Abstract
Workflows in urinary proteomics studies are often complex and require many steps to enrich, purify, deplete and separate the complex mixture. Many of these methods are laborious, time-consuming and have the potential for error. Although individual steps of these methods have been previously studied, their downstream compatibilities with fractionation technologies such as Off-Gel electrophoresis have not been investigated. We developed a One-Step sample preparation workflow that simultaneously i) concentrates proteins, ii) purifies by removing salts and other low molecular weight compounds and iii) depletes (albumin) from urine samples. This simple and robust workflow can be multiplexed and is compatible with a diverse range of downstream multidimensional separation technologies. Additionally, due to its high reproducibility and flexibility in processing samples with different volumes and concentrations, it has the potential to be used for standardization of urinary proteomics studies, as well as for studying other body fluids of similar complexity.
Keywords: Shotgun Proteomics, Off-Gel electrophoresis, Albumin Depletion, Urinary Proteomics
Introduction
The field of urinary proteomics presents an opportunity to discover clinically relevant diagnostic and prognostic biomarkers, and potential therapeutic targets of disease. Due to urine's importance as a biomarker source, numerous separation methods have been employed to reduce its complexity and to find potential low-abundance biomarkers1. Some of these include: Sodium Dodecyl Sulfate Poly-Acrylamide Gel Electrophoresis (SDS-PAGE)2, Two-Dimensional Gel Electrophoresis (2DE)3, 4, GeLC-MS5, 6, Size Exclusion Chromatography7, Gas Phase Fractionation8, Hydrophilic Interaction Liquid Chromatography-Reversed Phase Liquid Chromatography (HILIC-RPLC)9, protein Equalizer bead technology10, Strong Cation Exchange-Reversed Phase Liquid Chromatography (SCX-RPLC)11.
Methods are continuously being refined and improved in an effort to achieve maximum protein coverage. Off-Gel electrophoresis is a recent advance in separation technology that fractionates proteins or peptides according to their isoelectric point (pI) and facilitates the recovery of the focused species in the liquid phase12. The high resolution orthogonal separation, excellent reproducibility, and increased analytical throughput allowing multiple samples to be run in parallel, are among some of the advantages of the Off-Gel technology. The superiority of isoelectric focusing as the first dimension of separation has been shown in comparison with more widely used methods such as GeLC-MS13 and SCX14, 15.
The majority of methodology studies on urine have involved improving steps of gel separation1, 2, 16. We sought to apply the Off-Gel technology as the first dimension of separation of urinary peptides prior to the conventional LC-MS/MS step of the shotgun proteomics. Despite our initial success with low urine volumes (1 and 5 ml), the separation of higher volumes (> 5 ml) were precluded because of a sharp increase in conductivity during the focusing because of incompatible salt concentrations. Due to the concomitant effects of diluted protein concentration of urine samples (particularly for non-renal disease studies), high-salt content and interfering compounds (urea, metabolites, etc.), purification is an absolute prerequisite for efficient application of the Off-Gel to urinary proteomics. Moreover, depletion of human serum albumin may help reduce the complexity of urine to improve detection of low-abundance proteins16. Typically, albumin depletion is performed in series with purification steps, which further complicates workflows and increases the potential for sample loss.
With the rapidly growing interest in the human urine proteome, we developed a fast, easy and reproducible One-Step method to simultaneously concentrate urinary proteins, remove salts and other contaminates, and deplete albumin. We present the application of the One-Step method in combination with the Off-Gel technology on both adult and pediatric urine samples.
Methods
Urine Collection
Sample 1 was a clean catch midstream urine sample obtained as the second void of the day from a healthy 38 year old male. Samples 2 to 7 were catheterized urine samples collected from 6 infant males under the age of 2 years with different renal and urologic diseases. Samples were collected in sterile specimen containers (Falcon 4.5 oz / 110 ml # 35-4013) under sterile conditions. Samples were centrifuged at 3500 g in 4°C for 10 minutes to clear the debris. 5 ml aliquots were stored at −80 °C.
Protein Concentration
The concentration of the urine sample pre and post purification and depletion was estimated by Bradford assay (Bio-Rad) using a NanoDrop ND-1000 Spectrophotometer (Thermo). The Bradford assay was performed according to manufacturer's instructions. The measurements were performed in triplicate for each method and each biological replicate. The average value of the protein concentration is reported.
One dimensional gel electrophoresis (SDS-PAGE)
Samples from each experiment were re-suspended in 24 μl of water and 8 μl 4X LDS sample buffer (Invitrogen). Samples were heated at 70 °C for 10 minutes and then were loaded into a 4–12% Bis-Tris precast gel (Invitrogen). Gels were run at 100 V and stained overnight in colloidal Coomassie blue as per manufacturer protocol (Invitrogen).
