Abstract
Imatinib mesylate, currently marketed by Novartis as Gleevec in the US, has emerged as the leading compound to treat chronic phase of chronic myeloid leukemia (CML), through its inhibition of Bcr-Abl tyrosine kinase, and other cancers. However, resistance to imatinib develops frequently, particularly in late-stage disease. To identify new cellular pathways affected by imatinib treatment, we applied mass spectrometry together with SILAC (stable isotope labeling by amino acids in cell culture) for the comparative study of protein expression in K562 cells that were untreated or treated with a clinically relevant concentration of imatinib. Our results revealed that, among the 1344 quantified proteins, 73 had significantly altered levels of expression induced by imatinib and could be quantified in both forward and reverse SILAC labeling experiments. These included the down-regulation of thymidylate synthase, S-adenosylmethionine synthetase, and glycerol-3-phosphate dehydrogenase as well as the up-regulation of poly(ADP-ribose) polymerase 1, hemoglobins, and enzymes involved in heme biosynthesis. We also found, by assessing alteration in acetylation level in histone H4 upon imatinib treatment, that the imatinib-induced hemoglobinization and erythroid differentiation in K562 cells are associated with global histone H4 hyperacetylation. Overall, these results provided potential biomarkers for monitoring the therapeutic intervention of CML using imatinib and offered important new knowledge for gaining insights into the molecular mechanisms of action of imatinib.
Introduction
Chronic myeloid leukemia (CML) is a hematopoietic stem cell disorder, which arises from a translocation between the long arms of chromosomes 9 and 22 1, 2. The translocation renders the Bcr-Abl fusion gene where the Bcr gene on chromosome 22 is linked with the proto-oncogene Abl on chromosome 9 2. The fusion gene encodes the Bcr-Abl tyrosine kinase, which is constitutively active and leads to uncontrolled growth 2. The Bcr-Abl kinase activates several signaling pathways such as the Ras/mitogen-activated protein kinase, signal transducer and activator of transcription 5, and phosphatidylinositol 3 kinase/Akt pathways; enhances nuclear factor κB (NF-κB) activity; up-regulates the level of Bcl-XL; and suppresses the mitochondrial pathway of apoptosis 3.
Imatinib mesylate, which is marketed by Novartis as Gleevec in the US, has emerged as the leading compound to treat patients with CML 2. As a selective tyrosine kinase inhibitor, imatinib associates directly with the ATP-binding site and inhibits the kinase activity of Bcr-Abl. Upon imatinib treatment, the Bcr-Abl protein is rapidly dephosphorylated and becomes inactive, thereby interrupting the constitutive activation of signaling cascades, arresting cell cycle progression, and triggering apoptosis 4. Despite demonstrating remarkable clinical efficacy against chronic-phase CML, the outcome after imatinib therapy in the accelerated and blastic phases of CML is unacceptably poor 5, mostly owing to the emergence of mutations in the Bcr-Abl kinase domain that may inhibit binding of imatinib to the kinase domain. Thus, the discovery of novel targets of imatinib could contribute significantly to our understanding of the mechanisms of the anti-cancer functions of the drug and the development of resistance to imatinib among CML patients.
There have been a few studies on the imatinib-induced perturbation in global protein expression 6–8, in which 2-dimensional gel electrophoresis (2-DE) coupled with tandem mass spectrometry (MS/MS) was employed for protein identification and quantification. Other than 2-DE, several stable isotope-labeling strategies 9, especially stable isotope labeling by amino acids in cell culture (SILAC) 10, have been developed for MS-based differential protein expression analysis. SILAC is more efficient than 2-DE in the quantification of the whole proteome and in the detection of relatively small changes in protein abundance. In this context, Liang et al. 11 used SILAC together with LC-MS/MS and examined the imatinib-induced alterations of the Bcr-Abl kinase in CML cells.
