SUMMARY
Maize anther ontogeny is complex with expression of more than 30,000 genes over four days of cell proliferation, cell fate acquisition, and the start of meiosis. Although many male-sterile mutants disrupt these key steps, few have been investigated in detail. The terminal phenotypes of maize male-sterile 8 (ms8) are small anthers exhibiting meiotic failure. Here we document much earlier defects: ms8 epidermal cells are normal in number but fail to elongate, and there are fewer, larger tapetal cells that retain rather than secrete their contents. ms8 meiocytes separate early, have extra space between them occupied by excess callose, and the meiotic dyads abort. Thousands of transcriptome changes occur in ms8 including ectopic activation of genes not expressed in fertile siblings, failure to express some genes, differential expression compared to fertile siblings and about 40% of the differentially expressed transcripts appear precociously. There is a high correlation between mRNA accumulation assessed by microarray hybridization and qRT-PCR. Sixty-three differentially expressed proteins were identified after 2-D gel electrophoresis followed by LC/MS/MS (liquid chromatography tandem mass spectroscopy), including those involved in metabolism, plasmodesmatal remodeling, and cell division. The majority of these were not identified by differential RNA expression demonstrating the importance of proteomics to define developmental mutants.
Keywords: maize, ms8, anther, meiosis, microarray, proteomics
INTRODUCTION
In flowering plants, pollen production first requires cell proliferation and differentiation to produce an anther, then successful completion of meiosis in a subset of centrally-located anther locule cells, and finally the redifferentiation of anther somatic cells to support gametophyte maturation (Mascarenhas, 1990). These processes require coordinated gene expression among multiple cell types over 30 days in maize (Skibbe and Schnable, 2005). Maize anthers are bilaterally symmetrical with four lobes; cells divide and differentiate within each lobe to form four distinct somatic cell types (exterior epidermis, endothecium, middle layer, and innermost tapetum (Ma et al., 2008) surrounding the pollen mother cells (PMC). The tapetum plays a pivotal role in nutrient and enzyme secretion to the PMC (Goldberg et al., 1993). Nuclear male sterility has been reported in many flowering plants, and in most cases mutations affect primarily tapetal or microspore development (Juan et al., 2005; Xu et al., 2006; Wang et al., 2009).
Despite the existence of male-sterile mutants in Arabidopsis, tomato, rice, maize and other species, only a few genes that control the early phases of cell proliferation, cell fate setting, and initial differentiation have been identified. Furthermore, cloned genes represent diverse functions such as putative signaling pathways and metabolic enzymes rather than a cascade of transcription factors. For example, Arabidopsis BAM1 (BARELY ANY MERISTEM) and BAM2 encode Leu-rich repeat receptor-like kinases; bam1 and bam2 mutations affect cell division and differentiation patterns that result in the absence of endothecial, middle and tapetal layers (Hord et al. 2006). Similarly Arabidopsis EXS (EXTRA SPOROGENOUS CELLS) encodes a putative receptor thought to interact with a protein ligand encoded by TPD1 (TAPETUM DETERMINANT1 Canales et al., 2002; Yang et al., 2003). GhACS1 is a cotton anther-specific acyl-CoA synthetase (ACS) required for normal fatty acid metabolism in PMC and tapetal cells. Suppression of GhACS1 expression severely affected the tapetal cells and microsporogenesis in early anther development (Wang and Li, 2009). Male sterile converted anther 1 (msca1) encodes a maize (Zea mays) glutaredoxin (U. S. Patent # 20090038027) in which the anther has normal morphology, but all cells differentiate as leaf cell types (Chaubal et al., 2003); normally, early leaf development genes are expressed in very immature anthers and then turned off, however, expression of these genes persists in msca1 (Ma et al., 2007). Additional genes involved in early Arabidopsis and maize anther development are summarized in Table S1.
Despite the importance of pollen in crop yield and hybrid seed production, molecular analysis of early anther development exploiting maize mutants was initiated only recently (Ma et al., 2007). Maize has several advantages, primarily the large number of anthers per tassel and the lack of female floral parts in the tassel (Bedinger and Fowler, 2009). Relative to most angiosperms, maize anthers are larger, easier to dissect, and cellular events are highly synchronous within each anther. Male-sterile mutants that disrupt the normal pattern of cell proliferation and differentiation prior to meiosis can be highly instructive in charting steps in cell fate acquisition and function, because specification of anther cell types is completed by the onset of meiosis. ms8 was first recognized for meiotic failure and lack of pollen (Beadle, 1931; Albertsen and Phillips, 1981), however, anther locule cellular organization, gene expression, and biochemical features have yet to be investigated. The current study was designed to quantify ms8 defects cytologically and then to link the defects to transcriptome and proteome alterations with the additional goal of testing whether transcriptome changes were predictive of protein abundance changes.
RESULTS
ms8 anther development
ms8 plants are indistinguishable from fertile siblings during vegetative and ear development, however, ms8 tassels have fewer branches, anthers do not exert, and no viable pollen is produced (Figure 1a). When fertile siblings shed pollen, ms8 anthers are senescing. ms8 anthers often adhere and cease elongating at 3.0 mm, about 60% of normal final length. Dwarfing of ms8 could reflect early cessation of cell division and/or failure to sustain cell expansion. By scanning electron microscopy at 3.0 mm, ms8 epidermal cells were well organized and the number of cells was similar to fertile siblings. Normal epidermal cells elongate longitudinally after meiosis and reach 85 μm at the microspore stage, whereas ms8 epidermal cells arrest at an average length of 52 μm (Figure 2b). Therefore, smaller ms8 epidermal cells are one explanation for shorter anthers, but there is no substantial defect in longitudinal cell division.
Figure 1.
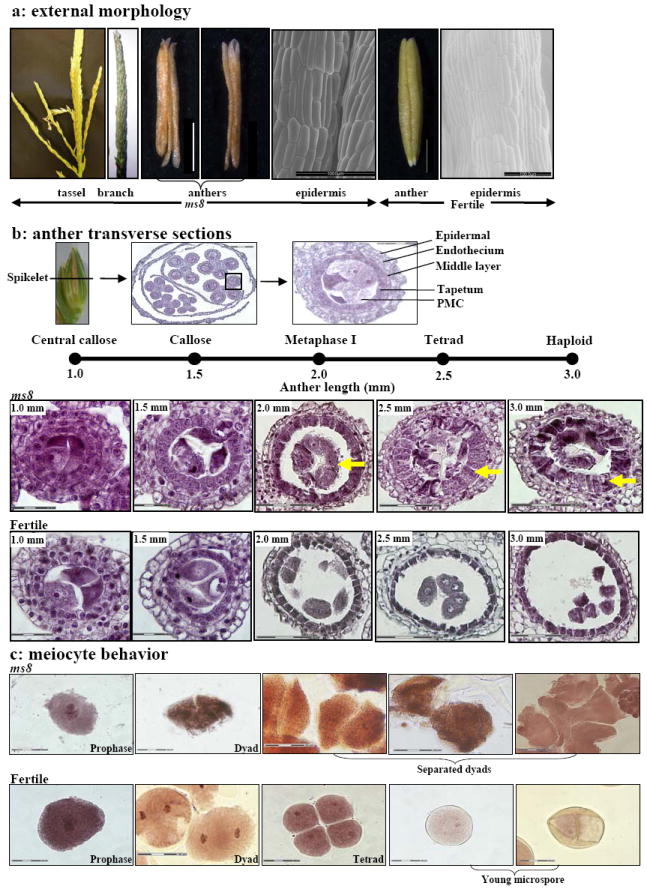
Phenotypic analysis of ms8 anther locule and cell morphology. (a) External morphology. Samples were collected from families segregating 1:1 for fertility (ms8//+): sterility (ms8//ms8). A typical ms8 tassel is thinner and has fewer tassel branches at maturity. Dissected sterile anthers are shriveled and brown, compared to fertile anthers, which are plump and pale yellow-green. Scanning electron microscopy was used to assess epidermal cell shape and number at the 3 mm stage of ms8 and 4 mm stage of fertile individuals. Bar = 1 mm (white), bar = 100 μm (black). (b) Each spikelet encloses two florets with three anthers each; lower floret anthers develop more slowly. There are four anther locule wall layers: epidermis, endothecium, middle layer and tapetum. PMCs undergo meiosis and proceed through the dyad (two cells) and tetrad (four cells) stages. During meiosis, the PMCs and product cells are normally connected by plasmodesmata. After meiosis, haploid cells normally undergo two mitotic divisions to form pollen, the microgametophyte. Anther progression is assessed by two measures: anther length and differentiation of the innermost (pre-meiotic, meiotic, and then post-meiotic) cells. Cytological staging relative to maize anther length is shown above the locule photographs. Transverse sections of locules from ms8 and from fertile plants at five developmental stages were assessed. Bar = 30 μm for 1.0 mm and 1.5 mm anther locules; bar = 75 μm for 2.0 – 3.0 mm anther locules. Arrows highlight the features of ms8 and its fertile counterpart. (c) Meiocytes from ms8 and from fertile anthers at different developmental stages were stained by hematoxylin-iron-aceto-carmine. ms8 dyads were smaller ellipsoid, separated from each other and collapsed soon after. Bar = 30 μm.
Figure 2.