One-Step (combined enrichment, purification and albumin depletion)
A 5 ml urine sample was reduced for 30 minutes with 30 μl of 20 mM tris [2-carboxyethyl] phosphine (TCEP) and then alkylated for 30 minutes in the dark with 30 μl of 150 mM iodoacetamide. Both reactions were carried out at room temperature. 200 μl of the Anti-HSA resin (Sartorius Stedim) and a 5 ml urine sample were added to a Vivaspin 6 spin-filter. After a 10-minute incubation step, the column is placed in the centrifuge (20 minutes at 7000 g) and then washed 2 times each with 5 ml pure water for 10 minutes at 7000 g. The supernatant (30 μl) was collected. In order to assure a good recovery of the sample from the resin, 100 μl water was then added and vortexed with the resin. The supernatant was then aspirated and combined with the initial supernatant.
In-Solution Digestion
After the One-Step method, the purified / depleted protein was dried by vacuum centrifugation and brought up to 100 μl with 100 mM triethyl ammonium bicarbonate (TEAB), pH 8.5. The proteins were digested by addition of 4 μl of 1 mg/ml trypsin (sequencing grade modified trypsin, Promega) to obtain a ratio of 1:50 w/w. Proteolysis was performed overnight at 37 °C.
Off-Gel Electrophoresis
After digestion of 300 to 400 μg of protein, peptides were separated using an Agilent 3100 Off-Gel Fractionator (Agilent). 24 cm, pH 3–10 IPG DryStrips (GE Healthcare) were used. Strips were rehydrated for 20 minutes with 20 μl/well of the IEF buffer containing 0.2% IPG buffer pH 3–10. Samples were dissolved in 3600 μl of the IEF buffer. 150 μl of this peptide solution was loaded into each well, the cover seal was set into place and Immobiline DryStrip cover fluid (GE Healthcare) was added to both ends of the strip. Focusing was performed according to manufacturer's preset program up to 50 kV-h with maximum current of 50 μA. Fractions were collected from each well. 100 μl of 0.1% formic acid was added to each well and extracted after 10 minutes. The extracted peptides were combined and dried down in a speed vac.
LC-MS/MS
All experiments were performed using an LTQ-Orbitrap mass spectrometer linked to a microautosampler and a nanoflow HPLC pump (both: Eksigent). The reversed phase columns were packed in-house by using Magic C18 particles (3 μm, 200 Å; Michrom Bioresource) and PicoTip Emitters (New Objective, Woburn, MA). The peptides were eluted with a 60-minute linear gradient and data acquired in a data dependent fashion, fragmenting the 6 most abundant peptide species, which were then dynamically excluded for 60 seconds. Buffer A was 0.2% formic acid; buffer B was AcN/0.2% formic acid, and loading buffer was 5% formic acid/5% AcN.
Database Searching and Validation
The 200 most intense fragment ions of each raw product ion spectrum were used for searches against the IPI human database (version 3.56) using Mascot version 3.217. The following search parameters were applied: default charge states of 1+, 2+, 3+, and 4+ were used; a maximum of one missed cleavage was allowed with an average peptide mass tolerance of 5 ppm. A fragment ion search tolerance of 0.8 Da was permitted. Fixed modification on cysteine was carbamidomethylation and variable modifications were deamidation, oxidation of methionine, and N-terminal pyro-glutamic acid formation. All data were searched against a merged target and decoy database. The peptide level score cutoff for each of the runs was automatically adjusted to ensure a 1% false discovery rate throughout the experiments. Peptides isoelectric points (pI) were calculated18. In each fraction, peptides with calculated pI values beyond the ±0.5 window around the average pI were discarded.
Results and Discussion
Challenges of shotgun urine proteomics
Although urine is abundant and easy to collect, it is a challenging sample for proteomics studies19, 20. The interfering salts and other contaminants can reduce the efficiency of digestion and effectiveness of multi-dimensional separation strategies that are critical for deep profiling using shotgun proteomics21. Obtaining pure protein samples from urine, without large sample loss, can be difficult and a major impediment to a successful gel-free approach.
Numerous protocols have been employed to concentrate and purify urinary proteins; e.g., lyophilization2, 8, 22, precipitation3, 23, ultracentrifugation3, 4 and centrifugal filtration2, 7, 16, 24. The majority of these methods were developed for gel-based analysis, rather than for in-solution digestion and shotgun proteomics, which typically require more pure samples. Some of these methods, such as ethanol precipitation, are biased towards hydrophilic proteins and do not remove salts from the precipitated sample and thus alone prohibit proper digestion and separation in the downstream steps. Other methods, such as reverse-phase trapping columns, process the samples in sequence and therefore are prone to carry-over contamination2.
