SYNOPSIS
ARHI (DIRAS3) is an imprinted tumor suppressor gene whose expression is lost in the majority of breast and ovarian cancers. Unlike its homologs Ras and Rap, ARHI functions as a tumor suppressor. Our previous study showed that ARHI can interact with the transcriptional activator Stat3 and inhibit its nuclear translocation in human breast and ovarian cancer cells. To identify proteins that interact with ARHI in nuclear translocation, we have performed proteomic analysis and identified several importins that can associate with ARHI. To further explore this novel finding, we have purified 10 GST-importin fusion proteins (importin 7, 8, 13, β1, α1, α3, α5, α6, α7 as well as mutant α1). Using a GST-pull down assay, we found that ARHI can bind strongly to most importins; however, its binding is significantly reduced with an importin α1 mutant which contains an altered nuclear localization signal (NLS) domain. In addition, an ARHI N-terminal deletion mutant (NTD) exhibits much less binding to all importins than does wild type ARHI. In nuclear import assays, addition of ARHI blocked nuclear localization of phosphorylated Stat3. ARHI protein also inhibits interaction of Ran-importin complexes with GFP fusion proteins that contain an NLS domain and a beta-like import receptor binding domain, blocking their nuclear localization. By GST-pull down assays, we found that ARHI could compete for Ran-impotin binding. Thus, ARHI-induced disruption of importin binding to cargo proteins including Stat3 could serve as an important regulatory mechanism that contributes to the tumor suppressor function of ARHI.
Keywords: ARHI, importin, nuclear import, Ran, Stat3, nuclear translocation
INTRODUCTION
Transport of macromolecules between the nucleus and cytoplasm is critical for the normal function of eukaryotic cells. Two groups of karyopherins - importins and exportins - mediate RanGTPase-dependent transport through the nuclear pore [1]. During malignant transformation, aberrant nucleocytoplasmic transport of transcription factors (such as Stat3 and E2F1) [2, 3] and their regulatory kinases (such as Sgk and Erk/MAPK) [4] occurs through impaired nuclear import, enhanced export, suppression of degradation, and sequestration in protein aggregates. Conversely, secreted factors such as Cysteine-rich protein 61, Connective tissue growth factor, and Nephroblastoma overexpressed protein (CCN) proteins, Epidermal Growth Factor (EGF), Fibroblast Growth Factors (FGFs) and their receptors are often detected in the nucleus of cancer cells. Nuclear localization of these molecules has been correlated with tumor progression and poor prognosis for patient survival [5, 6].
The classical nuclear import pathway consists of importin α and β. Whereas importin α interacts with nuclear localization signal (NLS) in the cargo, importin β binds to the autoinhibitory domain on importin α and mediates the transport of the trimeric complex from the cytoplasm to the nucleus through the nuclear pores. Once inside the nucleus, the small GTP binding protein Ran (RanGTP) dissociates the complex by interacting with importin β. Importin α and β are shuttled separately back to the cytoplasm [7]. Importin α family includes importin α1, α3, α4, α5, α6 and α7 [8]. There are 20 members in the importin β superfamily such as importin β1, 7, 8, 9, and 13 [9, 10, 11, 12, and 13]. Importin αs are composed of a flexible N-terminal importin-beta-binding (IBB) domain. The flexible IBB domain interacts either in trans with importin β or in cis with the classical NLS (cNLS)-binding groove [8]. Importin βs have in common an N-terminal Ran binding domain. Importins direct the import of various cargoes and may have different functions. For example, importin β/importin 7 heterodimer is a functional nuclear import receptor for histone H1 [10]; importin β, transportin, importin 7, and importin 9 promoted efficient import of c-Jun into the nucleus; importin alpha, by contrast, inhibited nuclear import of c-Jun in vitro [11]. Importin 13, a recently identified importin β family member, regulates nuclear import of the glucocorticoid receptor in airway epithelial cells [12, 13].
Ran is a small Ras-like GTP-binding protein that switches between a GTP- and a GDP-bound form by GTP hydrolysis and nucleotide exchange [14]. The GTPase Ran plays a crucial role in nucleo-cytoplasmic transport of tumor suppressors, proto-oncogenes, signaling molecules and transcription factors. The RanGTPase cycle provides directionality to nucleocytoplasmic transport, regulating interactions between cargoes and nuclear transport receptors of the importin-β family. The common principle underlying these diverse functions throughout the cell cycle is thought to be anisotropy of the distribution of RanGTP (the RanGTP gradient), driven by the chromatin-associated guanine nucleotide exchange factor RCC1 [15].
ARHI is a maternally imprinted tumor suppressor gene that encodes a 26 kD protein with 55–62% homology to Ras and Rap [16]. In contrast to Ras, ARHI contains a 34 amino acid N-terminal extension and inhibits growth, motility and invasion of cancer cells [16, 17]. Our recent research found that ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells by downregulating PI3K and Ras/MAP signaling, downregulating mTOR [18]. ARHI can also interact with transcription activator Stat3 and inhibit its nuclear translocation and transcription activity in human breast and ovarian cancer cells [19]. The ARHI N-terminal deletion mutant (NTD) has markedly reduced growth inhibitory activity, suggesting that this unique extension may contribute to ARHI’s inhibitory effects on Stat3-mediated transcriptional activities [19]. To identify additional ARHI-interacting proteins, we have performed proteomic analysis and found that ARHI is complexed with several importin proteins. We have explored the possibility that ARHI might displace cargo proteins, including Stat3, and inhibit their nuclear localization.