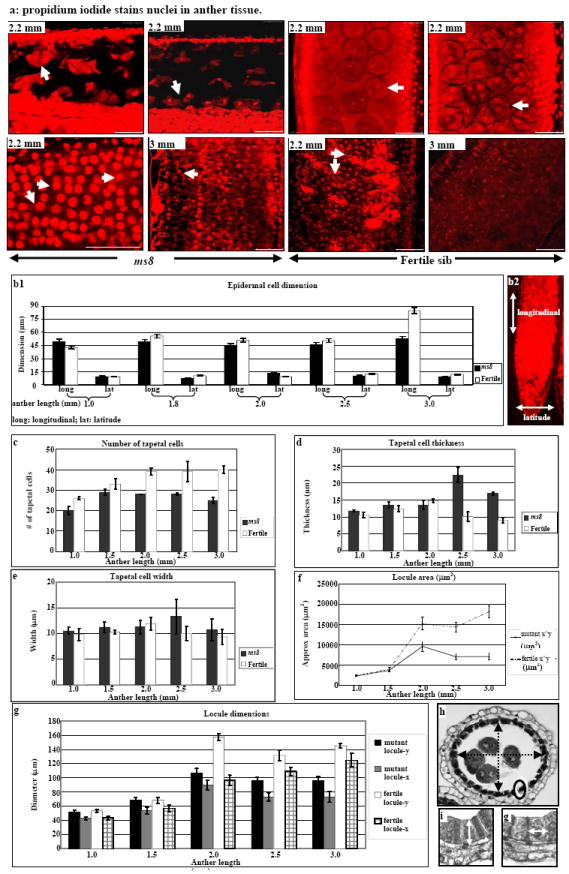
Morphometric analysis of ms8 and fertile anthers. (a) Propidium iodide stains nuclei in meiocytes (upper panels) and tapetal cells (lower panels). ms8 anther at 2.2 mm length, ellipsoid dyads with gaps between them (arrow); fertile locules were filled with round dyads and tetrads. A majority of ms8 tapetal cells have a single nucleus at 2.2 mm (arrow); they remain intact and reach maximum size when anthers are 3 mm. Most fertile tapetal cells are bi-nucleate at 2.5 mm, and they have collapsed and nuclear staining was faint at 3 mm. Bar = 50 μm. (b1) Epidermal cell dimension comparison of ms8 and fertile anthers from 1.0 mm to 3.0 mm. (b2) Longitudinal and latitude directions are shown by arrows. (c – g) Morphometric comparison of ms8 and fertile anthers. (c) The number of tapetal cells. (d) Tapetal cell thickness. (e) Tapetal cell width. (f) Locule area. (g) Dimensions of the central zone internal to the tapetum. (h) Illustrates the two measurements taken for the x (shorter diameter) and y (longer diameter) at five stages of ms8 and fertile siblings. The measurement of tapetal cell thickness and width are shown in (i) and (g), respectively. Error bars indicate standard deviation.
Key stages of anther ontogeny relative to fertile anther length are summarized in Figure 1b. At 1.0 mm during rapid mitotic proliferation, there were no apparent cytological differences between ms8 and fertile sibs (Figure1b). At this stage there are four somatic wall layers of typical cell morphology. The endothecium and middle layers contain flattened cells elongated periclinally; neighboring tapetal cells are cuboidal with a denser cytoplasm. The presumptive pre-meiotic cells (PMC) at the center of each locule are encased in callose (Figure 1b). At the 1.5 mm stage, pollen mother cells partially separate and advance into meiotic prophase I in normal anthers (Figure 1b). Within ms8 anthers, there was more space between the meiocytes, and this was occupied by excess callose (Figure 1b). Normal 2.0-2.5 mm anthers had meiocytes progressing through meiosis (Figure 1b); dyads were round and intact, with no gap between these two daughter cells (Figure 1c, 2a). In contrast, ms8 meiocytes completed the first meiotic division but the dyads were smaller ellipsoids, with gaps between cells; the two daughter cells separated and collapsed soon after (Figure 1b, c, 2a). In subsequent stages, when fertile tetrads released and normal young microspore developed an exine, intine and germ pore (Figure 1c), there was only cellular debris in the center of ms8 locules (Figure 1b, c), indicating that meiotic cells had disintegrated.
From transverse sections (Figure S1), ~26 tapetal cells are present in 1.0 mm fertile locules, gradually increasing to ~40 (Figure 2c); only a subset of tapetal precursors present at 1.0 mm undergo a subsequent mitosis. In terms of morphology, both cell thickness (diameter in the radial dimension, Figure 2d) and width (diameter within the tapetal ring, Figure 2e) were relatively constant in early stages. After meiosis, however, the normal tapetum thins and begins to degenerate (Figure 1b, 2d). Much of the dark-staining contents are secreted to coat pollen, and subsequently tapetal cells appear nearly empty. In contrast, ms8 had only ~20 tapetal cells at the 1.0 mm stage, increasing to ~28 at the post-mitotic 1.5 mm stage. Thus, there are fewer cell divisions and an early cessation of cell division (Figure 2c), indicating a different contributing factor to the dwarf size of ms8 anthers in this cell layer compared to the epidermis.
At the 1.0 and 1.5 mm stages, the external morphologies of ms8 and fertile anthers are virtually identical. Maintenance of anther girth appears to reflect excessive growth of the smaller number of ms8 tapetal cells (Figure S1, a1 – a3). Cell length (radial diameter) was twice that of normal and width was slightly greater when anther length was 2.5-3.0 mm (Figure 2d, e, i, g). Consequently, ms8 tapetal cells are very large, occupying much of the locule space, whereas the debris from degenerating meiotic cells occupies only a small area relative to meiotic cells in fertile siblings (Figure 1b). Unlike bi-nucleate normal tapetum (Figure 2a), a majority of ms8 tapetal cells have a single nucleus (Figure 2a). Normal tapetal cells degenerate during microspore maturation (Figure 1b, 2a), whereas mutant tapetal cells did not shrink (Figure 1b) and few degraded nuclei were found at the 3.0 mm stage (Figure 2a).
Locule transverse area was approximated by multiplying two diameter measurements. Despite the abnormalities of ms8, locule size (Figure 2f - h) was equivalent to fertile siblings before meiosis. Thereafter, fertile locules continued growing in girth whereas the ms8 mutant locule area decreased after the 2.0 mm stage. Reduced locule area was caused by extra thick tapetal cells and cell layers expansion failure. There is also a shape change: ms8 locules are oval but fertile locules are round (Fig 1b).
Transcriptome diversity quantified by microarray hybridization
RNA samples from fertile and sterile anthers were hybridized to custom Agilent 4×44 microarrays containing 42,034 probes (excluding controls) to 39,162 genes (Skibbe et al., 2009). Fertile anthers exhibit astonishing transcript complexity: there are 27,400 constitutively expressed genes, 2143 stage-specific, and 2484 expressed at two stages summing to 32,037 genes expressed over a 90 h period. This is more than 75% of the projected maize gene number. Highest diversity occurs at 1.5 mm (Figure 3a, b, S2a), confirming previous studies (Ma et al., 2006; Ma et al., 2008). Unlike these prior studies, we did not observe a 10% drop in transcript types at 2.0 mm. During the initial days of meiosis anther length is constant; hence size-based staging is not as accurate as cytology. We selected anthers at metaphase I of meiosis (Figure 1b); this narrower pooling allowed a greater depth of transcript detection than pooling 2.0 mm anthers.
Figure 3.
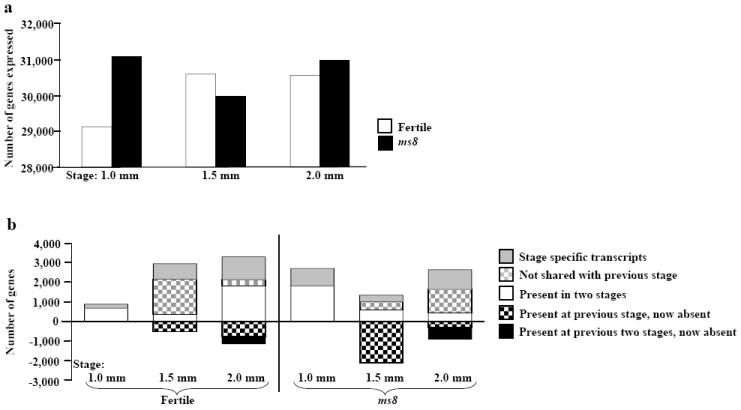
Transcriptome analysis of ms8 and fertile siblings anthers. (a) Transcriptome sizes at three stages of ms8 and fertile siblings anthers. (b) Analysis of transcriptome changes during fertile (left part) and sterile (right part) anther development; 27,400 transcripts shared by all stages of fertile anthers and 27,284 transcripts shared by all stages of ms8 anthers are not shown. The y-axis of (a) shows the number of probes expressed, the y-axes of (b) show the number of probes; the x-axes list the developmental stages.
As shown in Figures 3b and S2a, at 1.0 mm there are 197 stage-specific transcripts, 780 at 1.5 mm, and 1166 at 2.0 mm. During the 1.0 - 1.5 mm transition, which spans about 72 h (Ma et al., 2008), there is a major transcriptome change: 1799 new transcripts (grey checked bar in Figure 3b) and 523 transcripts expressed in the 1.0 mm anther are not detectable at 1.5 mm (black checked bar in Figure 3b). At the subsequent 1.5 - 2.0 mm transition, which spans about 18 h (Ma et al., 2008), 326 transcripts appeared (grey checked bar in Figure 3b), 780 transcripts (black checked bar in Figure 3b) found at 1.5 mm disappeared, and 359 transcripts (black bar in Figure 3b) present at both the 1.0 and 1.5 mm stages are no longer detectable.