We initially anticipated that a reverse-phase trapping column (C4 macrotrap, Michrom Bioresources) purification would permit separation using IEF. The C4 macrotrap is a large pore polymeric reversed-phase trapping column designed to desalt and concentrate protein mixtures. At low urine volumes (e.g. 1 and 5 ml), the C4 macrotrap purified proteins (peptides after in-solution digestion) were separated by IEF, but at high-currents and over-heating of the IPG strip (Data not shown). These findings are typically secondary to interfering compounds. At larger starting volumes (e.g. 20 ml) the C4 macrotrap purified urine proteins/peptides could not be separated by IEF. This implies that C4 purification did not clear enough of the interfering compounds from urine, and further highlights the challenges of urine.
As in serum and other discovery experiments, low-abundance urinary proteins are challenging to identify and quantify without appropriate depletion of abundant proteins. The ability to specifically and reproducibly remove high-abundant proteins may improve the identification of low abundant proteins25. Human Serum Albumin (HSA) is particularly troubling for urinary proteome analysis, especially in patients with proteinuria26. A variety of methods have been developed to remove albumin and other high abundance proteins from samples prior to analysis27, 28. However, most of these methods have been originally designed for blood-derived body fluids and add more steps to the workflow. Similarly, urine proteomics workflows typically require multiple steps to concentrate, purify, deplete and separate proteins or peptides. Our One-Step method permits simultaneous enrichment of urinary proteins, removal of salts and other contaminants, and depletion of albumin. Additionally, it is fast, reproducible, efficient, and allows parallel analysis of multiple samples with no risk of carryover.
One-Step (simultaneous concentration, purification and albumin depletion)
The overall One-Step integrated workflow is presented in Figure 1. Initially, we employed the Vivapure Anti-HSA columns due to their high recovery, effective albumin depletion and ease in processing multiple samples in parallel. However, Vivapure columns are intended for serum and have a small volume capacity of 500 μl. Because the protein concentration in urine is dramatically lower than serum and it varies significantly based on renal function and disease, spin columns that can accommodate a range of volumes are needed to recover the desired amount of protein.
Figure 1. One-Step method workflow.
The final workflow for preparation and analysis of urine samples. Urine is first spun down to remove debris and the pH is adjusted to neutral. Urine proteins are reduced and alkylated. This sample is then added to the Vivaspin 6 spin-filters. Anti-HSA resin is added. Samples are incubated and then spun down completing the protein concentration, purification and albumin depletion. Proteins are removed leaving the Anti-HSA resin behind. Proteins undergo in-solution digestion with trypsin. Samples could then be chemically labeled for quantitation and multiplexed*, prior to undergoing 1st dimension separation (Off-Gel electrophoresis is currently utilized**) and LC-MS/MS.
To accommodate larger volumes we used a Vivaspin column because of its variable loading capacity (500 μl to 20 ml). These filters have a low-binding poly-ether-sulfone vertical membrane, which affords high recovery and minimal membrane blockage. Another advantage is that they have an integrated dead-stop-volume of 30 μl, eliminating the risk of concentrating to dryness. Importantly, multiple samples can be processed at the same time with no risk of cross contamination or sample carryover. To integrate albumin depletion, we utilized the same Anti-HSA resin used in Vivapure columns. The Anti-HSA resin has a high specificity and recovery and can be added directly to the sample.
Protein recovery and albumin depletion
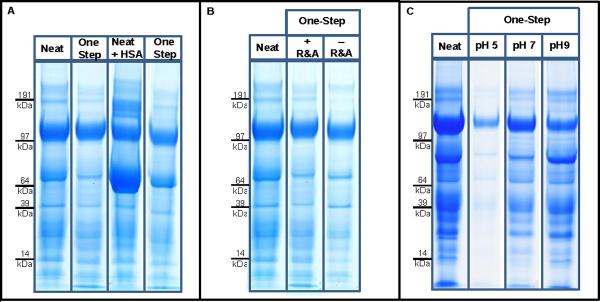
As shown in Figure 2A, efficient depletion and good recovery was obtained with the One-Step method. Sample 1 was used for this initial experiment. Both the neat urine sample and the neat sample with 20 μg spiked-in HSA were efficiently depleted. The concentration was reduced from 50.1 μg/ml of the neat urine to 38.6 μg/ml after the One-Step process. The sample with spiked-in HSA (72.5 μg/ml) also had excellent depletion (42.3 μg/ml). Measuring the concentration of three technical repeats of the same samples showed a variability of only 5% indicating good reproducibility.