MATERIALS AND METHODS
Cell lines and reagents
The SKBr3 human breast cancer cell line was maintained as described [17]. HeLa cells were grown at 37°C to near confluence in Dulbecco’s Modified Eagle’s Medium containing 10% fetal bovine serum. Constructs of Ran and importin α1 mutant that contained an altered NLS domain, as well as anti-importin β1 antibody were provided by Dr. Karsten Weis (University of California Berkeley). Constructs of GST, GST-importin 7, GST-importin 8, GST-GFP, GST-GFP-BIB (beta-like import receptor binding), GST-GFP-NLS were provided by Dr. Keith Yamamoto (University of California San Francisco) [20]. Anti-importin 7 and anti-importin 9 antibodies were obtained from Dr. Dirk Görlich (University of Heidelberg, Germany). Anti-Ran and anti-GST antibodies were purchased from Upstate Biotechnology Inc. (Billerica, MA). Anti-ARHI antibody was produced by our group as previously described [17]. Adenovirus constructs expressing LacZ, ARHI and NTD were prepared [21]. The reagents for a nuclear import assay (digitonin, ATP, GTP, creatine phosphate, creatine kinase) were purchased from Sigma (St. Louis, MO).
Proteomic analysis
Wild-type ARHI and its NTD mutant were re-expressed in the breast cancer cell line SKBr3 with a dual adenovirus system [21]. LacZ adenovirus-infected cells were used as a control. ARHI interacting proteins were immunoprecipitated using an ARHI monoclonal antibody and further purified using Protein G beads (Pierce, Rockfold, IL). Proteins were separated on a 15% SDS-PAGE gel and analyzed by mass spectrometry at the University of Texas M.D. Anderson Cancer Center Proteomics Facility.
Western blot analysis
Confluent SKBr3 cells were lysed and the cell lysates were clarified by centrifugation for 5 min at 14,000 rpm at room temperature. Equal amounts of total cellular proteins were electrophoresed in 15 % SDS/PAGE and transferred to polyvinylidene fluoride membrane (Millipore, Billerica, MA). The membranes were immunoblotted with antibodies and signals measured using an ECL system (Amersham, Piscataway, NJ).
Fusion protein expression and purification
GST, GST-importin α1, α3, α5, α6, α7, GST-importin α1 mutant, GST-importin β1, 7, 8, and 13 were all expressed in BL21 (DE3) Escherichiacoli (Stratagene). Overnight bacterial cultures in LB/Ampicillin werediluted 1/100 and grown to an OD595 of 0.6 to 0.8 at 37°C. Cultures were cooled, induced with 0.25 mM IPTG overnight at 16°C and harvested by centrifugation. Cell pellets were re-suspended in lysis buffer (50 mM Tris, pH 8.0,150 mM NaCl, 1 mM EDTA, 1 mM DTT, 1 μg/mlleupeptin, 1 μg/ml pepstatin, 1 μg/mlaprotinin, and 1 mM PMSF), sonicated, and clarified at 40,000 × g for 30 min at 4°C. After centrifugation, the supernatant fraction was further purified on an AKTA FPLC system using a glutathione sepharose column and a Superdex 200 gel filtration column. Tyrosine phosphorylated Stat3β (residues 127–722) (pStat3β) was purified as published [22], briefly; the core fragment of STAT3β was co-expressed with the Elk kinase in E. coli. The identity of the phosphoprotein was confirmed by using mass spectrometry.
The ARHI and NTD cDNA sequences were PCR amplified from pcDNA3-ARHI plasmid DNA and cloned into the pQE30 vector. The importin 7 coding sequence was retrieved from the GST-importin 7 plasmid by restriction digestion and inserted into the pQE30 vector. For the nuclear import substrates, GST-GFP-BIB and GST-GFP-NLS proteins were purified by glutathione sepharose beads (Amersham) according the manufacturer’s instructions. pQE-Ran, pQE-importin 7, pQE-ARHI, and pQE-NTD were expressed in SG13009 competent cells and purified as described by Nachury and Weis [23]. Importin β1 was cleaved from GST-β1 fusion by PreScission protease (GE Healthcare, Piscataway, NJ) and importin α7 was cleaved from GST-α7 fusion using TEV protease (Invitrogen, Carlsbad, CA), and the mixture was loaded on a HiTrap Q cartridge (Amerhsam) to separate GST from the importin proteins. All fusion proteins were concentrated using Centricon 30 concentrators, aliquoted, flash frozen in liquid nitrogen, and stored at −80 °C.
Labelling of pStat3β
pStat3β proteins were FITC-labeled using a FITC Protein Labeling kit (Pierce) according manufacturer’s instructions.
GST pull-down assay
GST-importin fusion proteins (2–4 μg) were mixed with 10–30 μg of cell lysates from SKBr3 cells in binding buffer (20 mM Hepes, pH 7.9, 150 mM NaCl, 0.5 mM EDTA, 10 % Glycerol, 0.1% Triton X-100, 1 mM DTT). The mixture was incubated with shaking at 4°C for 1 h. Fifteen microliters of glutathione-sepharose beads, pre-equilibrated in TEE buffer (50 mM Tris, pH 7.9, 1 mM EDTA, 1 mM EGTA) were added to the mixture and incubated with shaking at 4°C for another 1 h. The beads were collected and washed 4–5 times with Washing buffer (20 mM Hepes, pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 1% glycerol, 1% Triton X-100) and once with ice-cold PBS. The beads were resuspended in 20 μl of 2X SDS loading buffer and then boiled for 5 min. The soluble proteins were separated on 15% SDS-PAGE gels for Coomassie blue staining or western-blotting analysis.
