flow cytometry. Expression given is the mean fluorescent intensity minus the background signal in the absence of TNF. Triangles, 106344 treated; circles, control oligonucleotide. Inset: data normalized to maximal E-selectin expression.
In order to determine if processing could be controlled more specifically, oligonucleotides were designed to the upstream polyadenylation signals as well. Non-quantitative RT–PCR was used to distinguish between the three forms of the message. In the absence of oligonucleotide all three types of transcript were detected (Fig. 5A, lane 3). These results differ from those obtained by northern blot in which only the Type III message was observed in untreated cells. We attribute the difference to variation in the efficacy of PCR amplification of templates of disparate length, with the shorter Type I and II messages being more efficiently amplified than the Type III product. Differences in amplification may be exaggerated by the high number of cycles required to achieve a satisfactory signal.
Figure 5.
Anchored RT–PCR of E-selectin 3′-ends from antisense-treated HUVEC. (A) Ethidium bromide staining of 1.8% agarose gel containing the products of RT–PCR analysis. Oligonucleotides were administered for 4 h at 200 nM for single oligonucleotide treatment or 150 nM each for dual oligonucleotide treatment. Treated cells were incubated overnight prior to TNF induction of the E-selectin message: –, no TNF treatment; +, 5 ng/ml TNF for 2 h. Bold arrows, expected positions of the Types I, II and III transcripts; thin arrow, expected length of product for the variant poly(A) signal. (B) Diagrammatic representation of the oligonucleotide target sites and position of the PCR primers.
In agreement with the northern blot data, treatment with 200 nM 106344, targeting the Type III signal, resulted in a decrease of the Type III product, while the Type I and II species were both increased relative to their levels in untreated cells (Fig. 5A, lane 4). The antisense oligonucleotides designed to the Type I and II polyadenylation sites are shown schematically in Figure 5B. Oligonucleotide 135568, targeting the Type II site, resulted in a sharp decrease of the Type II message and corresponding increases of the Type I and III species (Fig. 5A, lane 5). The results were similar when 135570, which targets the Type I site, was used (Fig. 5A, lane 6); Type I message was greatly reduced, and Types II and III were increased.
To evaluate whether or not combined treatment with two oligonucleotides would result in the production of a single species of E-selectin message, HUVEC were co-treated with 106344 and either 135568 or 135570 at 150 nM of each oligonucleotide. In both cases, co-administration of oligonucleotides resulted in the primary production of the single species whose polyadenylation signal was not targeted (Fig. 5A, lanes 7 and 8). Cells were also co-treated with 135568 and 135570. The primary transcript detected was Type III (Fig. 5A, lane 9) with its levels increased above those seen in the untreated control (Fig. 5A, lane 3). Interestingly, a band also appears that migrates intermediately between the Type I and II species. Analysis of the mRNA sequence reveals a non-canonical polyadenylation site, AGUAAA, midway between sites I and II that is apparently utilized when these two signals are blocked by the oligonucleotides. A survey of variant polyadenylation signal usage recently published by Beaudoing et al. (24) identifies the AGUAAA hexamer as a common and functional polyadenylation signal.
DISCUSSION
Traditionally, antisense therapeutics have been designed with the intent of specifically reducing the target RNA for a gene whose expression is known to affect a given disease (25). Most commonly, oligonucleotides decrease mRNA levels in cells by an RNase H-mediated mechanism in which endogenous RNase H cleaves the duplex formed by specific binding of an antisense oligonucleotide to its targeted RNA. Additionally, oligonucleotides that do not support RNase H have been used to inhibit translation (26–28) and splicing (29,30) of certain mRNAs. We believe that this is the first demonstration of the use of oligonucleotides to increase levels of a targeted message. This was accomplished by redirecting polyadenylation from a site that results in an mRNA with many destabilization elements to sites resulting in a shorter message with fewer destabilizing elements.
In HUVEC the most abundant species of E-selectin mRNA detected is the full-length transcript that encompasses all eight 3′-UTR destabilizing elements (11). MOE antisense oligonucleotides were designed with complementarity to the 3′-most polyadenylation signal or site. Administration of either oligonucleotide resulted in a shift in polyadenylation site usage from the preferred 3′-site to either of two upstream sites (Fig. 2). One plausible mechanism is that the oligonucleotide covering the polyadenylation signal inhibits interaction of the CPSF with the AAUAAA signal on the message thereby preventing cleavage and subsequent poly(A) addition. Alternatively, the oligonucleotide bound at the polyadenylation site may directly inhibit cleavage by interfering with association of cleavage factor proteins with the message. In these experiments, this oligonucleotide also overlaps the GU/U sequence element that is thought to interact with the CstF. Since the CstF interacts with the CPSF to stimulate cleavage and poly(A) addition, disruption of binding of the CstF may also account for inhibition of polyadenylation.