Figure 2. Efficiency, Recovery and pH sensitivity of the One-Step Method.
A. The recovery and depletion efficiency of the One-Step enrichment, purification and depletion method applied to neat urine sample from a healthy individual with and without 20 μg spiked-in HSA. B. The effect of Reduction and Alkylation (R&A) on the depletion efficiency and recovery of the One-Step process. C. The effect of pH on the One-Step method recovery and depletion.
Since albumin is a common carrier of low-molecular weight proteins, we investigated whether depletion of albumin resulted in a co-depletion phenomena29. We examined the effect of reducing and alkylating our samples to disrupt the bonds between albumin and other proteins prior to albumin depletion and purification to determine if recovery would improve. Using our combined method, we measured the protein concentration of sample 1 with reduction & alkylation to shift from 50.1 μg/ml (before the depletion), to 38.6 μg/ml (after the depletion); and to 35.0 μg/ml without reduction & alkylation (Figure 2B). Although this does not definitively demonstrate the co-depletion phenomena, upfront reduction and alkylation does not diminish the effectiveness of the method. Additionally, the most beneficial advantage of performing reduction and alkylation prior to purification is that it allows removal of the excess of reduction/alkylation reagents along with other impurities. This modification reduces the overall number of sample handling steps and risk of sample loss.
Effect of urine pH
Since the pH of urine is highly variable as a function of normal physiology and diet30, we investigated the effect of variable pH on depletion and recovery. As shown in Figure 2C, in the acidic pH range (3–5), high depletion efficiency is achieved but the recovery is poor. In the basic pH range (9–11) zone, high recovery is achieved, with suboptimal depletion. Neutral pH (7–8) condition was the ideal condition to achieve both efficient depletion and high recovery. Consequently, we recommend that all urine samples be balanced to a pH of 7 prior to the initiation of the workflow.
Reproducibility
In order to evaluate the applicability of the One-Step method to urine samples of varying composition, six patients (samples 2–7) with varying renal and urologic diseases were chosen from our urine sample repository. As shown in Figure 3, despite differences in starting protein concentration, proteome composition, and concentration of albumin, all samples were effectively depleted and a good recovery was obtained. Albumin depletion appeared to significantly improve the intensity of other proteins within the sample.
Figure 3. Reproducibility and efficiency of the One-Step method.
The reproducibility and efficiency of the One-Step enrichment, purification and albumin depletion method is tested on urine samples 2 to 7. Protein concentrations before and after the One-Step method are presented.
Applying the One-Step urine sample preparation to Off-Gel electrophoresis
As a method of separation, Off-Gel has been demonstrated to be a high resolution and reproducible alternative to the traditional multi-dimensional LC separations employed in shotgun workflows12, 31, 32. It has the advantage of true orthogonality and providing complimentary pI information. However, because urine has a high amount of interfering salts and metabolites, it has been difficult to implement IEF technology towards the investigation of the urinary proteome. We investigated whether the One-Step workflow would provide a desalted sample that is compatible with Off-Gel separation regardless of the starting volume.
Since lower volumes of urine (1–5 ml) worked with Off-Gel, we first used 30 ml of sample 1 to evaluate the application of the One-Step method to Off-Gel electrophoresis. Urine protein concentration is often low and large volumes may be required for shotgun proteomic discovery studies. 30 ml provides an appropriate amount of protein for optimal IEF separation (400ug) and should adequately test the efficacy of the One-Step method. The run was completed in less than 12 hours with no current problems during the focusing. Peptides were separated into 24 fractions. In total, we identified 703 proteins corresponding to 2698 peptides using a 1% false discovery rate. 16% of these proteins were identified with a single peptide. The number of proteins and peptides identified is presented in Table 1 and their complete list is provided in Supplementary Data 1.
Table 1.
Number of unique proteins and peptides identified in each sample using the One-Step urine sample preparation method in combination with Off-Gel electrophoresis. The number of proteins identified with a single peptide is presented.
| Sample | Number of proteins | Number of peptides | Number of single hits |
|---|---|---|---|
| 1 | 703 | 2698 | 109 |
| 2 | 650 | 2093 | 155 |
| 3 | 666 | 2203 | 125 |
| 5 | 628 | 1988 | 145 |
| 7 | 695 | 2386 | 176 |
As shown in Figure 4A, the average standard deviation of the peptides pI values from the average pI per fraction indicates good focusing quality for the majority of the fractions. Only a few basic fractions demonstrated higher deviations. This can be partly explained by the well-known low focusing quality in the basic region of the IPG strips33 and partly attributed to the pI prediction algorithm used, which is specifically optimized for peptides in the acidic region18.