Nuclear import assay
HeLa cells were grown on glass cover slips for 40–42 h. Cells were then washed twice with ice-cold Transport buffer (20 mM HEPES, pH 7.3, 110 mM KOAc, 5 mM NaOAc, 2 mM MgOAc, 2 mM DTT, 1 mM EGTA), incubated with 40 μg/ml digitonin in Transport buffer for 6 min on ice, washed twice in cold Transport buffer, and kept on ice for 20 min. Cover slips containing permeabilized cells were incubated for 30 min at 30°C with a 30 μl nuclear import reaction mixture [2 μM GFP-tagged import substrate or 4 μM FITC labeled p-Stat3β and an ATP-regenerating system (1 mM ATP, 5 mM creatine phosphate, 10 U/ml creatine kinase, 0.5 mM GTP)]. Some experiments were conducted with 3 μM RanGDP, 3 μM importin α7, 2 μM importin β1, 3 μM ARHI or 3 μM NTD. After the import reactions were completed, the cells were rinsed once in PBS, fixed in 4% paraformaldehyde in PBS, and analyzed by fluorescence microscopy.
RESULTS
ARHI interacts with importins
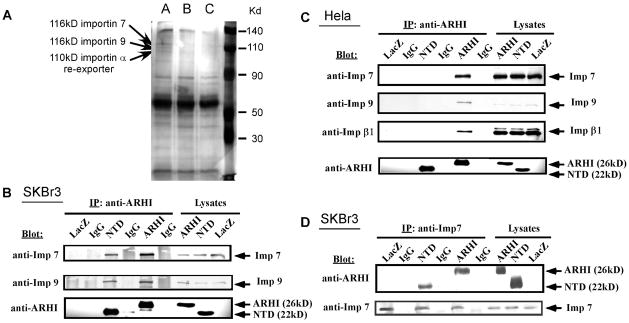
Our previous studies have shown that, when ARHI and Stat3 were both expressed in SKOv3 cells, ARHI formed a complex with Stat3 in the cytoplasm and prevented interleukin-6–induced Stat3 accumulation in the nucleus (19). To identify the protein(s) interacting with ARHI in the nuclear import process, we performed proteomic analysis. Wild-type ARHI and its NTD mutant ARHI were expressed in SKBr3 breast cancer cells with a dual adenovirus system [21]. Cells infected with the LacZ adenovirus served as a control. ARHI- or NTD-complexes were immunoprecipitated using a specific ARHI antibody and co-immunoprecipitated proteins were analyzed by mass spectrometry. Several novel interacting proteins were identified in complexes with wild type ARHI that bound less intensely to the NTD mutant. Three of these proteins (Fig. 1A) belong to the family of importins (importin 7, importin 9, importin α re-exporter). These results were further confirmed by co-immunoprecipitation assays. As shown in Figure 1B and 1C, in SKBr3 and HeLa cells, ARHI antibody could immunoprecipitate importin 7, 9 and β1, and improtin 7 antibody could also immunoprecipitate ARHI (Fig. 1D). NTD:importin binding was much reduced compared to wild-type ARHI:importin interaction.
Figure 1.
ARHI-importin form complexes. (A) Proteomic analysis. Wild-type ARHI and its NTD mutant were expressed in the SKBr3 breast cancer cells using a dual adenovirus system. LacZ adenovirus infected cells served as a control. A: ARHI complexes; B: NTD complexes; C: LacZ control. Arrows point to protein bands corresponding to the indicated novel binding proteins. (B–D) Immunoprecipitation of ARHI-importin complexes. Protein lysates from (B) SKBr3 or (C) HeLa cells infected with ARHI, NTD, or LacZ adenoviruses were incubated with anti-ARHI antibody. The immunoprecipitates were analyzed by Western blotting with anti-importin (Imp) 7, 9, β1 antibodies. Mouse IgG was used as control. To confirm the efficiency of the assays, the immunoprecipitates were also analyzed by Western blotting with anti-ARHI antibody. (D) Proteins immunoprecipitated with anti-importin 7 antibodies were analyzed by Western analysis with anti-ARHI antibody. To confirm the efficiency of the assays, the immunoprecipitates were also analyzed by Western blotting with anti-importin 7 antibody.
ARHI can bind to multiple importins
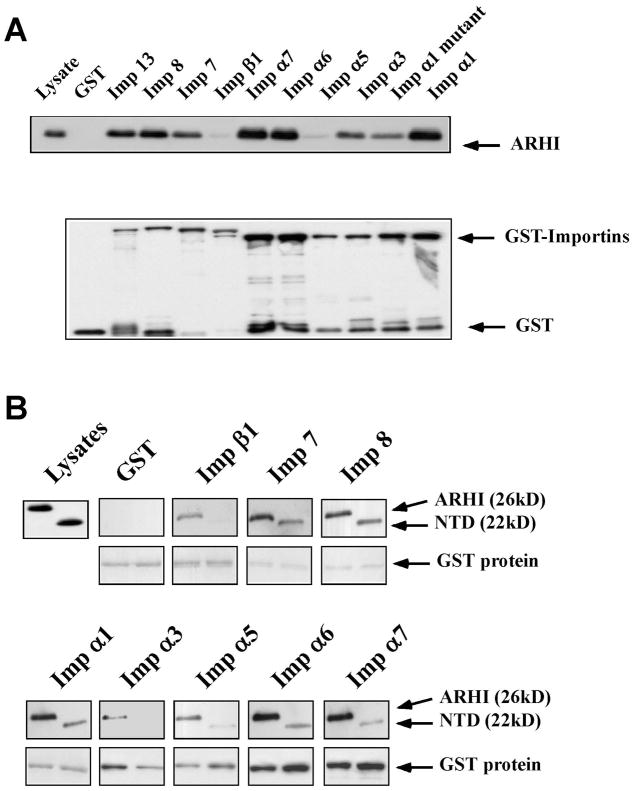
To investigate the ARHI:importin interaction, 10 GST-importins fusion proteins (importin α1, α1 mutant, α3, α5, α6, α7, importin β1, and importin 7, 8, and 13) were purified. GST protein was used as a negative control. ARHI bound strongly to most of importins in GST pull-down assays. Binding of ARHI to importin α1 was stronger than binding of ARHI to an importin α1 mutant that contained an altered NLS domain (Fig. 2A), suggesting that ARHI may bind to importins through the NLS domain.