The transcriptome of ms8 anthers is also very large: 27,284 (72%) transcripts are shared across all three ms8 anther stages, similar to fertile siblings (Figure 3a, b, S2b). Strikingly, the 1.0 mm ms8 anther expresses nearly 2000 more genes than fertile, and many of these are stage-specific within the context of ms8 development (Figure 3b). Although ms8 anthers are defective, the transcriptome is highly dynamic because new transcription factors (5% [113/2224] of the stage-specific genes) and predicted nucleic acid binding and protein binding classes are expressed. Differential expression between ms8 and fertile siblings was evaluated using a minimum fold change of 1.5 as calculated using the limma package in R (Smyth, 2005). As shown in Table S2, there are 3196 differentially regulated genes at 1.0 mm, representing reprogramming of 11% of the normal transcriptome (3196/29,124). The significant impact of the ms8 mutation persists through the subsequent two stages with 3748 (12%) genes differentially regulated at the 1.5 mm stage, and 3328 (11%) differentially regulated at the 2.0 mm meiotic stage. Among the differentially regulated transcripts at the 1.0 mm stage, ms8 anthers had fewer down-regulated (25%) than up-regulated (75%) genes whereas the opposite pattern occurs in the 1.5 and 2.0 mm stages (Table S2).
ms8 anthers develop precociously, achieving some developmental landmarks at a smaller size. Interestingly, among the 902 1.0 mm stage-specific transcripts of ms8 anthers (grey bar in Figure 3b), 416 (46%) are expressed at later stages in normal anthers (Table S3); more than 41% (133/322) of the 1.5 mm stage-specific ms8 transcripts are expressed at the 2.0 mm stage in fertile siblings (Table S4). Consequently, one aspect of differential gene expression in ms8 is temporal acceleration of some transcriptome programs. This likely disrupts the temporally coordinated growth and differentiation of ms8 anther cells resulting in the observed defects in cell division (tapetum) and cell elongation (epidermis).
Distinctive and shared features of transcriptome patterns of ms8 and fertile anthers
The relationship between ms8 and fertile was further explored by k-means clustering. For example, among fertile genes expressed constitutively at high levels (1000-fold above the median), 150 genes in ms8 were down-regulated dramatically (average 4-fold) in 1.5 mm anthers but expressed equivalently in the preceding and following stages (Figure 4a). Within this cluster are genes related to cell division, including cell division protein FtsH (TC286699) and T cytoplasm male sterility restorer factor2 (Rf2). Aniline blue staining and quantitative assays detected more callose around the mutant meiocytes at the 2.0 mm stage than in fertile siblings (Figure 5, compare c and d to g and h, i). Ninety-five genes are down-regulated in the 2.0 mm ms8 anther (Figure 4b), including β-D-glucosidase (Figure 5j), which is secreted by normal tapetum to dissolve callose (Bucciaglia and Smith, 1994). We hypothesize that excess callose accumulates because of a lack of degradation. There are 907 genes up-regulated in ms8 during the 1.5 mm stage (Figure 4c). MYB transcription factors have been identified as important in cell fate setting (Pastore et al., 2008), and 10 such genes show increased expression in 1.5 mm ms8 anthers relative to fertile during the final step in cell fate setting in maize anther development (Table S5).
Figure 4.
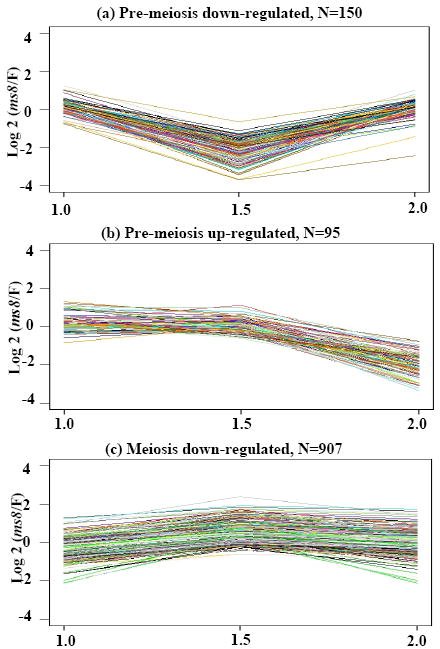
Differentially expressed transcripts grouped by k-means clustering. Transcripts constitutively expressed in fertile plants were clustered, resulting in the ms8/Fertile fold change patterns shown. (a) 150 probes were down-regulated at the 1.5 mm stage; (b) 95 probes were down-regulated at the 2.0 mm stage; (c) 907 probes were up-regulated at 1.5 mm. The y-axes are the log2 ratio fold change of ms8/Fertile. The x-axes show the developmental stages.
Figure 5.
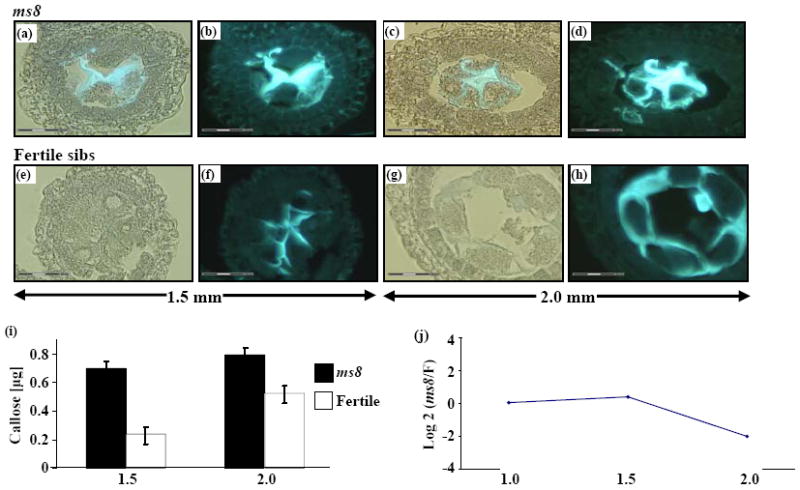
Aniline blue staining for callose in ms8 and fertile sibs. (a – d) Fluorescence in ms8 anthers under UV illumination. (e – h) Fertile counterparts under the same UV intensity. (a), (c), (e) and (g) Bright field and UV illumination. (b), (d), (f) and (h) UV only. (a), (b), (e) and (f) Anthers at 1.5 mm. More fluorescence was detected in ms8 locules (a, b) than in fertile counterparts (e, f), which means excess callose surrounds ms8 PMC. The callose occupied the extra space between separated ms8 cells. (c), (d), (g) and (h) Anthers at 2.0 mm. More callose surrounds the multinucleated syncytium in ms8 locules, and no separate meiocytes were observed (c and d). The normal meiocytes were well separated and covered with a thin coat of callose (g and h). (i) Callose comparison between ms8 anther and fertile counterpart. The y-axis shows the callose amount, and the x-axis shows two developmental stages. (j) The expression pattern of β-D-glucosidase, an enzyme that is secreted by the tapetum to dissolve callose and release the tetrads. The y-axis is the fold change of ms8/Fertile in log2 ratio, the x-axis shows the three developmental stages named by anther length.
Zinc finger transcription factors are among the best studied plant DNA binding proteins, and several play important roles in floral development. In Petunia hybrida, seven family members were activated sequentially during anther development, and it was proposed that they might act as a regulatory cascade (Kobayashi et al., 1998). Disruption of specific zinc finger genes causes floral abnormalities, i.e., mutations in Arabidopsis rabbit ear (rbe) (Takeda et al., 2004) and rice rid1 (Wu et al., 2008). Previously, 253 unique zinc finger-related probes were examined from the maize 1.0 mm anther stage through mature pollen (Ma et al., 2008), and both stage-specific and persistent expression patterns were observed. All but 28 of the 253 genes queried were expressed in at least 1 of the 3 stages examined here, and the majority (186/253 = 74%) are expressed constitutively in both ms8 and fertile anthers (cluster 1, Figure 6). A few family members are differentially expressed: TC314427 increases 10-fold from the 1.0 to 2.0 mm stages in both fertile and ms8 anthers (purple line, cluster 3, Figure 6). In comparing ms8 to fertile anthers, eight zinc finger family genes are differentially expressed in at least one stage (Table S6). Interestingly, zinc finger transcript abundances are very diverse; they range in expression from low to moderately abundant (~8-fold above the median, cluster 1, Figure 6) to ~10-12 fold above the median (clusters 2 and 3), and six express at exceptionally high levels (cluster 4).
Figure 6.