Figure 4. One-Step method and Off-Gel Electrophoresis.
The application of the One-Step sample preparation method to the Off-Gel technology. A. Demonstrates the average pI of the peptides identified in each fraction with their average standard deviations. B. Depicts the number of fractions each peptide has been identified in as a surrogate for the resolution. C. Shows the distribution of the peptides along the IPG strip for each sample.
The resolution obtained by analytical IEF is amongst the highest from present biochemical separation techniques33. Figure 4B shows the number of fractions each peptide was identified as a measure for resolution. 79% of the peptides were uniquely identified in one fraction followed by 9% in 2, 5% in 3 and 3% in 4 fractions. These results demonstrated good resolution of the Off-Gel separation, and imply that the One-Step methodology is providing a purified sample. The heterogeneous distribution of the identified peptides per fraction is demonstrated in Figure 4C. In some fractions more than 900 peptides have been identified, in others less than 50 peptides were detected. Unfortunately, the current fixed well-sizes of the Off-Gel system does not permit improvement in the distribution of collected fractions by adapting fraction size34. Nevertheless, the use of Off-Gel is advantageous due to the extra information provided by pI. The expected pI of a peptide can be used to support or refute its identification as determined by mass spectrometry, as well as detect possible post-translational modifications35.
Reproducibility
We applied the One-Step / Off-Gel workflow to 4 different biological samples with varying renal and urologic disease (Samples 2, 3, 5 and 7). Four IPG strips (24 cm, 3–10) were run in parallel and 24 samples were collected from each strip (96 total). Figure 5 demonstrates that the degree of separation and depth of interrogation obtained with our initial test sample (sample 1) were reproduced in these 4 diseased samples. All 4 samples showed a similar pattern of their average pI per fraction along the IPG strips. High resolution separation is shown in Figure 5B with 76%, 80%, 82% and 78% of peptides in samples 2, 3, 5 and 7 uniquely identified in a single fraction. Finally, the distribution of the peptides per fraction showed minor differences among the samples, as expected due to the high variation of the urinary proteome among different individuals. To verify the reproducibility of the workflow, a technical replicate of sample 2 was run from the first stage of the One-Step method. Four random fractions were chosen and the identified proteins were compared to the first experiment. As expected, new proteins were identified in each run36. 76%, 83%, 85% and 73% reproducibility was obtained for the identified proteins in fractions 4, 9, 12 and 22, respectively, confirming good reproducibility. Overall, using the One-Step method we were able to process a range of urine volumes from 1 ml to 30 ml and compositions without observing any problems with digestion efficiency, OFF-gel focusing, or LC-MS/MS.
Figure 5. Application of the One-Step method and Off-Gel Electrophoresis to different samples.
The comparison of four urine sample (samples 2, 3, 5 and 7) prepared with the One-Step sample preparation method and analyzed using the Off-Gel technology. A. Demonstrates the average pI of the peptides identified in each fraction. B. Depicts the number of fractions each peptide has been identified in as a surrogate for the resolution. C. Shows the distribution of the peptides along the IPG strip for each sample.
Applying the One-Step urine sample preparation to High pH reversed phase separation
The primary goal for the development of the One-Step method was to produce and develop a fast, reproducible and reliable method for urine sample preparation to be compatible with Off-Gel electrophoresis. To determine the compatibility of the One-Step method with other 1st dimension separation methods, we tested prepared samples using High pH reversed phase separation37. Without performing optimization of pH gradients and the High pH workflow, we identified 499 unique proteins corresponding to 1729 unique peptides. 74 of these proteins were detected with a single peptide. The complete list of the identified proteins and peptides is provided in Supplementary Data 2. This preliminary data demonstrates that the One-Step method can produce a sample that is applicable to various 1st dimension separation techniques.
Conclusion and Perspective
To identify clinically useful urinary biomarkers, it is essential to have a fast and reliable urinary proteomics workflow. Many methods of protein enrichment, purification and depletion are effective but not necessarily compatible with the downstream steps in a multi-dimensional shotgun proteomics workflow. We introduce a One-Step method that simultaneously concentrates urinary proteins, removes salts and other contaminates, and depletes albumin. The application of our workflow to the analysis of complex pathological urine samples showed the efficiency of this method in handling different urine samples independent of the volume and the amount of proteins and salts. Recently, in our laboratory, we have successfully applied chemical labeling methods (iTRAQ and TMT) to the samples prepared by the One-Step method (manuscript in preparation). Chemical labeling has allowed for multiplexing using different 1st dimension separation methods. In the near future we plan to expand this workflow to allow the quantitative analysis of post-translational modifications in the urinary proteome, and other biologic fluids of similar complexity.