Figure 2.
ARHI-importin binding analyzed by GST pull-down assays. (A) SKBr3 cells were infected with ARHI adenovirus and lysed. Ten GST-importin (Imp) fusion proteins and ARHI antibody were used to detect ARHI-importin binding. GST protein served as a negative control. Loading was measured with anti-GST antibody. (B) Wild type ARHI binds more strongly to most importins than does the NTD mutant. Equal amounts of ARHI or NTD protein lysates were used in GST-pull down assays. Nine GST-importin fusion proteins and ARHI antibody were used to detect the binding. GST proteins in the complexes were detected by SDS-PAGE.
N-terminal of ARHI may mediate the binding with importins
Our previous studies demonstrated that deletion of the unique 34 amino acid N-terminal extension of ARHI nearly abolished its inhibitory effect on cancer cell growth [17] and its ability to block the DNA binding activity of Stat3 [19]. Thus, the ARHI N-terminal extension could be important for its biological function. In the proteomic analysis and immunoprecipitation assays (Fig. 1), NTD protein exhibited much less binding to importins. To test further the possible role of the N-terminus of ARHI in binding importins, equal amounts of ARHI complexes and NTD complexes were tested in GST-pull down assays. As presented in Figure 2B, wild type ARHI bound more strongly to most importins than did the NTD mutant, consistent with the possibility that the 34 amino acid N-terminal extension of ARHI mediates ARHI-importin binding.
ARHI can block Stat3 protein nuclear localization signal
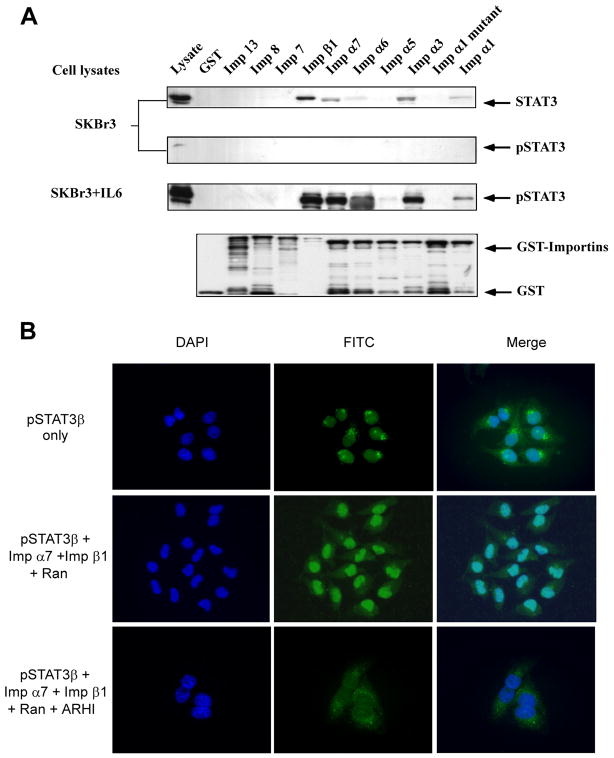
Using a similar GST pull-down assay, Stat3 protein was shown to bind importins β1, α1, α3, α6, and α7, but not the other importins and importin α1 mutant (Fig. 3A). After IL-6 stimulation, the level of pStat3 greatly increased. Importins β1, α1, α3, α6, and α7 bound both to phosphorylated Stat3 (after IL-6 stimulation) and non-phosphorylated Stat3 (without IL-6 stimulation), but these bindings to pStat3 were much stronger. We also produced the phosphorylated Stat3β (pStat3β) core fragment as published [22]. Using native gel electrophesis we confirmed that it can bind to a subset of the importins: α3, α6, and α7 (K.Z and X.C., unpublished data). To investigate if ARHI could block Stat3 protein nuclear translocation, pStat3β was labeled with FITC and evaluated in the nuclear import assay. As shown in Figure 3B, pStat3β protein alone was concentrated at nuclear pores; when importins and Ran were added to the system, pSTAT3β was found in the nucleus. Addition of purified ARHI protein blocked Ran-dependent pSTAT3β nuclear translocation.
Figure 3.
ARHI can block pStat3β nuclear translocation. (A) Stat3-importin binding analyzed by GST-pull down assays. SKBr3 cells were treated with or without IL-6 (10 ng/mL) for 30 minutes before cells were harvested for cell lysate preparation. Ten GST-importin (Imp) fusion proteins and Stat3/pStat3 antibodies were used to detect binding between Stat3 and importins. GST protein was used as a control. Loading was measured with anti-GST antibody. (B) Nuclear import assay for pStat3β-GFP. Nuclear import assays were performed as described in Materials and Methods. Purified pStat3β and RanGDP, importin α7, and importin β1 were incubated with permeabilized HeLa cells. Import assays were also performed by adding ARHI protein.