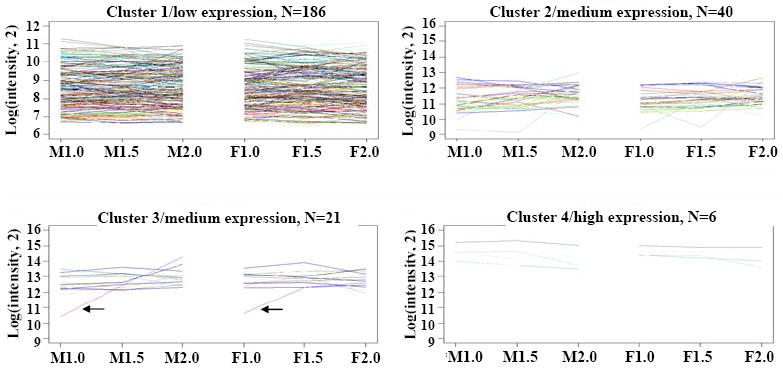
Clustering of zinc finger related gene expression. Genes are sorted after k-means clustering (k = 4). Arrows in cluster 3 show TC314427 increases 10-fold from the 1.0 to 2.0 mm stages in both fertile and ms8 anthers. M1.0, 1.0 mm stage ms8 anther; M1.5, 1.5 mm stage ms8 anther; M2.0, 2.0 mm stage ms8 anther. F1.0, 1.0 mm stage fertile anther; F1.5, 1.5 mm stage fertile anther; F2.0, 2.0 mm stage fertile anther.
Is there mis-expression of marker genes in specific anther cell types?
Previously msca1, mac1 (multiple archesporial cells1), and ms23 (male sterile23) were compared to fertile siblings in 1.0 to 2.0 mm anthers (Ma et al., 2007). All three mutants lack a cytologically normal tapetum and were missing ten transcripts present in normal siblings; these 10 were designated tapetal marker genes. Six of these probes were queried in the current study: five had the 1.5 mm valley pattern, with a dramatic (>4-fold) decrease in ms8 compared to normal followed by an increase when meiosis began (Figure 7). Among them, only gene TC290478 was confirmed to be expressed in epidermal and tapetal cells by laser micro-dissection and microarray analysis (D. S. Skibbe, pers. communication); it decreased dramatically in ms8 at the 1.5 mm stage. A sixth probe (TC314200) was low at the 1.0 mm stage in ms8 relative to fertile sibs and increased at subsequent stages, exceeding wild type levels at the 2.0 mm stage. The ms8 mutant has a recognizable tapetal layer albeit with fewer cells (Figure 2c), and these cells accumulate but never secrete dark-staining material (likely protein, Figure S1, a1 - a3). The array data indicate that tapetal cellular differentiation is disrupted, and the transcriptome pattern of 6 tapetal marker genes is abnormal at the 1.5 mm stage.
Figure 7.
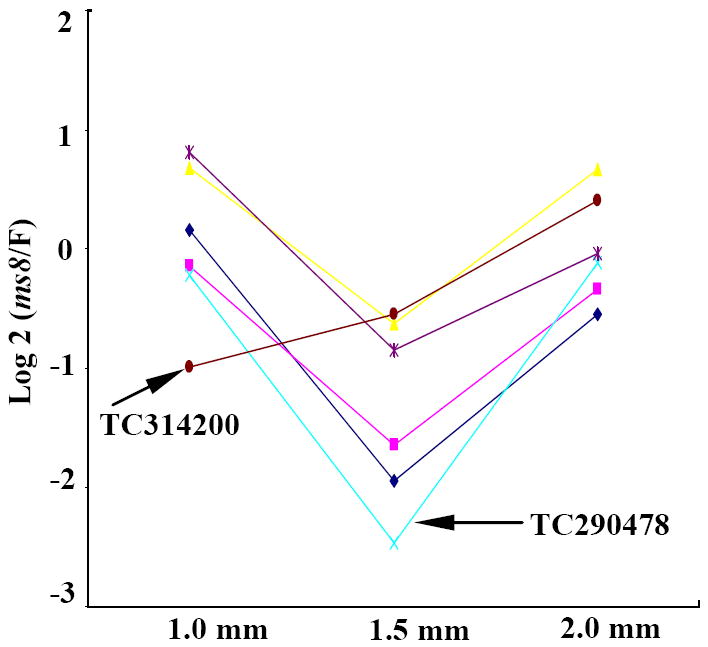
Expression comparison of potential cell layer markers. Fold change pattern of previously published potential tapetum markers (Ma et al., 2007), N=6. The y-axis is log2 (fold change). The x-axis shows the anther developmental stages.
To address the contribution of an individual cell type to the distinctness of the ms8 transcriptome, the sets of cell-type specific genes from three laser-microdissected cell types (i.e., the epidermis, tapetum and meiocytes) from 2.0 mm fertile anthers (D. S. Skibbe, pers. communication) were compared to the ms8 transcriptome. The ms8 mutant is 96 - 98% similar quantitatively to a normal anther for the cell type-specific transcripts assessed (epidermis = 632 out of 659; tapetum = 451 out of 459; meiocytes = 1944 out of 2007); most differences between ms8 and fertile siblings reflect mis-expression of transcripts expressed in more than one cell type at 2.0 mm rather than cell type-specific transcripts. Considered along with the measured impacts on growth in epidermal and tapetal cells it is clear that the ms8 mutation affects multiple cell types.
qRT-PCR with primers for a panel of 22 genes was used to determine the validity of specific conclusions such as constitutive expression or differential regulation of ms8 relative to fertile. Sixteen constitutive genes that maintained the same quantitative expression levels across all three stages were confirmed by qRT-PCR analysis of fertile and ms8 samples. Six genes differentially expressed in at least one stage also were consistent with the microarray data (Figure S3, Table S7).
Identification of proteins with altered abundance by 2-D DIGE and LC/MS/MS
Anthers from the same family collected for array hybridization were used for 2D-DIGE, comparing ms8 to normal siblings. For each stage, IEF was performed using two overlapping pH gradients, pH 4-7 and pH 6-11, analyzed on two different gels to increase the probability of separating proteins with similar isoelectric points. Typically, for pH 4-7 strips (24 cm), about 2700 spots were analyzed plus about 2200 spots for pH 6-11 strips (18 cm, data not shown). In the comparisons, virtually all resolved spots are yellow indicating a similar intensity (Figure S4). Eighty-five protein spots with differential intensities of more than 1.5-fold in ms8 to fertile comparisons were detected across the three stages: 68 were higher in ms8 anthers and 17 were lower (data not shown). For some spots, differential expression at several stages was detected (Table S8). After MS/MS peptide sequencing, 63 unique gene products were identified (Table S8) and functional annotations were summarized in Figure S5; proteins associated with meiosis and tapetal activities in other plants were found to be differentially expressed in ms8 (Table S9).
As observed in transcriptome studies, the highest representation of differentially expressed proteins is in metabolism: 24 proteins are up-regulated and 5 are down-regulated in ms8 anthers (Table S8). During microsporogenesis, the tapetum supplies nutrients to sporogenous tissue (Regan and Moffatt, 1990) preceded by synthesis of free fatty acids, polar lipids, and neutral esters stored in elaioplasts and lipid bodies (Piffanelli and Mudrphy, 1998). ATP citrate lyase (Elshourbagy et al., 1992) is critical in fatty acid synthesis and is elevated in ms8 anthers. Two down-regulated proteins are involved in amino acid metabolism: homocysteine S-methyltransferase-3 and cysteine synthase. Several tapetum-specific markers are cysteine-rich proteins proposed to be components of a regulatory network in young anthers (Tzeng et al., 2009); synthesis of such anther-specific regulators might be affected by altered amino acid levels. Starch is synthesized in anthers before meiosis and subsequently hydrolyzed to provide energy for lipid synthesis in both tapetum and microspores (Vizcay-Barrena and Wilson, 2006). Aldolase1 is also increased in ms8 anthers, which could alter levels of sugar and starch, two molecules key to biosynthesis and energy balance. UDP-glucose-4-epimerase is higher in ms8 anthers; this enzyme catalyzes the reversible epimerization of UDP-galactose and UDP-glucose. Elevated enzyme levels in ms8 anthers might cause low UDP-galactose in vivo and affect the biosynthesis of galactose-containing cell-wall carbohydrates (Dormann and Benning, 1998). Phospholipase D increases 2.82 fold at the 1.5 mm stage in ms8 anthers, when mutant meiocytes had already entered prophase I. Activation of this enzyme triggers plant microtubule reorganization in interphase and during preprophase of mitotic cells (Dhonukshe et al., 2003), hence the up-regulation of this protein in ms8 at the 1.5 mm stage could contribute to abnormal proliferation in the tapetum or aberrant features of meiosis.
Besides metabolic pathways, proper anther development requires diverse regulatory processes. 14-3-3 proteins are phosphoserine-binding proteins that regulate targets via direct protein-protein interactions, modulating among other processes plasma membrane H-ATPase, nitrate reductase, sucrose phosphate synthase, and diverse cytoplasmic enzymes (Chung et al., 1999). A 14-3-3 like protein (GI: 162458469) is down-regulated in ms8 1.5-mm-stage anthers (Table S8), and another 14-3-3 like protein (GI: 226499592) was up-regulated from the 1.0-mm to 1.5-mm stage (Table S8, Figure S6). The aberrant abundances of such 14-3-3 factors could contribute directly to ms8 defects. In ms8 anthers, methyl binding domain protein expression was increased before and during meiotic prophase I, and this may be related to the incipient dissolution of the PMC as developmentally-regulated DNA methylation is crucial in plants (Zemach and Grafi, 2007). Prohibitins are evolutionarily conserved proteins (Takahashi et al., 2003) that play key roles in cell cycle regulation, receptor-mediated signaling at the cell surface, senescence, tumor suppression, mitochondrial function and morphology (Narasimhan et al., 1997; Nadimpalli et al., 2000; Chen et al., 2005). There are 4 distinct full-length prohibitin-like genes in maize (Nadimpalli et al., 2000) and both prohibitin 1 and prohibitin 4 are up-regulated at the meiosis stage in ms8 anthers (Table S8), another hallmark of dysfunction.