In the present study, we employed LC-MS/MS, along with SILAC, to assess quantitatively the imatinib-induced alteration in protein expression in the Bcr-Abl-positive human K562 cells. We were able to quantify a total of 1344 proteins, among which 73 were significantly changed upon imatinib treatment in both forward and reverse SILAC measurements. The identification of proteins perturbed by imatinib treatment sets a stage for understanding the biological pathways affected by imatinib.
Material and Methods
Materials
Heavy lysine and arginine ([13C6,15N2]-L-lysine and [13C6,15N4]-L-arginine) were purchased from Cambridge Isotope Laboratories (Andover, MA). All reagents unless otherwise noted were from Sigma (St. Louis, MO).
Cell Culture
K562 cells (ATCC, Manassas, VA) were cultured in Iscove’s modified minimal essential medium (IMEM) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA) and penicillin (100 IU/mL). Cells were maintained in a humidified atmosphere with 5% CO2 at 37°C, with medium renewal at every 2–3 days. For SILAC experiments, the IMEM medium without L-lysine or L-arginine was custom-prepared following the ATCC formulation. The complete light and heavy IMEM media were prepared by the addition of light or heavy lysine and arginine, together with dialyzed FBS, to the above lysine, arginine-depleted medium. The K562 cells were cultured in heavy IMEM medium for at least 5 cell doublings to achieve complete isotope incorporation.
Imatinib Treatment and Cell Lysate Preparation
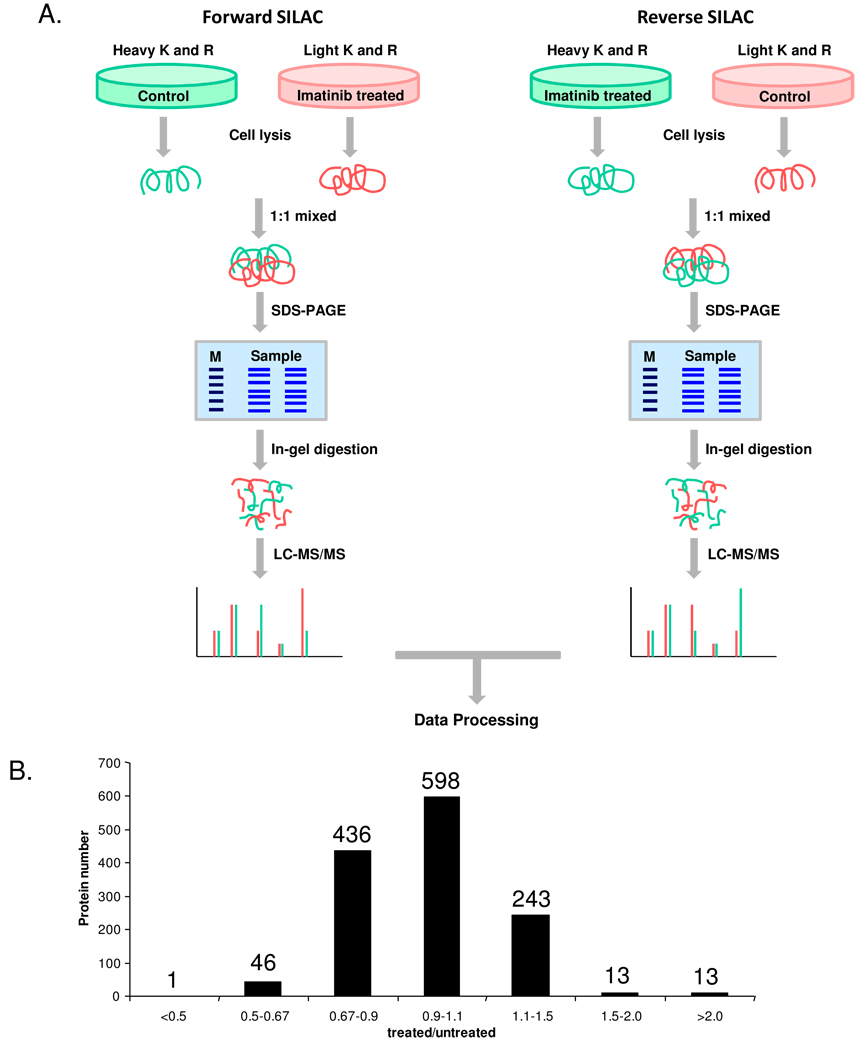
K562 cells, at a density of approximately 7.5×105 cells/mL, were collected by centrifugation at 300 g and at 4°C for 5 min, washed twice with ice-cold phosphate-buffered saline (PBS) to remove FBS, and resuspended in the fresh serum-free heavy or light media. In forward SILAC experiment, the cells cultured in light medium were treated with 1 µM imatinib for 24 hrs and the cells cultured in heavy medium were untreated. Reverse SILAC experiments were also performed where the cells cultured in the heavy and light media were treated with imatinib and mock-treated, respectively (Figure 1). After 24 hrs, the light and heavy cells were collected by centrifugation at 300 g, and washed three times with ice-cold PBS.