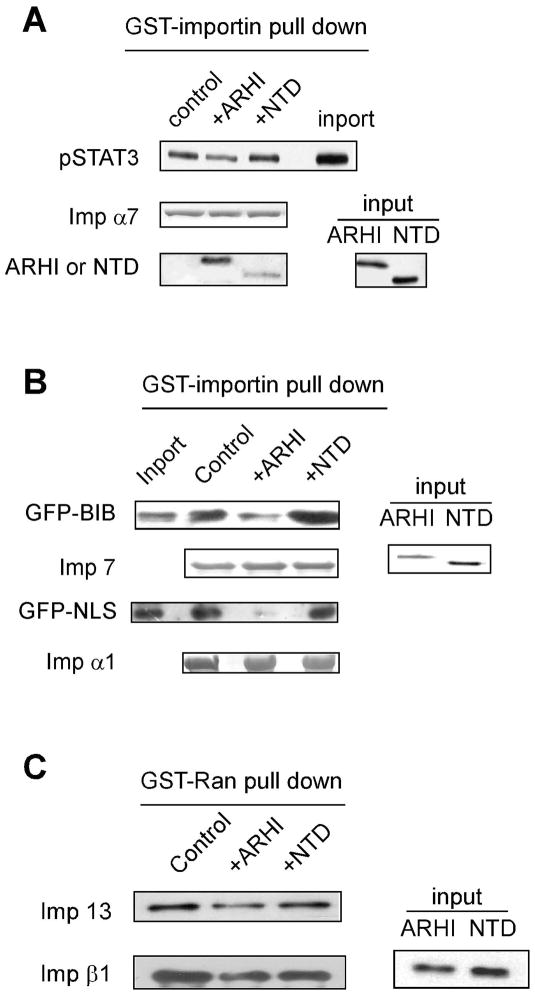
ARHI can block the protein nuclear localization signal
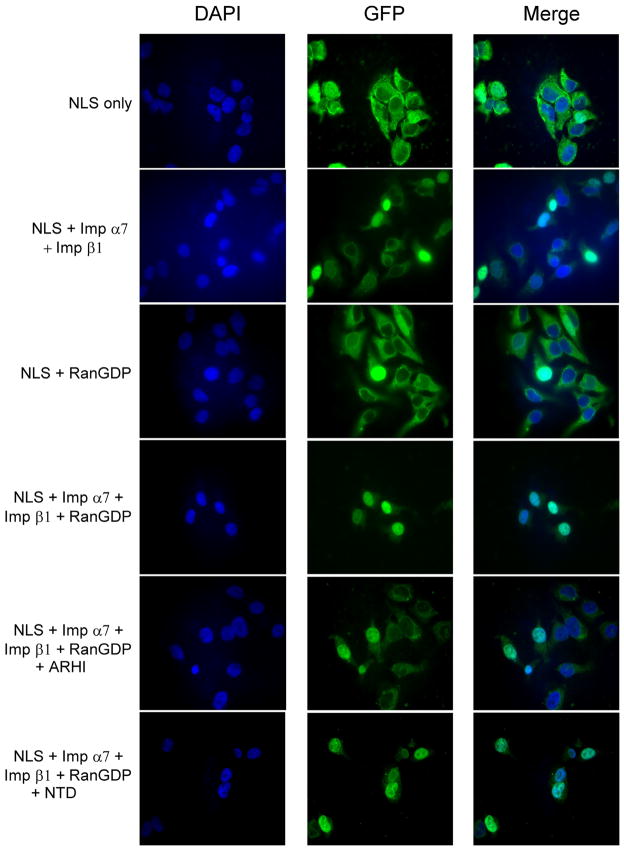
Several studies have shown that interaction of importins with the Ran protein is required to facilitate transport of cargo proteins into the nucleus [14]. Since ARHI and Ran belong to the same family of small G-proteins, ARHI therfore might antagonize the importins’ interaction with Ran. To test this hypothesis, we have purified ARHI and NTD proteins and assessed their effects on nuclear transport of cargo proteins in HeLa cells. In nuclear import assays, an NLS-GFP fusion protein could be imported into the nucleus in association with importin α7, importin β1 and RanGDP (Fig. 4), whereas the BIB-GFP fusion protein only needed help from importin 7 and RanGDP for nuclear localization (Fig. 5). ARHI protein blocked the interaction of NLS with Ran-importin β and α complexes, and also the interaction of BIB with Ran-importin 7 complexes, preventing their nuclear translocation (Figs. 4, 5). In comparison, NTD protein did not block these interactions or nuclear transport (Figs. 4, 5).
Figure 4.
Nuclear import assay for NLS-GFP. Nuclear import assay was performed as described in Materials and Methods. NLS-GFP protein was mixed with RanGDP, importin α7 (Imp α7), importin β1, and incubated with permeabilized HeLa cells. Import assays were also performed by adding equal amount of ARHI or NTD proteins.
Figure 5.