Both of the short transcripts resulting from upstream polyadenylation lack six of the mRNA destabilizing elements found in the full-length message. In experiments in which rabbit reticulocyte lysates were used to evaluate mRNA stability, Chu et al. (11) found that the half-lives of Type I and Type III messages were 43 and 1 min, respectively. In repeated experiments in cells, we never observed a >3-fold difference in stability between the Type I/II and the Type III message as assayed by northern blot. It is likely that there are major differences in mRNA processing between intact cells and lysates. The difference in findings may also be explained by the use, by Chu et al., of GST–E-selectin fusion constructs instead of the native RNA to obtain half-life results (11).
Duration of protein cell surface expression was also slightly increased by administration of the oligonucleotide targeting the Type III polyadenylation signal (Fig. 4). The effect was not as dramatic as that seen at the mRNA level, most likely due to the low relative abundance (<20%) of the short message compared to the full-length message. It is interesting to note that, in addition to a longer duration of protein expression, there is also an increase in overall E-selectin cell surface expression in the cells treated with 106344. It is possible that the short message may be more efficiently polyadenylated than the long message. It has been demonstrated that polyadenylation site strength can directly influence the amount of cytoplasmic RNA produced from a transcript (5). Once in the cytoplasm, the poly(A) tail on the message can affect the stability of the message and its ability to be translated (31). Therefore, use of cryptic sites may affect not only the stability of the RNA, but also the translation efficiency of the message.
Upstream polyadenylation signals were also successfully targeted, shifting the relative abundance of the three message forms (Fig. 5). Targeting of two sites simultaneously lead to almost exclusive usage of the non-targeted site. Surprisingly, when the upstream canonical polyadenylation signals were blocked, a non-canonical signal was used. A recent large-scale EST clustering analysis, in which over 8700 human 3′-untranslated sequences were compared, identified the alternate polyadenylation signal AGUAAA, as present in 2.7% of the polyadenylation sites (24). The authors speculate that this and other non-canonical signals are processed less efficiently than the canonical signal, AAUAAA. Our data seem to support this hypothesis as the E-selectin AGUAAA signal is used only when the canonical signals are suppressed by hybridization with antisense oligonucleotides.
Although upregulation of E-selectin is not therapeutically useful, the E-selectin mRNA was an ideal target to demonstrate the use of oligonucleotides as a mechanism to redirect polyadenylation and enhance gene expression. A large number of genes with alternative polyadenylation sites, many resulting in messages with variable numbers of destabilization sequences, have been identified (1,4). It is likely that many other such messages, some with therapeutic potential, will be identified as cDNA databases continue to expand.
Acknowledgments
ACKNOWLEDGEMENTS
We are most grateful to Prof. Jim Manley and Tom Condon for helpful scientific discussions.
References
- 1.Gautheret D., Poirot,O., Lopez,F., Audic,S. and Claverie,J.M. (1998) Alternate polyadenylation in human mRNAs: a large-scale analysis by EST clustering. Genome Res., 8, 524–530. [DOI] [PubMed] [Google Scholar]
- 2.Shaw G. and Kamen,R. (1986) A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediated selective mRNA degradation. Cell, 46, 659–667. [DOI] [PubMed] [Google Scholar]
- 3.Wahle E. and Keller,W. (1996) The biochemistry of polyadenylation. Trends Biochem. Sci., 21, 247–250. [PubMed] [Google Scholar]
- 4.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denome R.M. and Cole,C.N. (1988) Patterns of polyadenylation site selection in gene constructs containing multiple polyadenylation signals. Mol. Cell. Biol., 8, 4829–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Proudfoot N. (1996) Ending the message is not so simple. Cell, 87, 779–781. [DOI] [PubMed] [Google Scholar]
- 7.Wahle E. and Ruegsegger,U. (1999) 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol. Rev., 23, 277–295. [DOI] [PubMed] [Google Scholar]
- 8.Bevilacqua M.P., Pober,J.S., Mendrick,D.L., Cotran,R.S. and Gimbrone,M.A. (1987) Identification of an inducible endothelial-leukocyte adhesion molecule. Proc. Natl Acad. Sci. USA, 84, 9238–9242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sachs A.B. (1993) Messenger RNA degradation in eukaryotes. Cell, 74, 413–421. [DOI] [PubMed] [Google Scholar]
- 10.Belasco J., Greenberg,M. and Zubizgz,A. (1995) The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol., 15, 2219–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu W., Presky,D.H., Swerlick,R.A. and Burns,D.K. (1994) Alternatively processed human E-selectin transcripts linked to chronic expression of E-selectin in vivo. J. Immunol ., 153, 4179–4189. [PubMed] [Google Scholar]
- 12.Crooke S.T. (1995) Antisense therapeutics. In Cuello,A. and Collier,B. (eds), Pharmacol. Sci.: Perspect. Res. Ther. Late 1990s, 12th International Congress on Pharmacology. Birkhauser, Basel, Switzerland, pp. 393–399.