In eukaryotes, the spindle assembly checkpoint monitors the interaction between chromosomes and microtubules at the kinetochores, an elaborate mechanism required for correct chromosome segregation during mitosis (Caillaud et al., 2009). We speculate that increased accumulation of mitotic checkpoint protein BUB3 (Table S8) in young ms8 anthers subsequently affects spindle morphology/function in meiocytes and leads to abnormal meiosis. Phragmoplastin is a dynamin-like protein associated with cell plate formation in plants, and it is encoded by 8 types of maize ESTs (data not shown); in addition to mis-expression of mitotic checkpoint protein BUB3 that might affect phragmoplast expansion, accumulation of excess phragmoplastin (Table S8) may also disrupt meiotic progression because these cells are normally interconnected.
Congruence of ms8 anthers array data to proteome analysis
We compared the ms8-regulated proteins identified by 2-D DIGE with the ms8-regulated RNAs pinpointed by microarray studies (Figure S3b). Probes corresponding to only eight of the 63 identified differentially expressed proteins had differential RNA accumulation: 14-3-3 like protein (226499592), aspartate aminotransferase, translation initiation factor 5A, protein disulfide isomerase, adenosylhomocysteinase, GTP-binding protein SAR1A, histone H2A, and elongation factor 1-alpha. For the remaining proteins, there was no significant change in RNA abundance. The small overlap between proteomics and microarray data highlights the inability of transcriptome profiling to identify changes in translational regulation, protein turnover kinetics, and post-translational modification. Therefore, the proteomics assessment yielded critical additional information augmenting understanding of the ms8 phenotypes.
DISCUSSION
Previous studies classified ms8 as defective in tetrad formation (Albertsen and Phillips, 1981). Based on cytological and morphometric analysis we find that ms8 causes a dwarf anther with a cell expansion defect in the epidermis and cell proliferation and differentiation defects in the tapetum evident before meiosis initiates. Because excess callose is retained around the PMC and tapetal cells remain filled with material, it is likely that defective tapetal secretion starves meiotic cells resulting in abortive meiosis. A striking feature of anther development is high transcriptome diversity, observed in maize (Ma et al., 2006; this study), Arabidopsis (Wijeratne et al., 2007), and rice (Jung et al., 2005). The progression of gene expression programs is profoundly disrupted in ms8, because mutants express genes atypical of normal anthers and express many genes precociously.
Analysis of cell type-specific expression indicates that most epidermal, tapetal, and meiotic cell marker genes are expressed at quantitatively normal levels in ms8 at the 2.0 mm stage. Therefore, the majority of differences between ms8 and normal reflect mis-expression of transcripts expressed in more than one cell type at the 2.0 mm stage. ms23 and ms32 are defective because presumptive tapetal cells undergo a periclinal division forming two aberrant layers (Chaubal et al., 2000); ms8 is very distinctive from these strictly tapetal mutants in that a much higher fraction (>10%) of the ms8 transcriptome is altered than in either tapetal mutant (~1%, Ma et al., 2007; C. A. Young and V. Walbot, unpublished data). Furthermore, more than 40% of mis-expressed genes in ms8 represent acceleration of developmental programs relative to fertile siblings. This insight indicates that precise temporal modulation is key to the progression of early anther development.
The 1.5 mm stage of ms8 had the most significant changes compared to fertile in that transcriptome diversity was decreased, and there were the largest number of differentially expressed genes. ms8 meiocytes cease meiosis and the tapetum is highly abnormal by the 2.0 mm stage -- phenotypic consequences of mis-expression at the 1.5 mm stage 18 h earlier. We propose that the 1.5 mm stage represents a decision point for anther developmental processes, which if aberrant result in meiotic failure.
Mutations affecting tapetal cell size and number have been reported across a broad spectrum of flowering plants (Ma, 2005), and almost all lead to an abortive meiosis and male sterility. Arabidopsis TDF1 (defective in tapetal development and function 1) expresses an anther-specific transcript that encodes a putative R2R3 MYB transcription factor, which is indirectly required for callose breakdown. From the tetrad stage, tdf1 tapetum cells are abnormally vacuolated and enlarged with excessive cell division (Zhu et al., 2008). Mutation in rice TDR (tapetum degeneration retardation) encoding a putative basic helix-loop-helix protein caused tapetal cells to grow abnormally large, ultimately occupying the majority of the locule (Li et al., 2006) as found in maize ms8. Both tdf1 and tdr mutants have small locules of irregular shape (Li et al., 2006; Zhu et al., 2008), a feature shared by many other mutants (Sorensen et al., 2003; Yang et al., 2003; Albrecht et al., 2005; Zhang et al., 2006); however, locule dimension and cell count assays are available only for ms8, pinpointing specific defects. Meiotic failure is a routine feature among Arabidopsis mutants with tapetum developmental defects (Sorensen et al., 2003; Yang et al., 2003; Albrecht et al., 2005; Zhang et al., 2006; Zhu et al., 2008); ms8 dyads broke down and meiosis aborted, another example connecting tapetal abnormality to meiotic failure. Transcription profiling and proteomics data are not available for tdf and tdr, or other maize anther mutants in early stages for comparison to ms8. Therefore, this study provides a compilation of in-depth transcriptome and proteome changes during early anther development and will be a reference for future studies of additional mutants of maize and other species.
Callose, a β-1, 3-glucan polymer with β-1, 6-branches, is an insoluble polysaccharide, deposited onto the PMC at the start of meiotic prophase I. It reaches a maximum at the tetrad stage (Popova et al., 2008). By aniline blue staining and quantitative analysis, ms8 anthers accumulate more callose at 1.5 mm, which occupied the extra space between PMC, while at the 2.0 mm stage callose is so excessive that meiocytes are often crowded to one side of the locule. Callose accumulation appears to reflect failure of the tapetum to degrade and remodel this polysaccharide, because β -D-glucosidase is down-regulated in ms8 mutants and no differentially regulated callose synthase genes were detected. Based on sequence homology, twelve CALLOSE SYNTHASE (CalS) genes have been identified in Arabidopsis, and five of them are required for microgametogenesis (Chen et al., 2009). For example, CalS5 encodes a callose synthase protein responsible for the temporary deposition of the callose wall surrounding microspores. CalS5 gene knockouts severely disrupted callose deposition and reduced fertility (Dong et al., 2005). AtMYB103 is an R2R3 MYB protein that controls callose dissolution in Arabidopsis. In knockout mutants, callose persisted into the final stages of pollen maturation; such lines are male-sterile (Zhang et al., 2007). Several studies have also described mutants in callose wall formation and dissolution in petunia (Izhar and Frankel, 1971; Warmke and Overman, 1972) and tobacco (Worrall et al., 1992) that disrupt fertility. Collectively, the evidence indicates that the timing of callose formation and dissolution are critical for normal fertility, and ms8 is a new example in maize of a callose defect.
Post-meiotic tapetal cells nourish microspores and deposit complex lipoidal mixtures in tryphine or pollenkitt onto pollen (Bedinger, 1992). Several genes involved in lipid metabolism play important roles in Arabidopsis pollen development (Aarts et al., 1997; Mou et al., 2000). As was found in ms8, this class of proteins was also up-regulated in the tomato 7B-1 male-sterile mutant (Sheoran et al., 2009). Aberrant regulation of enzymes involved in polysaccharide (plastid starch synthase I precursor), sugar (UDP-glucose-4-epimerase, Aldolase1), and lipid metabolism are hallmarks of ms8 and other early-acting male-sterile mutants in flowering plants. Given the significant impact of the ms8 mutation on the transcriptome and proteome during early maize anther development, ms8 appears to be involved in the regulation of multiple metabolic pathways.
Experimental procedures
Plant material and microscopy
ms8 was obtained from P. Bedinger (Colorado State University, Ft. Collins CO) in the ND101 inbred and backcrossed four times into W23 and maintained in 1:1 segregating families (ms8//ms8 × ms8//+) in summer fields at Stanford University or in greenhouses equipped with lights to replicate 50% of summer solar fluence. Samples were removed from 3-6 cm immature tassels by cutting through the leaf whorl; at maturity, dissected plants were scored for fertility. Only upper floret anthers were pooled from several spikelets after careful developmental staging and then sections prepared (Wang et al., 2009) for bright field microscopy (Axioskop 40 Pol, Carl Zeiss MicroImaging, Inc, Germany) or stained with aniline blue (Aldrich Chemical Company, Milwaukee, WI) to visualize callose under UV illumination (Axioskop 40 Pol, DAPI channel). Callose was also quantified fluorimetrically in anther extracts (Kohle et al., 1985). Meiocytes were observed by hematoxylin-iron-aceto-carmine staining (Chang and Neuffer, 1989). Scanning electron microscopy (FEI Quanta 200 scanning electron microscope, FEI company, Hillsboro, OR) images were obtained using a published protocol (Ohashi-Ito and Bergmann, 2006). Morphometric data were collected from four independent samples; four locules for each stage were examined, averaged, and the data analyzed in Excel (Microsoft, Redmond, WA). Propidium iodide (CalBiochem, San Diego, CA) was used at 20 μg/mL in PBS solution (pH 7.4) for 30 min, with 3 infiltrations for 1-2 min each to stain anther nuclei for laser-scanning confocal microscopy using a Leica SP5 (Leica Microsystem Inc, U.S.). Anther samples were imaged in water on a glass slide with a cover slip using 520 to 550 nm excitation and 600 to 650 nm emission. Cell dimension measurements were obtained from propidium iodide images and calculated by volocity software (PerkinElmer, MA, U.S.A.).