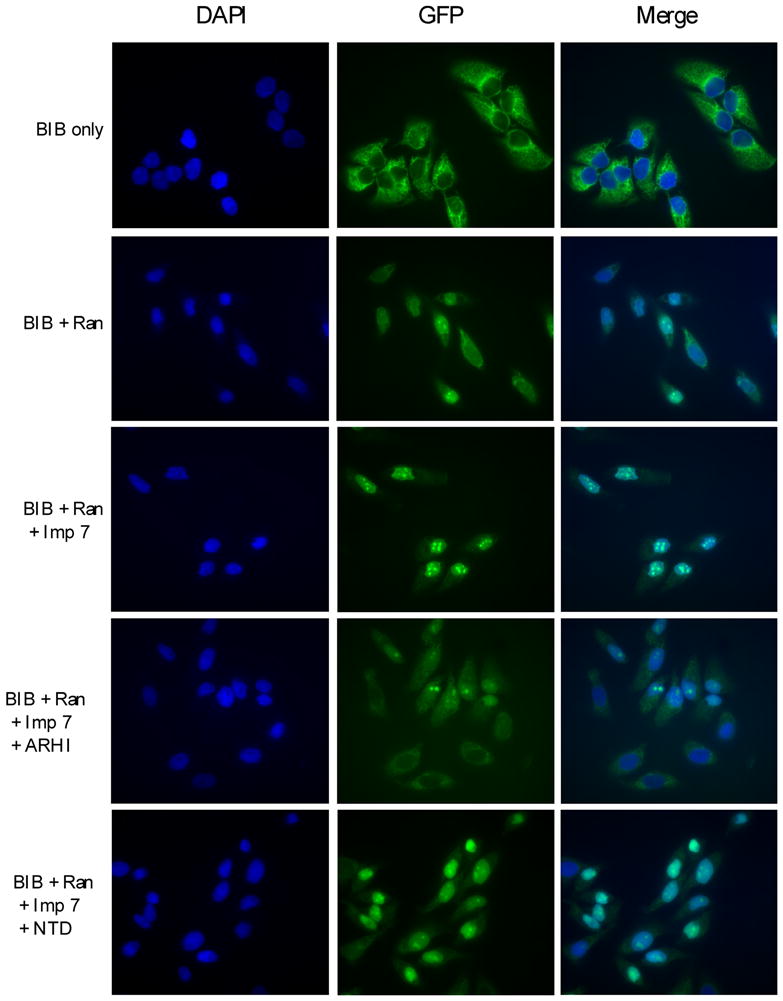
Nuclear import assay for BIB-GFP. Nuclear import assay was performed as described in Materials and Methods. BIB-GFP protein was mixed with RanGDP, importin 7 (Imp 7) and incubated with permeabilized HeLa cells. Import assays were also performed by adding equal amount of ARHI or NTD proteins.
ARHI can compete for importin-Ran binding
To determine whether ARHI binds to the same region of importins as Ran and could compete for importin-Ran binding, we have performed GST-pull down competition assays using GST-importin fusion proteins. pSTAT3, GFP-NLS and GFP-BIB proteins were bound to these GST proteins, ARHI protein was added for competition. NTD protein served as a control. As Figure 6A and 6B show, ARHI can significantly block the binding of NLS-importin α1, as well as partially block the binding of BIB-importin 7 and pSTAT3-importin α7. Using GST-Ran fusion proteins, importin 13 and β1 were strongly bound to Ran, ARHI protein could reduce at least 50% of these binding abilities (Fig. 6C). NTD protein did not have competitive activity in these assays.
Figure 6.
GST pull-down competition for Ran-importins binding. (A) ARHI competes for pSTAT3-importin α7 binding. GST-importin α7 (Imp α7) fusion proteins were bound to glutathione-Sepharose 4B, then incubated with p-STAT3 protein, treated with equal amount of ARHI or NTD proteins, then blotted with monoclonal antibodies to p-STAT3 and ARHI. The loading of GST-importin α7 and ARHI input was determined by Coomassie staining. (B) ARHI competes for NLS-importin α1 and BIB-importin 7 binding. GST-importin α1 (Imp α1) and GST-importin 7 (Imp 7) fusion proteins were bound to glutathione-Sepharose 4B, then incubated with GFP-NLS or GFP-BIB proteins, treated with equal amount of ARHI or NTD proteins, then blotted with anti-GFP monoclonal antibody. The loading of GST-importins and ARHI input was determined by Coomassie staining. (C) ARHI competes for importin-Ran binding. GST-Ran fusion proteins were bound to glutathione-Sepharose 4B, then incubated with importin β1 (Imp β1)-His or importin 13 (Imp 13)-His, treated with ARHI or NTD proteins, then blotted with monoclonal antibodies to His or polyclonal antibody to importin 13.
DISCUSSION
Stat3, a latent transcription factor, transduces signals from the cell surface to the nucleus and activates gene transcription, triggering proliferation, resistance to apoptosis, motility, invasion and angiogenesis [24, 25]. Recent studies clearly demonstrate that Stat3 is required for both tumor initiation and promotion [24]. Stat3 is frequently phosphorylated and activated in the majority of breast and ovarian cancers [26], where cytokines and growth factors such as IL-6 bind to specific receptors and activate JAK2 which, in turn, phosphorylates Stat3 and prompts translocation of pStat3 to the nucleus. The classic NLS-importin pathway has been reported to mediate the nuclear translocation of Stat3. Liu et al. reported that the Stat3 nuclear import is mediated by importin-α3 [27], whereas other studies have shown that the regulation of Stat3 nuclear import is through importins α5 and α7 [28]. Few studies investigated factors which regulate translocation of pStat3.
From our previous studies, re-expression of the putative tumor suppressor gene ARHI in cancer cells markedly inhibited binding of pStat3 to Stat response elements in target gene promoters and downregulated Stat3-dependent promoter activity without significantly affecting Stat3 phosphorylation [19]. When ARHI and Stat3 were co-expressed in SKOv3 ovarian cancer cells, ARHI formed a complex in the cytoplasm with Stat3 and prevented IL-6-induced Stat3 translocation to the nucleus [19]. Our present study has elucidated the mechanism by which ARHI prevents nuclear translocation of pStat3. Nuclear import assays have shown that pStat3 can be translocated to the nucleus in the presence of Ran and importins (Fig 3B), whereas non-phosphorylated Stat3 could not (data not shown). ARHI protein bound to importins (Figs. 1–2) and blocked nuclear translocation of pStat3 (Fig. 3B). ARHI also blocked nuclear translocation of NLS-GFP and BIB-GFP in the presence of Ran and appropriate importins (Figs 4–5). Thus, ARHI might affect nuclear localization of Stat3, but might also affect transport of other proteins that are required for oncogenesis.