- 13.Stein C.A., Subasinghe,C., Shinozuka,K. and Cohen,J.S. (1988) Physicochemical properties of phosphorothioate oligodeoxynucleotides. Nucleic Acids Res., 16, 3209–3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toulme J.J. and Helene,C. (1988) Antimessenger oligodeoxyribonucleotides: an alternative to antisense RNA for artificial regulation of gene expression – a review. Gene, 72, 51–58. [DOI] [PubMed] [Google Scholar]
- 15.Monia B.P., Johnston,J.F., Ecker,D.J., Zounes,M.A., Lima,W.F. and Freier,S.M. (1992) Selective inhibition of mutant Ha-ras mRNA expression by antisense oligonucleotides. J. Biol. Chem., 267, 19954–19962. [PubMed] [Google Scholar]
- 16.Lima W.F. and Crooke,S.T. (1997) Binding affinity and specificity of Escherichia coli RNase H1: impact on the kinetics of catalysis of antisense oligonucleotide–RNA hybrids. Biochemistry, 36, 390–398. [DOI] [PubMed] [Google Scholar]
- 17.Cook P.D. (1993) Medicinal chemistry strategies for antisense research. In Crooke,S.T. and Lebleu,B. (eds), Antisense Research and Applications. CRC Press, Boca Raton, FL, pp. 149–187.
- 18.Kogoma T., Subia,N.L. and von Meyenburg,K. (1985) Function of ribonuclease H in initiation of DNA replication in Escherichia coli K-12. Mol. Gen. Genet., 200, 103–109. [DOI] [PubMed] [Google Scholar]
- 19.Sproat B.S. and Lamond,A.I. (1993) 2′-O-alkyloligoribonucleotides. In Crooke,S.T. and Lebleu,B. (eds), Antisense Research and Applications. CRC Press, Boca Raton, FL, pp. 351–362.
- 20.Cook P.D. (1998) Second generation antisense oligonucleotides: 2′-modifications. Ann. Rep. Med. Chem., 33, 313–325. [Google Scholar]
- 21.Chiang M.Y., Chan,H., Zounes,M.A., Freier,S.M., Lima,W.F. and Bennett,C.F. (1991) Antisense oligonucleotides inhibit intercellular adhesion molecule 1 expression by two distinct mechanisms. J. Biol. Chem., 266, 18162–18171. [PubMed] [Google Scholar]
- 22.Vickers T.A., Wyatt,J.R. and Freier,S.M. (2000) Effects of RNA secondary structure on cellular antisense activity. Nucleic Acids Res., 28, 1340–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kluger M.S., Johnson,D.R. and Pober,J.S. (1997) Mechanism of sustained E-selectin expression in cultured human dermal microvascular endothelial cells. J. Immunol., 158, 887–896. [PubMed] [Google Scholar]
- 24.Beaudoing E., Freier,S., Wyatt,J., Claverie,J.M. and Gautheret,D. (2000) Patterns of variant polyadenylation signal usage in human genes. Genome Res., 10, 1001–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crooke S. (1992) Therapeutic applications of oligonucleotides. Ann. Rev. Pharmacol. Toxicol., 32, 329–376. [DOI] [PubMed] [Google Scholar]
- 26.Baker B.F. and Monia,B.P. (1999) Novel mechanisms for antisense-mediated regulation of gene expression. Biochim. Biophys. Acta, 1489, 3–18. [DOI] [PubMed] [Google Scholar]
- 27.Mologni L., Nielsen,P.E. and Gambacorti-Passerini,C. (1999) In vitro transcriptional and translational block of the bcl-2 gene operated by peptide nucleic acid. Biochem. Biophys. Res. Commun., 264, 537–543. [DOI] [PubMed] [Google Scholar]
- 28.Baker B.F., Lot,S.S., Condon,T.P., Cheng-Flournoy,S., Lesnik,E.A., Sasmor,H.M. and Bennett,C.F. (1997) 2′-O-(2-methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem., 272, 11994–12000. [DOI] [PubMed] [Google Scholar]
- 29.Kole R., Shukla,R.R. and Akhtar,S. (1991) Pre-mRNA splicing as a target for antisense oligonucleotides. Adv. Drug Del. Rev., 6, 271–286. [Google Scholar]
- 30.Taylor J.K., Zhang,Q.Q., Wyatt,J.R. and Dean,N.M. (1999) Induction of endogenous Bcl-xS through the control of Bcl-x pre-mRNA splicing by antisense oligonucleotides. Nat. Biotechnol., 17, 1097–1100. [DOI] [PubMed] [Google Scholar]
- 31.Sachs A.B., Sarnow,P. and Hentze,M.W. (1997) Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell, 89, 831–838. [DOI] [PubMed] [Google Scholar]