Microarray analysis and qRT-PCR assays
Total RNA was extracted with TRIzol (Invitrogen, Carlsbad, CA) from pools of staged anthers, and labeled cRNA was prepared as described previously (Ma et al., 2006) for hybridization on slides with 4 arrays each of 44K elements (Agilent Technologies, Santa Clara, CA) using the Two-Color Microarray-Based Gene Expression Analysis protocol (Agilent). Four independent biological replicates at each developmental stage were analyzed with balanced dye labeling to minimize systematic variances (Table S10, Ma et al., 2006; Kerr and Churchill, 2001). After hybridization according to the manufacturer’s protocols, slide scanning, feature extraction, signal criterion, statistical and graphical criteria, and GO annotation were performed as described previously (Skibbe et al., 2009). Normalized intensities for each probe were averaged for the 4 biological replicates and the resulting final expression data and gene annotation were summarized in Table S11. The log base 2 of the ratio of ms8 versus fertile sibling expression was used for clustering differentially-expressed genes as described previously (Ma et al., 2008). The log base 2 of the normalized expression values was used for clustering the zinc finger genes. Primer pairs (Table S7) were designed for selected genes, then qRT-PCR (quantitative real-time reverse transcriptase polymerase chain reaction) reactions and amplification conditions followed published protocols (Ma et al., 2008).
2D-DIGE
Protein extraction and quality assessment were performed as reported previously (Skibbe et al., 2009). Three biological replicates of each developmental stage were used to identify differentially abundant spots between ms8 and fertile siblings after labeling protein aliquots with either Cy3 or Cy5 (GE Healthcare, NJ). Four replicates were used for microarrays, because a balanced dye swap design is required for maximal statistical power; this design feature is not required for the proteomics analysis because an internal reference containing the same proteins was included in every gel. A mixture of all samples was labeled with Cy2 (GE Healthcare) as this internal reference on all gels to improve the accuracy of quantitative comparisons (Alban et al., 2003). An example of reproducibility is shown in Figure S4, utilizing a dye swap experiment of pairs of biological replicates. Gels were scanned using a Typhoon 8600 (GE Healthcare) with appropriate wavelengths for excitation and emission (Sheoran et al., 2009). Images were processed with PROGENESIS PG220 software (Non-linear USA, http://www.nonlinear.com) with statistical analyses in this software. Spots with fold change ≥ 1.5 or ≤ -1.5 and p value < 0.05 were considered to be differentially expressed. 200 μg aliquots of protein from ms8 and its fertile counterpart plus 25 μg of each sample labeled with Cy3 or Cy5 DIGE fluor were separated on a 2D-DIGE preparative gel. The gel was fixed and stained with Deep Purple (Fluorotechnics, http://www.fluorotechnics.com), spots of interest were edited with DeCyder software (GE Healthcare) and picked by an Ettan spot picker (GE Healthcare). In-gel tryptic digestion was done as described previously (Rosenfeld et al., 1992, donatello.ucsf.edu/ingel.html).
Reversed-phase LC-electrospray tandem mass spectrometry (LC-MS/MS) analysis
Digests were separated by nanoflow LC as described (Oses-Prieto et al., 2007) with minor revisions. LTQ-FT and LTQ-Orbitrap raw data were converted to ASCII peak lists by Mascot Distiller software v2.1.0.0 (Matrix Science, Boston, MA); the parameters for MS processing were used as described previously (Oses-Prieto et al., 2007). The peak lists were searched in ProteinProspector version 5.3.2 (http://prospector2.ucsf.edu/prospector/html/misc/revhist.htm) (public version at http://prospector.ucsf.edu) using an existing protocol with minor updates (Deng et al., 2007). The peak lists were searched using the NCBInr database as of July 10, 2009, using all entries for Zea, Oryza, Hordeum, Triticum, Secale, and Arabidopsis (249,067 entries searched). Because the maize genome is not yet completely annotated, in some cases we relied on identification of peptides conserved in predicted proteins of plants with completely sequenced genomes (Arabidopsis, rice) or in cereals (Triticum, Hordeum and Secale) (Table S9). The acceptance criteria were described previously (Skibbe et al., 2009); only proteins with at least 2 peptides identified were considered further. When several accessions matched the same set of peptides, the entries with the most descriptive name were reported. Individual protein isoforms were reported when unique peptides were found; if isoforms shared the peptides, they were all reported.
Supplementary Material
Figure S1. Tapetal cell morphology. Transverse sections were taken at several developmental stages. (a1) ms8 at 2 mm, tapetal cell thickness = 14 μm; (a2) ms8 at 2.5 mm, tapetal cell thickness = 20 μm; (a3) ms8 at 3 mm, tapetal cell thickness = 18 μm; (a4) fertile at 3 mm, tapetal cell thickness = 9 μm.
Figure S2. Venn diagrams of transcriptome changes during fertile (a) and sterile (b) anther development; stage-specific transcripts and transcripts shared by two or three stages are shown respectively.
Figure S3. qRT-PCR validation of microarray and proteome data. (a) Validation of a set of 22 genes using qRT-PCR. Assays were performed in triplicates on four biological replicates of both the fertile and ms8 samples for three developmental stages. Cyanase was used as the internal standard to calculate ΔCt=Ct(sample)-Ct(standard) (Ct: cycle threshold). The ΔCt value is a normalized relative expression value, parallel to a log2 ratio because of the doublings in PCR reactions. Those genes labeled with “◆” are differential expressed in ms8 anthers, based on microarray data. The red line-dot-line brackets the two-fold cutoff for up-regulated and down-regulated expression. (b) Congruence of eight genes that had same patterns in microarray, qRT-PCR, and proteomic data. The proteomic fold change, the log 2 ratio of microarray and qRT-PCR fold changes are shown.
Figure S4. 2-D DIGE analysis of ms8 anthers. (a) Proteins of ms8 (Cy5, red) were compared with those fertile sib samples (Cy3, green) by 2-D DIGE using 24-cm pH 4-7 IPG strips and a 12.5% SDS-PAGE gel. Proteins increased in ms8 appear red, those decreased appear green, whereas those unaffected appear yellow. (b) Cyanine dyes swap for (a). The molecular masses of marker proteins are indicated on the right of (a) and (b), and pH values on the edges of the gel are indicated on the top of (a).
Figure S5. Classification of ms8 regulated proteins based on their putative functions. Proteins with ratios at least 1.5-fold enhanced or decreased are included in the diagram. The categories proceed clockwise.
Figure S6. Increased spot I6-35 is found at both the 1.0 mm (left panel) and 1.5 mm stages (right panel) during ms8 anther development.
Table S1. Genes involved in early anther development of Arabidopsis or maize with detailed phenotypic descriptions.
Table S2. Differential expressed genes, comparing ms8 to fertile.
Table S3. Four hundred and sixteen 1.0 mm stage-specific genes are expressed at a later stage in normal anthers.
Table S4. One hundred and thirty-three 1.5 mm stage specific genes in ms8 are expressed at the 2.0 mm stage in normal anthers.
Table S5. Ten MYB transcription factors were up-regulated in ms8 during the 1.5 mm stage.
Table S6. Eight zinc finger family genes are differentially expressed in at least one stage.
Table S7. Primers used for qRT-PCR assays.
Table S10. Microarray experiment design.
Table S11. Average probe intensity, final processed data, probe sequences and samples annotation.
Table S8. ms8 regulated proteins with maize identifiers. Average protein level ratios of ms8/fertile sibs are shown. The t test p values were calculated from three biological samples and three repeat experiments.
Table S9. Proteins regulated by ms8. Using functional annotation from rice proteins, which have high identity to predicted maize proteins.
Acknowledgments
Research was supported by the National Science Foundation (07-01880) and by the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF (A.L. Burlingame, Director) supported by the Biomedical Research Technology Program of the NIH National Center for Research Resources, NIH NCRR P41RR001614 and NIH NCRR RR019934.
Contributor Information
Dongxue Wang, Email: wangdx@stanford.edu.
Juan A. Oses-Prieto, Email: joses@cgl.ucsf.edu.
Kathy H. Li, Email: kathyli@cgl.ucsf.edu.
John F. Fernandes, Email: jfernand@stanford.edu.
Alma L. Burlingame, Email: alb@cgl.ucsf.edu.
Virginia Walbot, Email: walbot@stanford.edu.