The precise mechanism by which ARHI disrupts the interaction of cargo proteins, Ran and importins remains to be elucidated. ARHI is a member of Ras superfamily, but contains a unique 34 residue extension at the N-terminus. Like Ras, ARHI can bind to GTP with high affinity, but has low intrinsic GTPase activity [17]. ARHI associates with the cell membrane after prenylation at the C-terminal cysteine residue. Mutation of the conserved CAAX box at the C-terminus led to a loss of its membrane association ability and a modest decrease in its ability to inhibit cell growth [16]. Most strikingly, deletion of ARHI’s unique N-terminal extension nearly abolishes its inhibitory effect on cell growth [17, 19]. In the present study, the NTD mutant lost much of the ability to bind importins and did not block the nuclear translocation of NLS-GFP or BIB-GFP. This suggests that interaction of the N-terminal extension of ARHI to importin may be required to prevent effective interaction of Ran-importin complexes with NLS and BIB. Interestingly, C-terminal deletion mutants of ARHI localize in the nucleus (data not shown), raising the possibility that accumulation of pStat3 in the cytoplasm in the presence of wild type ARHI protein could result from direct binding of ARHI to Stat3, which we have demonstrated [19], or to lack of an effective nuclear transport mechanism. As wild type ARHI exhibits a nuclear localization signal on its N-terminal extension, the ARHI might compete with cargo for binding to importins in the presence of Ran. Once bound, ARHI could trap importins in the cytoplasm as the small G protein is tethered to cell membranes through its prenylated C-terminus.
ARHI (DIRAS3) and Ran share a 22% sequence identity and are expected to have a similar overall structure. Although the crystal structure of ARHI is not yet available, those of the closest neighbors in the Ras family, DIRAS1 and DIRAS2, have been solved. We obtained a homology model for ARHI via the SWISS-MODEL server (http://swissmodel.expasy.org/). The model was generated using the DIRAS2 structure (PDB code: 2ERX) as the template and it covers residues 37 to 206 of the protein’s 229 residues. When the ARHI model is superimposed onto the crystal structure of Ran (PDB code: 1IRB, chain A) [29], the structural conservancy is clear. A DaliLite (http://www.ebi.ac.uk/DaliLite/) pairwise comparison resulted in a Z-score of 23.4 and Cα RMSD of 2.4 Å, over 165 aligned residues. In the crystal structure of the importin β:Ran complex [29], Ran utilizes an extensive surface in binding to importin β. In the present study, by GST-pull competition assay, ARHI protein could significantly block the binding of NLS-importin, partially block the binding of BIB-importin and pSTAT3-importin. ARHI protein could also partially block importin-Ran binding ability (Fig. 6), suggesting that ARHI binds to the same region of importins as Ran does and ARHI-induced disruption of Ran-importin binding could serve as an important regulatory mechanism that contributes to the tumor suppressor function of ARHI.
Acknowledgments
FUNDING
This work has been supported by grants from the National Caner Institute, CA 80957 (Y. Yu), CA 64602 (R.C. Bast) and GM68556 (X. Chen); from the National Foundation for Cancer Research LF 2004-00009224HM (R.C. Bast); from the National Natural Science Foundation of China, 3047085 and 90608007 (T.T) and from the Ministry of Science & Technology of China, 2006AA02A310 (T.T).
Abbreviations
- NTD
N-terminal deletion mutant
- NLS
nuclear localization signal
- p-Stat3
phosphorylated Stat3
- IBB
importin-beta-binding
- BIB
beta-like import receptor binding
References
- 1.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318:1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 2.Herrmann A, Vogt M, Mönnigmann M, Clahsen T, Sommer U, Haan S, Poli V, Heinrich PC, Müller-Newen G. Nucleocytoplasmic shuttling of persistently activated STAT3. J Cell Sci. 2007;120:3249–3261. doi: 10.1242/jcs.03482. [DOI] [PubMed] [Google Scholar]
- 3.Ivanova IA, Vespa A, Dagnino L. A novel mechanism of E2F1 regulation via nucleocytoplasmic shuttling: determinants of nuclear import and export. Cell Cycle. 2007;6:2186–2195. doi: 10.4161/cc.6.17.4650. [DOI] [PubMed] [Google Scholar]
- 4.Buse P, Maiyar AC, Failor KL, Tran S, Leong ML, Firestone GL. The stimulus-dependent co-localization of serum- and glucocorticoid-regulated protein kinase (Sgk) and Erk/MAPK in mammary tumor cells involves the mutual interaction with the importin-alpha nuclear import protein. Exp Cell Res. 2007;313:3261–3275. doi: 10.1016/j.yexcr.2007.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Planque N. Nuclear trafficking of secreted factors and cell-surface receptors: new pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun Signal. 2006;4:7. doi: 10.1186/1478-811X-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lo HW, Ali-Seyed M, Wu Y, Bartholomeusz G, Hsu SC, Hung MC. Nuclear-cytoplasmic transport of EGFR involves receptor endocytosis, importin beta1 and CRM1. J Cell Biochem. 98:1570–1583. doi: 10.1002/jcb.20876. [DOI] [PubMed] [Google Scholar]