Supplementary Material
Figure 6. Reproducibility of the One-Step method and Off-Gel Electrophoresis.
Technical reproducibility of the One-Step urine sample preparation method in combination with Off-Gel separation and data-dependent LC-MS/MS. Blue bars represent the number of proteins identified only in technical replicate 1 and red bars indicate the number of proteins identified exclusively in technical replicate 2. Green bars show the number of proteins identified in both technical replicates.
Abbreviations
- MS
Mass Spectrometry
- MS/MS
Tandem Mass Spectrometry
- LC
Liquid Chromatography
- 2DE
Two-Dimensional Gel Electrophoresis
- CE
Capillary Electrophoresis
- FFE
Free Flow Electrophoresis
- HILIC
Hydrophilic Interaction Liquid Chromatography
- RPLC
Reversed-Phase LC
- SCX
Strong Cation Exchange
- IPG
Immobilized pH Gradients
- IEF-AF4
Isoelectric Focusing and Asymmetrical Flow field-flow fractionation
- SDS-PAGE
Sodium Dodecyl Sulfate Poly-Acrylamide Gel Electrophoresis
- kV-h
kiloVolt-hour
- TEAB
triethyl ammonium bicarbonate
Footnotes
Supporting Information Supporting Information is Available: This material is available free of charge via the Internet at http://pubs.acs.org.
Supplementary Data The complete list of all proteins and peptides identified in samples 1, 2, 3, 5 and 7 using the One-Step method and Off-Gel electrophoresis are listed in individual sheets. The technical repeat data is listed in a separate sheet. The complete list of all proteins and peptides identified in sample 1 using the One-Step method and High pH separation is listed in the final sheet.
References
- 1.O'Riordan E, Gross SS, Goligorsky MS. Technology Insight: renal proteomics--at the crossroads between promise and problems. Nat Clin Pract Nephrol. 2006;2(8):445–58. doi: 10.1038/ncpneph0241. [DOI] [PubMed] [Google Scholar]
- 2.Lee RS, Monigatti F, Briscoe AC, Waldon Z, Freeman MR, Steen H. Optimizing sample handling for urinary proteomics. J Proteome Res. 2008;7(9):4022–30. doi: 10.1021/pr800301h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orenes-Pinero E, Corton M, Gonzalez-Peramato P, Algaba F, Casal I, Serrano A, Sanchez-Carbayo M. Searching urinary tumor markers for bladder cancer using a two-dimensional differential gel electrophoresis (2DDIGE) approach. J Proteome Res. 2007;6(11):4440–8. doi: 10.1021/pr070368w. [DOI] [PubMed] [Google Scholar]
- 4.Lafitte D, Dussol B, Andersen S, Vazi A, Dupuy P, Jensen ON, Berland Y, Verdier JM. Optimized preparation of urine samples for two-dimensional electrophoresis and initial application to patient samples. Clin Biochem. 2002;35(8):581–9. doi: 10.1016/s0009-9120(02)00362-4. [DOI] [PubMed] [Google Scholar]
- 5.Lee RS, Monigatti F, Lutchman M, Patterson T, Budnik B, Steen JA, Freeman MR, Steen H. Temporal variations of the postnatal rat urinary proteome as a reflection of systemic maturation. Proteomics. 2008;8(5):1097–112. doi: 10.1002/pmic.200700701. [DOI] [PubMed] [Google Scholar]
- 6.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7(9):R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pieper R, Gatlin CL, McGrath AM, Makusky AJ, Mondal M, Seonarain M, Field E, Schatz CR, Estock MA, Ahmed N, Anderson NG, Steiner S. Characterization of the human urinary proteome: a method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4(4):1159–74. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 8.Spahr CS, Davis MT, McGinley MD, Robinson JH, Bures EJ, Beierle J, Mort J, Courchesne PL, Chen K, Wahl RC, Yu W, Luethy R, Patterson SD. Towards defining the urinary proteome using liquid chromatography-tandem mass spectrometry. I. Profiling an unfractionated tryptic digest. Proteomics. 2001;1(1):93–107. doi: 10.1002/1615-9861(200101)1:1<93::AID-PROT93>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 9.Loftheim H, Nguyen TD, Malerod H, Lundanes E, Asberg A, Reubsaet L. 2-D hydrophilic interaction liquid chromatography-RP separation in urinary proteomics - Minimizing variability through improved downstream workflow compatibility. J Sep Sci. 2009 doi: 10.1002/jssc.200900554. [DOI] [PubMed] [Google Scholar]