Figure 1.
(A) Forward- and reverse-SILAC combined with LC-MS/MS for the comparative analysis of protein expression in K562 cells upon imatinib treatment. (B) The distribution of expression ratios (treated/untreated) for the quantified proteins.
The cell pellets were resuspended in the CelLytic™ M cell lysis buffer for 30 min with occasional vortexing. Cell lysates were centrifuged at 12,000 g at 4°C for 30 min, and the resulting supernatants were collected. To the supernatant was subsequently added a protease inhibitor cocktail, and the protein concentrations of the cell lysates were determined by using Quick Start Bradford Protein Assay kit (Bio-Rad, Hercules, CA).
SDS-PAGE Separation and In-gel Digestion
The light and heavy cell lysates were combined at 1:1 ratio (w/w), denatured by boiling in Laemmli loading buffer for 5 min and separated by a 12% SDS-PAGE with 4% stacking gel. The gel was stained with Coomassie blue; after destaining, the gel was cut into 20 slices, reduced in-gel with dithiothreitol (DTT) and alkylated with iodoacetamide (IAM). The proteins were digested in-gel with trypsin (Promega, Madison, WI) for overnight, after which peptides were extracted from the gels with 5% acetic acid in H2O and subsequently with 5% acetic acid in CH3CN/H2O (1:1, v/v). The resultant peptide mixtures were dried and stored at −20°C for further analysis.
Benzidine Staining
The untreated and imatinib-treated K562 cells were collected without removing the media and mixed, at 1:1 (v/v) ratio for 4 min, with an aqueous solution containing 0.2% benzidine dihydrochloride, 0.6% H2O2 and 0.5 M acetic acid. The cells were spotted to a hemocytometer and pictures were taken using a Nikon Eclipse TI microscope (Melville, NY).
Histone Extraction
Core histones were obtained following previously reported procedures 12. In brief, the untreated and imatinib-treated 562 cells were harvested by centrifugation at 500 g. The cell pellets were subsequently washed with a 5-mL lysis buffer containing 0.25 M sucrose, 0.01 M MgCl2, 0.5 mM PMSF, 50 mM Tris (pH 7.4), and 0.5% Triton X-100, then resuspended in 5 mL of the same buffer and kept at 4°C overnight. The histones were extracted from the cell lysate with 0.4 M sulfuric acid by incubating at 4°C for 4 hr with continuous vortexing, precipitated with cold acetone, centrifuged, dried and redissolved in water.
HPLC Purification and Protease Digestion
Core histones were purified by HPLC on an Agilent 1100 system (Agilent Technologies, Santa Clara, CA) as described previously 12. A 4.6×250 mm C4 column (Grace Vydac, Hesperia, CA) was used. The wavelength for the UV detector was set at 220 nm. The flow rate was 0.8 mL/min, and a 60-min linear gradient of 30–60% acetonitrile in 0.1% trifluoroacetic acid (TFA) was employed.