- 7.Otis KO, Thompson KR, Martin KC. Importin-mediated nuclear transport in neurons. Curr Opin Neurobiol. 2006;16:329–335. doi: 10.1016/j.conb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 8.Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 9.Harel A, Forbes DJ. Importin beta: conducting a much larger cellular symphony. Mol Cell. 2004;16:319–330. doi: 10.1016/j.molcel.2004.10.026. [DOI] [PubMed] [Google Scholar]
- 10.Jäkel S, Albig W, Kutay U, Bischoff FR, Schwamborn K, Doenecke D, Görlich D. The importin beta/importin 7 heterodimer is a functional nuclear import receptor for histone H1. EMBO J. 1999;18:2411–2423. doi: 10.1093/emboj/18.9.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Waldmann I, Wälde S, Kehlenbach RH. Nuclear import of c-Jun is mediated by multiple transport receptors. J Biol Chem. 2007;282:27685–27692. doi: 10.1074/jbc.M703301200. [DOI] [PubMed] [Google Scholar]
- 12.Tao T, Lan J, Lukacs GL, Haché RJ, Kaplan F. Importin 13 regulates nuclear import of the glucocorticoid receptor in airway epithelial cells. Am J Respir Cell Mol Biol. 2006;35:668–680. doi: 10.1165/rcmb.2006-0073OC. [DOI] [PubMed] [Google Scholar]
- 13.Quan Y, Ji Z, Wang X, Tartakoff AM, Tao T. Evolutionary and Transcriptional Analysis of Karyopherin β Superfamily Proteins. Mol Cell Proteomics. 2008;7:1254–1269. doi: 10.1074/mcp.M700511-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moore MS, Blobel G. The GTP-binding protein Ran/TC4 is required for protein import into the nucleus. Nature. 1993;365:661–663. doi: 10.1038/365661a0. [DOI] [PubMed] [Google Scholar]
- 15.Kaláb P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- 16.Yu Y, Xu F, Peng H, Fang X, Zhao S, Li Y, Cuevas B, Kuo WL, Gray JW, Siciliano M, Mills GB, Bast RC., Jr NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc Natl Acad Sci USA. 1999;96:214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo RZ, Fang X, Marquez R, Liu SY, Mills GB, Liao WS, Yu Y, Bast RC. ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene. 2003;22:2897–2909. doi: 10.1038/sj.onc.1206380. [DOI] [PubMed] [Google Scholar]
- 18.Lu Z, Luo RZ, Lu Y, Zhang X, Yu Q, Khare S, Kondo S, Kondo Y, Yu Y, Mills GB, Liao WS, Bast RC., Jr The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J Clin Invest. 2008;118:3917–3929. doi: 10.1172/JCI35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishimoto A, Yu Y, Lu Z, Mao X, Ren Z, Watowich SS, Mills GB, Liao WS, Chen X, Bast RC, Jr, Luo RZ. ARHI Directly Inhibits STAT3 Translocation and Activity in Human Breast and Ovarian Cancer Cells. Cancer Res. 2005;65:6701–6710. doi: 10.1158/0008-5472.CAN-05-0130. [DOI] [PubMed] [Google Scholar]
- 20.Freedman ND, Yamamoto KR. Importin 7 and importin alpha/importin beta are nuclear import receptors for the glucocorticoid receptor. Mol Biol Cell. 2004;15:2276–2286. doi: 10.1091/mbc.E03-11-0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bao JJ, Le XF, Wang RY, Yuan J, Wang L, Atkinson EN, LaPushin R, Andreeff M, Fang B, Yu Y, Bast RC., Jr Reexpression of the tumor suppressor gene ARHI induces apoptosis in ovarian and breast cancer cells through a caspase-independent calpain-dependent pathway. Cancer Res. 2002;62:7264–7272. [PubMed] [Google Scholar]
- 22.Becker S, Corthals GL, Aebersold R, Groner B, Müller CW. Expression of a tyrosine phosphorylated, DNA binding Stat3beta dimer in bacteria. FEBS Lett. 1998;441:141–147. doi: 10.1016/s0014-5793(98)01543-9. [DOI] [PubMed] [Google Scholar]
- 23.Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haura EB, Turkson J, Jove R. Mechanisms of disease: Insights into the emerging role of signal transducers and activators of transcription in cancer. Nat Clin Pract Oncol. 2005;2:315–324. doi: 10.1038/ncponc0195. [DOI] [PubMed] [Google Scholar]
- 25.Xie TX, Wei D, Liu M, Gao AC, Ali-Osman F, Sawaya R, Huang S. Stat3 activation regulates the expression of matrix metalloproteinase-2 and tumor invasion and metastasis. Oncogene. 2004;23:3550–3560. doi: 10.1038/sj.onc.1207383. [DOI] [PubMed] [Google Scholar]
- 26.Alvarez JV, Febbo PG, Ramaswamy S, Loda M, Richardson A, Frank DA. Identification of a genetic signature of activated signal transducer and activator of transcription 3 in human tumors. Cancer Res. 2005;65:5054–5462. doi: 10.1158/0008-5472.CAN-04-4281. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, McBride KM, Reich NC. STAT3 nuclear import is independent of tyrosine phosphorylation and mediated by importin-alpha3. Proc Natl Acad Sci USA. 2005;102:8150–8155. doi: 10.1073/pnas.0501643102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma J, Cao X. Regulation of Stat3 nuclear import by importin alpha5 and importin alpha7 via two different functional sequence elements. Cell Signal. 2006;18:1117–1126. doi: 10.1016/j.cellsig.2005.06.016. [DOI] [PubMed] [Google Scholar]
- 29.Vetter IR, Arndt A, Kutay U, Görlich D, Wittinghofer A. Structural view of the Ran-Importin beta interaction at 2.3 A resolution. Cell. 1999;97:635–646. doi: 10.1016/s0092-8674(00)80774-6. [DOI] [PubMed] [Google Scholar]