- 10.Castagna A, Cecconi D, Sennels L, Rappsilber J, Guerrier L, Fortis F, Boschetti E, Lomas L, Righetti PG. Exploring the hidden human urinary proteome via ligand library beads. J Proteome Res. 2005;4(6):1917–30. doi: 10.1021/pr050153r. [DOI] [PubMed] [Google Scholar]
- 11.Ru QC, Katenhusen RA, Zhu LA, Silberman J, Yang S, Orchard TJ, Brzeski H, Liebman M, Ellsworth DL. Proteomic profiling of human urine using multi-dimensional protein identification technology. J Chromatogr A. 2006;1111(2):166–74. doi: 10.1016/j.chroma.2005.06.081. [DOI] [PubMed] [Google Scholar]
- 12.Ros A, Faupel M, Mees H, Oostrum J, Ferrigno R, Reymond F, Michel P, Rossier JS, Girault HH. Protein purification by Off-Gel electrophoresis. Proteomics. 2002;2(2):151–6. doi: 10.1002/1615-9861(200202)2:2<151::aid-prot151>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 13.Krijgsveld J, Gauci S, Dormeyer W, Heck AJ. In-gel isoelectric focusing of peptides as a tool for improved protein identification. J Proteome Res. 2006;5(7):1721–30. doi: 10.1021/pr0601180. [DOI] [PubMed] [Google Scholar]
- 14.Essader AS, Cargile BJ, Bundy JL, Stephenson JL., Jr. A comparison of immobilized pH gradient isoelectric focusing and strong-cation-exchange chromatography as a first dimension in shotgun proteomics. Proteomics. 2005;5(1):24–34. doi: 10.1002/pmic.200400888. [DOI] [PubMed] [Google Scholar]
- 15.Waller LN, Shores K, Knapp DR. Shotgun proteomic analysis of cerebrospinal fluid using off-gel electrophoresis as the first-dimension separation. J Proteome Res. 2008;7(10):4577–84. doi: 10.1021/pr8001518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thongboonkerd V, Chutipongtanate S, Kanlaya R. Systematic evaluation of sample preparation methods for gel-based human urinary proteomics: quantity, quality, and variability. J Proteome Res. 2006;5(1):183–91. doi: 10.1021/pr0502525. [DOI] [PubMed] [Google Scholar]
- 17.Renard BY, Kirchner M, Monigatti F, Ivanov AR, Rappsilber J, Winter D, Steen JA, Hamprecht FA, Steen H. When less can yield more - Computational preprocessing of MS/MS spectra for peptide identification. Proteomics. 2009;9(21):4978–84. doi: 10.1002/pmic.200900326. [DOI] [PubMed] [Google Scholar]
- 18.Cargile BJ, Sevinsky JR, Essader AS, Eu JP, Stephenson JL., Jr. Calculation of the isoelectric point of tryptic peptides in the pH 3.5–4.5 range based on adjacent amino acid effects. Electrophoresis. 2008;29(13):2768–78. doi: 10.1002/elps.200700701. [DOI] [PubMed] [Google Scholar]
- 19.Vaezzadeh AR, Steen H, Freeman MR, Lee RS. Proteomics and opportunities for clinical translation in urological disease. J Urol. 2009;182(3):835–43. doi: 10.1016/j.juro.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thongboonkerd V. Urinary proteomics: towards biomarker discovery, diagnostics and prognostics. Mol Biosyst. 2008;4(8):810–5. doi: 10.1039/b802534g. [DOI] [PubMed] [Google Scholar]
- 21.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–7. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 22.Rasmussen HH, Orntoft TF, Wolf H, Celis JE. Towards a comprehensive database of proteins from the urine of patients with bladder cancer. J Urol. 1996;155(6):2113–9. [PubMed] [Google Scholar]
- 23.Marshall T, Williams K. Two-dimensional electrophoresis of human urinary proteins following concentration by dye precipitation. Electrophoresis. 1996;17(7):1265–72. doi: 10.1002/elps.1150170716. [DOI] [PubMed] [Google Scholar]
- 24.Sigdel TK, Lau K, Schilling J, Sarwal M. Optimizing protein recovery for urinary proteomics, a tool to monitor renal transplantation. Clin Transplant. 2008;22(5):617–23. doi: 10.1111/j.1399-0012.2008.00833.x. [DOI] [PubMed] [Google Scholar]