References
- Aarts MGM, Hodge R, Kalantidis K, Florack D, Wilson ZA, Mulligan BJ, Stiekema WJ, Scott R, Pereira A. The Arabidopsis MALE STERILITY 2 protein shares similarity with reductases in elongation/condensation complexes. Plant J. 1997;12:615–623. doi: 10.1046/j.1365-313x.1997.00615.x. [DOI] [PubMed] [Google Scholar]
- Alban A, David SO, Bjorkesten L, Andersson C, Sloge E, Lewis S, Currie I. A novel experimental design for comparative two-dimensional gel analysis: two dimensional difference gel electrophoresis incorporating a pooled internal standard. Proteomics. 2003;3:36–44. doi: 10.1002/pmic.200390006. [DOI] [PubMed] [Google Scholar]
- Albrecht C, Russinova E, Hecht V, Baaijens E, Vries S. The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES 1 and 2 control male sporogenesis. Plant Cell. 2005;17:3337–3349. doi: 10.1105/tpc.105.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen MC, Phillips RL. Developmental cytology of 13 genetic male sterile loci in maize. Genome. 1981;23:195–208. [Google Scholar]
- Beadle GW. Genes in maize for pollen sterility. Genetics. 1931;17:413–431. doi: 10.1093/genetics/17.4.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P. The remarkable biology of pollen. Plant Cell. 1992;4:879–887. doi: 10.1105/tpc.4.8.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger PA, Fowler JE. The maize male gametophyte. In: Bennetzen JL, Hake SC, editors. Handbook of Maize: Its Biology. New York: Springer; 2009. pp. 57–77. [Google Scholar]
- Bucciaglia PA, Smith AG. Cloning and characterization of Tag 1, a tobacco anther beta-1,3-glucanase expressed during tetrad dissolution. Plant Mol Biol. 1994;24:903–914. doi: 10.1007/BF00014444. [DOI] [PubMed] [Google Scholar]
- Caillaud MC, Paganelli L, Lecomte P, Deslandes L, Quentin M, Pecrix Y, Bris ML, Marfaing N, Abad P, Favery B. Spindle assembly checkpoint protein dynamics reveal conserved and unsuspected roles in plant cell division. PLoS ONE. 2009;4:1–9. doi: 10.1371/journal.pone.0006757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canales C, Bhatt AM, Scott R, Dickinson H. EXS, a putative LRR receptor kinase, regulates male germline cell number and tapetal identity and promotes seed development in Arabidopsis. Current Biol. 2002;12:1718–1727. doi: 10.1016/s0960-9822(02)01151-x. [DOI] [PubMed] [Google Scholar]
- Chang MT, Neuffer MG. A simple method for staining nuclei of mature and germinated maize pollen. Biotechnic Histochemistry. 1989;64:181–184. doi: 10.3109/10520298909106996. [DOI] [PubMed] [Google Scholar]
- Chaubal R, Anderson JR, Trimnell MR, Fox TW, Albertsen MC, Bedinger P. The transformation of anthers in the msca1 mutant of maize. Planta. 2003;216:778–788. doi: 10.1007/s00425-002-0929-8. [DOI] [PubMed] [Google Scholar]
- Chaubal R, Zanella C, Trimnell MR, Fox TW, Albertsen MC, Bedinger P. Two male-sterile mutants of Zea mays (Poaceae) with an extra cell division in the anther wall. Am J Bot. 2000;87:1193–1201. [PubMed] [Google Scholar]
- Chen JC, Jiang CZ, Reid MS. Silencing a prohibitin alters plant development and senescence. Plant J. 2005;44:16–24. doi: 10.1111/j.1365-313X.2005.02505.x. [DOI] [PubMed] [Google Scholar]
- Chen XY, Liu L, Lee E, Han X, Rim Y, Chu H, Kim SW, Sack F, Kim JY. The Arabidopsis callose synthase gene GSL8 is required for cytokinesis and cell patterning. Plant Physiol. 2009;150:105–113. doi: 10.1104/pp.108.133918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HJ, Sehnke PC, Ferl RJ. The 14-3-3 proteins: cellular regulators of plant metabolism. Trends Plant Sci. 1999;4:367–371. doi: 10.1016/s1360-1385(99)01462-4. [DOI] [PubMed] [Google Scholar]
- Deng Z, Zhang X, Tang W, Oses-Prieto JA, Suzuki N, Gendron JM, Chen H, Guan S, Chalkley RJ, Peterman TK, Burlingame AL, Wang Z. A proteomics study of brassinosteroid response in Arabidopsis. Mol Cell Proteomics. 2007;6:2058–2071. doi: 10.1074/mcp.M700123-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Laxalt AM, Goedhart J, Gadella TWJ, Munnik T. Phospholipase D activation correlates with microtubule reorganization in living plant cells. Plant Cell. 2003;15:2666–2679. doi: 10.1105/tpc.014977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DPS. Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J. 2005;42:315–328. doi: 10.1111/j.1365-313X.2005.02379.x. [DOI] [PubMed] [Google Scholar]
- Dormann P, Benning C. The role of UDP-glucose epimerase in carbohydrate metabolism of Arabidopsis. Plant J. 1998;13:641–652. doi: 10.1046/j.1365-313x.1998.00067.x. [DOI] [PubMed] [Google Scholar]
- Elshourbagy NA, Near JC, Kmetz PJ, Wells TN, Groot PH, Saxty BA, Hughes SA, Franklin M, Gloger IS. Cloning and expression of a human ATP-citrate lyase cDNA. Eur J Biochem. 1992;204:491–499. doi: 10.1111/j.1432-1033.1992.tb16659.x. [DOI] [PubMed] [Google Scholar]
- Goldberg RB, Beals TP, Sanders PM. Anther development: basic principles and practical applications. Plant Cell. 1993;5:1217–1229. doi: 10.1105/tpc.5.10.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hord CLH, Chen C, DeYoung BJ, Clark SE, Ma H. The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell. 2006;18:1667–1680. doi: 10.1105/tpc.105.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izhar S, Frankel R. Mechanism of male sterility in Petunia: The relationship between pH, callase activity in the anthers, and the breakdown of the microsporogenesis. Theoret Appl Gen. 1971;41:104–108. doi: 10.1007/BF00277751. [DOI] [PubMed] [Google Scholar]
- Juan KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim Y, Nahm BH, An G. Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell. 2005;17:2705–2722. doi: 10.1105/tpc.105.034090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr MK, Churchill GA. Statistical design and the analysis of gene expression microarray data. Genet Res. 2001;77:123–128. doi: 10.1017/s0016672301005055. [DOI] [PubMed] [Google Scholar]
- Kobayashi A, Sakamoto A, Kubo K, Rybka Z, Kanno Y, Takatsuji H. Seven zinc-finger transcription factors are expressed sequentially during the development of anthers in petunia. Plant J. 1998;13:571–576. doi: 10.1046/j.1365-313x.1998.00043.x. [DOI] [PubMed] [Google Scholar]
- Kohle H, Jeblick W, Poten F, Blaschek W, Kauss H. Chitosan-elicited callose synthesis in soybean cells as a Ca2+-dependent process. Plant Physiol. 1985;77:544–551. doi: 10.1104/pp.77.3.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, Wen TQ, Huang H, Luo D, Ma H, Zhang DB. The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell. 2006;18:2999–3014. doi: 10.1105/tpc.106.044107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. Molecular genetic analyses of microsporogenesis and microgametogenesis in flowering plants. Annu Rev Plant Biol. 2005;56:393–434. doi: 10.1146/annurev.arplant.55.031903.141717. [DOI] [PubMed] [Google Scholar]
- Ma J, Duncan D, Morrow DJ, Fernandes J, Walbot V. Transcriptome profiling of maize anthers using genetic ablation to analyze pre-meiotic and tapetal cell type. Plant J. 2007;50:637–648. doi: 10.1111/j.1365-313X.2007.03074.x. [DOI] [PubMed] [Google Scholar]
- Ma J, Morrow DJ, Fernandes J, Walbot V. Comparative profiling of the sense and antisense transcriptome of maize lines. Genome Biol. 2006;7 doi: 10.1186/gb-2006-7-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Skibbe DS, Fernandes J, Walbot V. Male reproductive development: gene expression profiling of maize anther and pollen ontogeny. Genome Biol. 2008;9 doi: 10.1186/gb-2008-9-12-r181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP. Gene activity during pollen development. Annu Rev Plant Physiol. 1990;41:317–338. [Google Scholar]
- Mou Z, He Y, Dai Y, Liu X, Li J. Deficiency in fatty acid synthase leads to premature cell death and dramatic alterations in plant morphology. Plant Cell. 2000;12:405–417. doi: 10.1105/tpc.12.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadimpalli R, Yalpani N, Johal GS, Simmons CR. Prohibitins, stomatins, and plant disease response genes compose a protein superfamily that controls cell proliferation, ion channel regulation, and death. J Biol Chem. 2000;275:29579–29586. doi: 10.1074/jbc.M002339200. [DOI] [PubMed] [Google Scholar]
- Narasimhan S, Armstrong M, McClung JK, Richards FF, Spicer EK. Prohibitin, a putative negative control element present in Pneumocystic carinii. Infect Immun. 1997;65:5125–5130. doi: 10.1128/iai.65.12.5125-5130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi-Ito K, Bergmann DC. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell. 2006;18:2493–2505. doi: 10.1105/tpc.106.046136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oses-Prieto JA, Zhang X, Burlingame AL. Formation of Є-formyllysine on silver-stained proteins. Mol Cell Proteomics. 2007;6:181–192. doi: 10.1074/mcp.M600279-MCP200. [DOI] [PubMed] [Google Scholar]
- Pastore J, Chavdaroff N, Wagner D. LMI2, a MYB transcription factor involved in the vegetative to reproductive transition in Arabidopsis thaliana. Dev Biol. 2008;319:565–575. [Google Scholar]