Histones H4 and H3 were digested with Asp-N and Arg-C in buffers containing 50 mM sodium phosphate (pH 8.0) and 100 mM NH4HCO3 (pH 8.0), respectively. The digestion was carried out with a protein/enzyme ratio of 20:1 (w/w) at 37°C overnight. The peptide mixtures were subjected directly to LC-MS/MS analysis, or to a further peptide fractionation by HPLC and then analyzed by MALDI-MS (See below).
Mass Spectrometry
MALDI mass spectra were acquired on a Voyager DE STR MALDI-TOF mass spectrometer (Applied Biosystems, Framingham, MA) in positive reflection mode. The mass spectrometer was equipped with a pulsed nitrogen laser operated at 337 nm with 3 ns duration pulses. The acceleration voltage, grid voltage, and delayed extraction time were set as 20 kV, 65%, and 190 ns, respectively. Each mass spectrum was acquired from an average of 100 laser shots.
Online LC-MS/MS analysis was carried out on an Agilent 6510 Q-TOF system with an Agilent HPLC-Chip Cube MS interface (Agilent Technologies, Santa Clara, CA). The sample injection, enrichment, desalting, and HPLC separation were carried out automatically on the Agilent HPLC Chip with an integrated trapping column (160 nL) and a separation column (Zorbax 300SB-C18, 75 µm×150 mm, 5 µm in particle size). The peptide or protein sample was first loaded onto the trapping column with a solvent mixture of 0.1% formic acid in CH3CN/H2O (2:98, v/v) at a flow rate of 4 µL/min, which was delivered by an Agilent 1200 capillary pump. The peptides were then separated using a 120-min linear gradient of 2–35% acetonitrile, while intact histones were eluted with a 30-min linear gradient of 40–70% acetonitrile in 0.1% formic acid and at a flow rate of 300 nL/min, which was delivered by an Agilent 1200 Nano pump.
The Chip spray voltage (VCap) was set at 1950 V and varied with chip conditions. The temperature and flow rate of the drying gas were 325°C and 4 L/min, respectively. Nitrogen was employed as the collision gas, and the collision energy followed an equation with a slope of 3 V/100 Da and an offset of 2.5 V. MS/MS experiments were carried out in the data-dependent scan mode where a maximum of five MS/MS scans were acquired following each MS scan. The m/z ranges for MS and MS/MS were 300–2000 and 60–2000, and the data acquisition rates were 6 and 3 spectra/s, respectively.
Data Processing
The LC-MS/MS raw data were searched against human IPI protein database (version 3.21) and its reverse complement using TurboSEQUEST with Bioworks 3.2 (Thermo Fisher Scientific, San Jose, CA) for protein identification. Cysteine carbamidomethylation was set as a fixed modification. Methionine oxidation (+16 Da) as well as lysine (+8 Da) and arginine (+10 Da) mass shifts introduced by heavy isotope labeling were considered as variable modifications. Peptide filters with appropriate cross-correlation (Xcorr ≥ 1.9, ≥ 2.4, ≥ 3.5 for peptide ions that are singly, doubly, and triply charged) and delta correlation (ΔCn ≥ 0.1) scores were used to sort the search results. The protein false discovery rate was less than 1%.
Census, developed by Yates and coworkers 13, 14, was employed for protein quantification. The TurboSEQUEST search results were first filtered using DTASelect 15 and ion chromatograms were generated for peptide ions based on their m/z values. Peptide ion intensity ratios were subsequently calculated in Census from peak areas found in each pair of extracted-ion chromatograms. The ratio measurement results were filtered by setting thresholds of Determinant Factor as 0.5 and Outlier p-Value as 0.01.