- 25.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5(13):3304–13. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 26.Magistroni R, Ligabue G, Lupo V, Furci L, Leonelli M, Manganelli L, Masellis M, Gatti V, Cavazzini F, Tizzanini W, Albertazzi A. Proteomic analysis of urine from proteinuric patients shows a proteolitic activity directed against albumin. Nephrol Dial Transplant. 2009;24(5):1672–81. doi: 10.1093/ndt/gfp020. [DOI] [PubMed] [Google Scholar]
- 27.Roche S, Tiers L, Provansal M, Seveno M, Piva MT, Jouin P, Lehmann S. Depletion of one, six, twelve or twenty major blood proteins before proteomic analysis: the more the better? J Proteomics. 2009;72(6):945–51. doi: 10.1016/j.jprot.2009.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Dekker LJ, Bosman J, Burgers PC, van Rijswijk A, Freije R, Luider T, Bischoff R. Depletion of high-abundance proteins from serum by immunoaffinity chromatography: a MALDI-FT-MS study. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;847(1):65–9. doi: 10.1016/j.jchromb.2006.09.038. [DOI] [PubMed] [Google Scholar]
- 29.Shen Y, Kim J, Strittmatter EF, Jacobs JM, Camp DG, 2nd, Fang R, Tolie N, Moore RJ, Smith RD. Characterization of the human blood plasma proteome. Proteomics. 2005;5(15):4034–45. doi: 10.1002/pmic.200401246. [DOI] [PubMed] [Google Scholar]
- 30.Thongboonkerd V, Mungdee S, Chiangjong W. Should urine pH be adjusted prior to gel-based proteome analysis? J Proteome Res. 2009;8(6):3206–11. doi: 10.1021/pr900127x. [DOI] [PubMed] [Google Scholar]
- 31.Elschenbroich S, Ignatchenko V, Sharma P, Schmitt-Ulms G, Gramolini AO, Kislinger T. Peptide Separations by On-Line MudPIT Compared to Isoelectric Focusing in an Off-Gel Format: Application to a Membrane-Enriched Fraction from C2C12 Mouse Skeletal Muscle Cells. J Proteome Res. 2009;8(10):4860–9. doi: 10.1021/pr900318k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michel PE, Reymond F, Arnaud IL, Josserand J, Girault HH, Rossier JS. Protein fractionation in a multicompartment device using Off-Gel isoelectric focusing. Electrophoresis. 2003;24(1–2):3–11. doi: 10.1002/elps.200390030. [DOI] [PubMed] [Google Scholar]
- 33.Rabilloud T, Vaezzadeh AR, Potier N, Lelong C, Leize-Wagner E, Chevallet M. Power and limitations of electrophoretic separations in proteomics strategies. Mass Spectrom Rev. 2009;28(5):816–43. doi: 10.1002/mas.20204. [DOI] [PubMed] [Google Scholar]
- 34.Vaezzadeh AR, Hernandez C, Vadas O, Deshusses JJ, Lescuyer P, Lisacek F, Hochstrasser DF. PICarver: a software tool and strategy for peptides isoelectric focusing. J Proteome Res. 2008;7(10):4336–45. doi: 10.1021/pr8002672. [DOI] [PubMed] [Google Scholar]
- 35.Cargile BJ, Stephenson JL., Jr. An alternative to tandem mass spectrometry: isoelectric point and accurate mass for the identification of peptides. Anal Chem. 2004;76(2):267–75. doi: 10.1021/ac0352070. [DOI] [PubMed] [Google Scholar]
- 36.Tabb DL, Vega-Montoto L, Rudnick PA, Variyath AM, Ham AJ, Bunk DM, Kilpatrick LE, Billheimer DD, Blackman RK, Cardasis HL, Carr SA, Clauser KR, Jaffe JD, Kowalski KA, Neubert TA, Regnier FE, Schilling B, Tegeler TJ, Wang M, Wang P, Whiteaker JR, Zimmerman LJ, Fisher SJ, Gibson BW, Kinsinger CR, Mesri M, Rodriguez H, Stein SE, Tempst P, Paulovich AG, Liebler DC, Spiegelman C. Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J Proteome Res. 9(2):761–76. doi: 10.1021/pr9006365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Delmotte N, Lasaosa M, Tholey A, Heinzle E, Huber CG. Two-dimensional reversed-phase x ion-pair reversed-phase HPLC: an alternative approach to high-resolution peptide separation for shotgun proteome analysis. J Proteome Res. 2007;6(11):4363–73. doi: 10.1021/pr070424t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.