- Piffanelli P, Murphy DJ. Novel organelles and targeting mechanisms in the anther tapetum. Trends Plant Sci. 1998;3:250–253. [Google Scholar]
- Popova AF, Ivanenko GF, Ustinova AY, Zaslavsky VA. Localization of callose in microspores and pollen grains in Sium latifolium L. plants in different water regimes. Cytol Gen. 2008;42:363–368. [PubMed] [Google Scholar]
- Regan SM, Moffatt BA. Cytochemical analysis of pollen development in wild-type Arabidopsis and a male-sterile mutant. Plant Cell. 1990;2:877–889. doi: 10.1105/tpc.2.9.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J, Capdeveille J, Guillemot JC, Ferrara P. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal Biochem. 1992;203:173–179. doi: 10.1016/0003-2697(92)90061-b. [DOI] [PubMed] [Google Scholar]
- Sheoran IS, Ross ARS, Olson DJH, Sawhney VK. Differential expression of proteins in the wild type and 7B-1 male-sterile mutant anthers of tomato (Solanum lycopersicum): A proteomic analysis. J Proteomics. 2009;71:624–636. doi: 10.1016/j.jprot.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Skibbe DS, Schnable PS. Male sterility in maize. Maydica. 2005;50:367–376. [Google Scholar]
- Skibbe DS, Fernandes JF, Medzihradszky KF, Burlingame AL, Walbot V. Mutator transposon activity reprograms the transcriptomes and proteomes of developing maize anthers. Plant J. 2009;59:622–633. doi: 10.1111/j.1365-313X.2009.03901.x. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- Sorensen AM, Kröber S, Unte US, Huijser P, Dekker K, Saedler H. The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 2003;33:413–423. doi: 10.1046/j.1365-313x.2003.01644.x. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Kawasaki T, Wong HL, Suharsono U, Hirano H, Shimamoto K. Hyperphosphorylation of a mitochondrial protein, prohibitin, is induced by calyculin A in a rice lesion-mimic mutant cdr1. Plant Physiol. 2003;132:1861–1869. doi: 10.1104/pp.103.021733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S, Matsumoto N, Okada K. RABBIT EARS, encoding a superman-like zinc finger protein, regulates petal development in Arabidopsis thaliana. Development. 2004;131:425–434. doi: 10.1242/dev.00938. [DOI] [PubMed] [Google Scholar]
- Tzeng JD, Hsu SW, Chung MC, Yeh FL, Yang CY, Liu MC, Hsu YF, Wang CS. Expression and regulation of two novel anther-specific genes in Lilium longiflorum. J Plant Physiol. 2009;166:417–427. doi: 10.1016/j.jplph.2008.06.019. [DOI] [PubMed] [Google Scholar]
- Vizcay-Barrena G, Wilson ZA. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J Exp Bot. 2006;57:2709–2717. doi: 10.1093/jxb/erl032. [DOI] [PubMed] [Google Scholar]
- Wang D, Li C, Zhao Q, Zhao L, Wang M, Zhu D, Ao G, Yu J. Zm401p10, encoded by an anther-specific gene with short open reading frames, is essential for tapetum degeneration and anther development in maize. Functional Plant Biol. 2009;36:73–85. doi: 10.1071/FP08154. [DOI] [PubMed] [Google Scholar]
- Wang XL, Li XB. The GhACS1 gene encodes an acyl-CoA synthetase which is essential for normal microsporogenesis in early anther development of cotton. Plant J. 2009;57:473–486. doi: 10.1111/j.1365-313X.2008.03700.x. [DOI] [PubMed] [Google Scholar]
- Warmke HE, Overman MA. Cytoplasmic male sterility in sorghum. I. Callose behavior in fertile and sterile anthers. J Hered. 1972;63:103–108. [Google Scholar]
- Wijeratne AJ, Zhang W, Sun Y, Liu W, Albert R, Zheng Z, Oppenheimer D, Zhao D, Ma H. Differential gene expression in Arabidopsis wild-type and mutant anthers: insights into anther cell differentiation and regulatory networks. Plant J. 2007;52:14–29. doi: 10.1111/j.1365-313X.2007.03217.x. [DOI] [PubMed] [Google Scholar]
- Worrall D, Hird DL, Hodge R, Paul W, Draper J, Scott R. Premature dissolution of the microsporocyte callose wall causes male sterility in transgenic tobacco. Plant Cell. 1992;4:759–771. doi: 10.1105/tpc.4.7.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, You C, Li C, Long T, Chen G, Byrne ME, Zhang Q. RID1, encoding a Cys2/His2-type zinc finger transcription factor, acts as a master switch from vegetative to floral development in rice. Proc Nat Acad Sci USA. 2008;105:12915–12920. doi: 10.1073/pnas.0806019105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu SX, Liu GS, Chen RD. Characterization of an anther- and tapetum-specific gene and its highly specific promoter isolated from tomato. Plant Cell Rep. 2006;25:231–240. doi: 10.1007/s00299-005-0056-7. [DOI] [PubMed] [Google Scholar]
- Yang SL, Xie LF, Mao HZ, Puah CS, Yang WC, Jiang L, Sundaresan V, Ye D. The TAPETUM DETERMINANT1 gene is required for cell specialization in the Arabidopsis anther. Plant Cell. 2003;15:2792–2804. doi: 10.1105/tpc.016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemach A, Grafi G. Methyl-CpG-binding domain proteins in plants: interpreters of DNA methylation. Trends Plant Sci. 2007;12:1380–1385. doi: 10.1016/j.tplants.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Zhang W, Sun Y, Timofejeva L, Chen C, Grossniklaus U, Ma H. Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development. 2006;133:3085–3095. doi: 10.1242/dev.02463. [DOI] [PubMed] [Google Scholar]
- Zhang ZB, Zhu J, Gao JF, Wang C, Li H, Li H, Zhang HQ, Zhang S, Wang DM, Wang QX, Huang H, Xia HJ, Yang ZN. Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 2007;52:528–538. doi: 10.1111/j.1365-313X.2007.03254.x. [DOI] [PubMed] [Google Scholar]
- Zhu J, Chen H, Li H, Gao JF, Jiang H, Wang C, Guan YF, Yang ZN. Defective in tapetal development and function 1 is essential for anther development and tapetal function for microspore maturation in Arabidopsis. Plant J. 2008;55:266–277. doi: 10.1111/j.1365-313X.2008.03500.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Tapetal cell morphology. Transverse sections were taken at several developmental stages. (a1) ms8 at 2 mm, tapetal cell thickness = 14 μm; (a2) ms8 at 2.5 mm, tapetal cell thickness = 20 μm; (a3) ms8 at 3 mm, tapetal cell thickness = 18 μm; (a4) fertile at 3 mm, tapetal cell thickness = 9 μm.
Figure S2. Venn diagrams of transcriptome changes during fertile (a) and sterile (b) anther development; stage-specific transcripts and transcripts shared by two or three stages are shown respectively.
Figure S3. qRT-PCR validation of microarray and proteome data. (a) Validation of a set of 22 genes using qRT-PCR. Assays were performed in triplicates on four biological replicates of both the fertile and ms8 samples for three developmental stages. Cyanase was used as the internal standard to calculate ΔCt=Ct(sample)-Ct(standard) (Ct: cycle threshold). The ΔCt value is a normalized relative expression value, parallel to a log2 ratio because of the doublings in PCR reactions. Those genes labeled with “◆” are differential expressed in ms8 anthers, based on microarray data. The red line-dot-line brackets the two-fold cutoff for up-regulated and down-regulated expression. (b) Congruence of eight genes that had same patterns in microarray, qRT-PCR, and proteomic data. The proteomic fold change, the log 2 ratio of microarray and qRT-PCR fold changes are shown.
Figure S4. 2-D DIGE analysis of ms8 anthers. (a) Proteins of ms8 (Cy5, red) were compared with those fertile sib samples (Cy3, green) by 2-D DIGE using 24-cm pH 4-7 IPG strips and a 12.5% SDS-PAGE gel. Proteins increased in ms8 appear red, those decreased appear green, whereas those unaffected appear yellow. (b) Cyanine dyes swap for (a). The molecular masses of marker proteins are indicated on the right of (a) and (b), and pH values on the edges of the gel are indicated on the top of (a).
Figure S5. Classification of ms8 regulated proteins based on their putative functions. Proteins with ratios at least 1.5-fold enhanced or decreased are included in the diagram. The categories proceed clockwise.
Figure S6. Increased spot I6-35 is found at both the 1.0 mm (left panel) and 1.5 mm stages (right panel) during ms8 anther development.
Table S1. Genes involved in early anther development of Arabidopsis or maize with detailed phenotypic descriptions.
Table S2. Differential expressed genes, comparing ms8 to fertile.
Table S3. Four hundred and sixteen 1.0 mm stage-specific genes are expressed at a later stage in normal anthers.
Table S4. One hundred and thirty-three 1.5 mm stage specific genes in ms8 are expressed at the 2.0 mm stage in normal anthers.
Table S5. Ten MYB transcription factors were up-regulated in ms8 during the 1.5 mm stage.
Table S6. Eight zinc finger family genes are differentially expressed in at least one stage.
Table S7. Primers used for qRT-PCR assays.
Table S10. Microarray experiment design.
Table S11. Average probe intensity, final processed data, probe sequences and samples annotation.
Table S8. ms8 regulated proteins with maize identifiers. Average protein level ratios of ms8/fertile sibs are shown. The t test p values were calculated from three biological samples and three repeat experiments.
Table S9. Proteins regulated by ms8. Using functional annotation from rice proteins, which have high identity to predicted maize proteins.