The ratio obtained for each individual protein was then normalized against the average ratio for all quantified proteins. In this “multi-point” normalization strategy, it was assumed that the ratios for the majority of proteins were not perturbed by imatinib treatment, facilitating the use of the average ratio of all quantified proteins to re-scale the data. This has been widely employed to remove the inaccuracy during sample mixing introduced by protein quantification with the Bradford assay 16, 17. Some peptides identified by TurboSEQUEST for only 1 or 2 sets of SILAC samples could also be manually found, in the LC-MS/MS data for the remaining set(s) of SILAC samples, and quantified. In this context, the Chip HPLC provided excellent reproducibility in retention time, and most of the time the difference in elution time for a peptide among different runs was within 2 min, though occasionally the difference could be up to 5 min. In addition, the mass accuracy afforded by the Q-TOF mass spectrometer is within 20 ppm with external calibration. Therefore, the accurate m/z values of peptide ions (within 20 ppm) and HPLC retention time (within 5 min variation) were employed as criteria to locate the light/heavy peptide pairs for the quantification. Only those proteins with fold changes >1.5 and quantified in at least 2 sets (including both forward and reverse) of SILAC measurements were reported as significantly changed proteins.
Results and Discussion
Imatinib Treatment, Protein Identification and Quantification
To gain insights into the molecular pathways perturbed by imatinib treatment, we used SILAC in combination with LC-MS/MS to assess the imatinib-induced differential expression of the whole proteome of K562 cells. The standard dose of imatinib used in the clinical treatment of CML is 400 mg/day, which gives plasma concentrations at baseline and 3 hr after administration at 0.5–4 and 1.5–8 µM, respectively 18, 19. We observed, based on trypan blue exclusion assay, a less than 5% cell death after a 24-hr treatment of K562 cells with 1 µM imatinib, whereas greater than 20% cells were dead if treated with 1.5 µM imatinib for 24 hr. Thus, we chose 1 µM imatinib in subsequent experiments to minimize the apoptosis-induced alteration in protein expression.
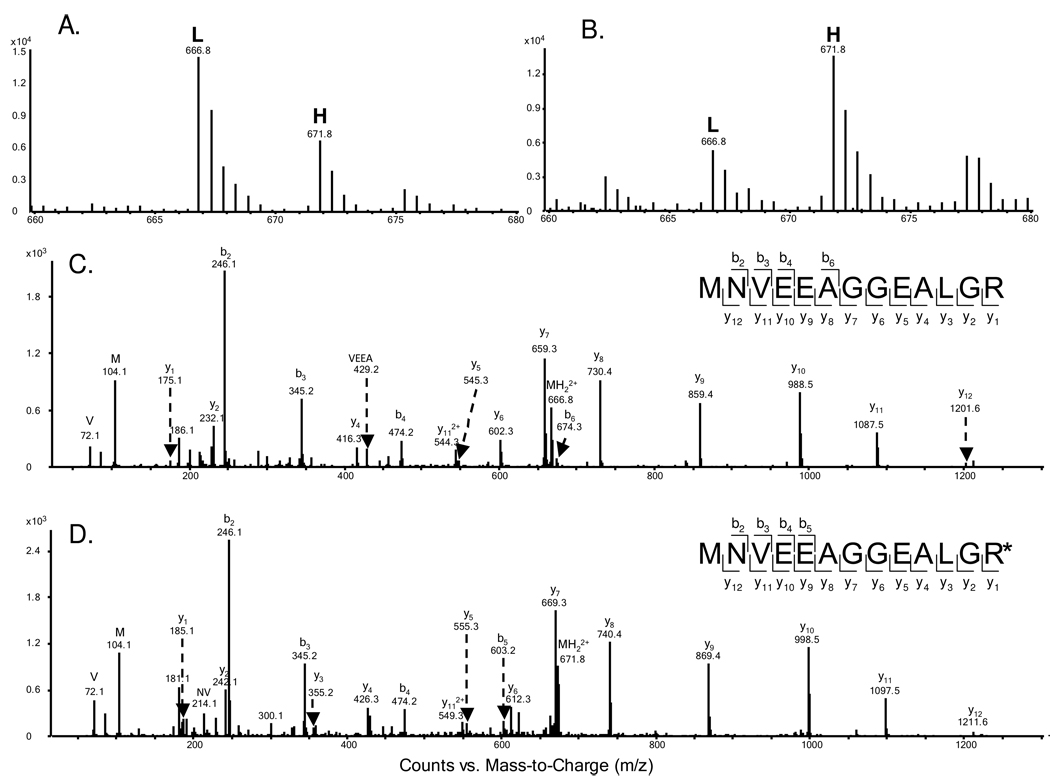
To obtain reliable results, we carried out the SILAC experiments in triplicate and both forward and reverse SILAC labeling was performed (Figure 1A, see also Materials and Methods). Figure 2 shows representative mass spectrometric results for the identification and quantification of the peptide MNVEEAGGEALGR from hemoglobin ε subunit, which reveals clearly the up-regulation of this protein in both forward and reverse SILAC experiments (Figure 2A & B). The peptide sequence was confirmed by MS/MS analysis (Figure 2C & D).
Figure 2.
Representative ESI-MS and MS/MS data revealing the imatinib-induced up-regulation of hemoglobin ε subunit. Shown are the MS for the [M+2H]2+ ions of hemoglobin ε peptide MNVEEAGGEALGR and MNVEEAGGEALGR* (‘R*’ designates the heavy arginine) from the forward (A) and reverse (B) SILAC samples. Light and heavy peptides are labeled as “L” and “H”, respectively. The ratios determined from the forward and reverse SILAC experiments were 2.17 and 2.45 (treated/untreated), respectively. Depicted in (C) and (D) are the MS/MS for the [M+2H]2+ ions of MNVEEAGGEALGR and MNVEEAGGEALGR*, respectively.
We were able to quantify a total of 1344 proteins (Table S1), and the distribution of changes in protein expression levels induced by imatinib treatment is shown in Figure 1B. The average relative standard deviation in the expression ratios determined for all quantified proteins based on different tryptic peptides of the protein from a single SILAC experiment was within 9%. In addition, the average relative standard deviation in the expression ratios for all the quantified proteins from the results of three sets of SILAC measurements was 15%. Thus, we considered those proteins with expression levels being altered by more than 1.5 fold as significantly changed proteins induced by imatinib treatment. Those significantly changed proteins that could be quantified in at least two, which encompassed at least one forward and one reverse, sets of SILAC experiments were considered for the subsequent discussion. Together, this gives quantifiable results for 73 proteins being significantly altered upon imatinib exposure, with 26 and 47 being up- and down-regulated, respectively (Table 1, and the detailed information about the quantified peptides and ratios for each measurement is listed in Table S2).
Table 1.
Proteins quantified with greater than 1.5 fold changes, with IPI numbers, protein names, average ratios and S.D. The listed S.D. values were calculated based on the ratios obtained from three biological replicates. Peptides used for the quantification of individual proteins are listed in Table S2.
| IPI Number | Protein name | Peptide number |
Average±S.D. |
|---|---|---|---|
| Erythroid differentiation-related proteins | |||
| IPI00410714.4 | Hemoglobin α subunit | 6 | 2.36±0.76 |
| IPI00220706.9 | Hemoglobin γ1 subunit | 4 | 2.21±0.83 |
| IPI00217471.2 | Hemoglobin ε subunit | 5 | 2.75±0.77 |
| IPI00554676.1 | Hemoglobin γ2 subunit | 1 | 2.84±0.58 |
| IPI00217473.4 | Hemoglobin ζ subunit | 12 | 3.02±1.18 |
| IPI00030809.1 | γ-G globin | 6 | 2.34±0.55 |
| IPI00027776.6 | Ferrochelatase, mitochondrial precursor | 5 | 1.54±0.22 |
| IPI00093057.6 | Coproporphyrinogen III oxidase, mitochondrial precursor |
5 | 1.61±0.31 |
| IPI00028160.1 | Splice isoform 1 of porphobilinogen deaminase | 8 | 1.83±0.07 |
| IPI00008475.1 | Hydroxymethylglutaryl-CoA synthase, cytoplasmic |
12 | 0.67±0.21 |
| IPI00642144.2 | Aldehyde dehydrogenase 1 family, member A1 | 2 | 1.59±0.24 |
| Translation-related proteins | |||
| IPI00021266.1 | 60S ribosomal protein L23a | 9 | 0.63±0.13 |
| IPI00299573.11 | 60S ribosomal protein L7a | 14 | 0.65±0.15 |
| IPI00013415.1 | 40S ribosomal protein S7 | 8 | 0.65±0.19 |
| IPI00219155.4 | 60S ribosomal protein L27 | 2 | 0.66±0.09 |
| IPI00030179.3 | 60S ribosomal protein L7 | 6 | 0.66±0.15 |
| IPI00037619.2 | Ribosomal protein L39 variant | 1 | 0.67±0.05 |
| IPI00176692.7 | Heterogeneous nuclear ribonucleoprotein A1 | 4 | 0.63±0.24 |
| IPI00029266.1 | Small nuclear ribonucleoprotein E | 2 | 0.64±0.23 |
| IPI00013068.1 | Eukaryotic translation initiation factor 3 subunit 6 | 2 | 0.57±0.17 |
| IPI00016910.1 | Eukaryotic translation initiation factor 3 subunit 8 | 11 | 0.66±0.07 |
| IPI00219678.2 | Eukaryotic translation initiation factor 2 subunit 1 | 2 | 0.67±0.10 |
| Histones | |||
| IPI00021924.1 | Histone H1x | 4 | 1.59±0.20 |
| IPI00219038.8 | Histone H3.3 | 1 | 2.11±0.22 |
| IPI00217465.4 | Histone H1.2 | 10 | 2.26±0.47 |
| IPI00003935.5 | Histone H2B | 7 | 2.35±0.70 |
| IPI00171611.5 | Histone H3.1 | 4 | 2.61±0.41 |
| IPI00026272.1 | Histone H2A | 4 | 2.75±0.70 |
| IPI00453473.5 | Histone H4 | 8 | 3.06±0.82 |
| Other Enzymes | |||
| IPI00221108.4 | Thymidylate synthase | 2 | 0.52±0.04 |
| IPI00010349.1 | Alkyldihydroxyacetonephosphate synthase, peroxisomal precursor |
2 | 0.56±0.08 |
| IPI00291669.3 | Ubiquitin-like domain containing CTD phosphatase 1 |
2 | 0.62±0.07 |
| IPI00305166.1 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial precursor |
1 | 0.62±0.04 |
| IPI00017895.2 | Splice isoform 1 of glycerol-3-phosphate dehydrogenase, mitochondrial precursor |
3 | 0.64±0.07 |
| IPI00026260.1 | Nucleoside diphosphate kinase B | 5 | 0.65±0.22 |
| IPI00017617.1 | Probable ATP-dependent RNA helicase DDX5 | 13 | 0.66±0.10 |
| IPI00220373.3 | Insulin-degrading enzyme | 2 | 0.67±0.05 |
| IPI00010157.1 | S-adenosylmethionine synthetase isoform type-2 | 4 | 0.67±0.01 |
| IPI00000728.3 | Splice isoform 1 of ubiquitin carboxyl-terminal hydrolase 15 |
5 | 0.67±0.10 |
| IPI00295741.4 | Cathepsin B precursor | 6 | 1.50±0.22 |
| IPI00218568.6 | Pterin-4-α-carbinolamine dehydratase | 1 | 1.58±0.11 |
| IPI00337541.3 | NAD(P) transhydrogenase, mitochondrial precursor |
1 | 1.91±0.03 |
| IPI00449049.4 | Poly [ADP-ribose] polymerase 1 | 7 | 2.43±0.42 |
| Other Proteins | |||
| IPI00032561.1 | Calcium-binding protein 39 | 2 | 0.40±0.06 |
| IPI00002214.1 | Importin α2 subunit | 6 | 0.52±0.02 |
| IPI00011631.5 | Centromere/kinetochore protein zw10 homolog | 1 | 0.55±0.08 |
| IPI00005045.1 | ATP-binding cassette sub-family F member 2 | 5 | 0.57±0.04 |
| IPI00026182.4 | F-actin capping protein α2 subunit | 2 | 0.58±0.10 |
| IPI00218505.6 | Protein FAM112B | 2 | 0.59±0.03 |
| IPI00337602.4 | 56 kDa protein | 1 | 0.59±0.09 |
| IPI00218292.2 | Splice isoform short of ubiquitin fusion degradation protein |
4 | 0.60±0.23 |
| IPI00306043.1 | Splice isoform 1 of YTH domain protein 2 | 1 | 0.60±0.01 |
| IPI00009010.3 | UPF0315 protein AD-001 | 2 | 0.61±0.06 |
| IPI00743544.1 | 15 kDa protein | 1 | 0.61±0.02 |
| IPI00220113.1 | Splice isoform 2 of microtubule-associated protein 4 |
11 | 0.61±0.04 |
| IPI00183294.3 | Nuclear pore complex protein Nup214 | 7 | 0.61±0.16 |
| IPI00221035.3 | Splice isoform 1 of transcription factor BTF3 | 3 | 0.61±0.08 |
| IPI00329625.2 | Cell cycle progression 2 protein isoform 1 | 2 | 0.62±0.24 |
| IPI00009057.1 | Splice isoform A of Ras-GTPase-activating protein-binding protein 2 |
6 | 0.63±0.04 |
| IPI00012535.1 | DnaJ homolog subfamily A member 1 | 3 | 0.64±0.16 |
| IPI00027493.1 | 4F2 cell-surface antigen heavy chain | 5 | 0.65±0.09 |
| IPI00186290.5 | Elongation factor 2 | 31 | 0.66±0.03 |
| IPI00012479.1 | Putative nascent polypeptide-associated complex subunit α-like protein |
1 | 0.66±0.06 |
| IPI00013495.1 | Splice isoform 2 of ATP-binding cassette sub-family F |
2 | 0.66±0.04 |
| IPI00023748.3 | Nascent polypeptide-associated complex α subunit | 7 | 0.67±0.04 |
| IPI00397828.2 | 20 kDa protein | 1 | 0.67±0.02 |
| IPI00444704.2 | Splice isoform 2 of G-rich sequence factor 1 | 1 | 0.67±0.01 |
| IPI00219229.2 | U6 snRNA-associated Sm-like protein LSm3 | 2 | 0.67±0.10 |
| IPI00156689.3 | Synaptic vesicle membrane protein VAT-1 homolog |
6 | 1.50±0.10 |
| IPI00217357.2 | Cell division cycle and apoptosis regulator protein 1 |
2 | 1.51±0.11 |
| IPI00024145.1 | Splice isoform 1 of voltage-dependent anion-selective channel 2 |
4 | 1.57±0.03 |
| IPI00220835.6 | Protein transport protein Sec61 β subunit | 1 | 1.57±0.03 |
| IPI00744503.1 | 17 kDa protein | 1 | 1.60±0.17 |
Imatinib induced erythroid differentiation in K562 cells
Among the differentially expressed proteins, all the identified hemoglobins as well as γ-globin were markedly up-regulated in the drug-treated K562 cells (Table 1). Hemoglobinization is the overarching feature of erythroid differentiation, during which less specialized blood cells are differentiated into erythrocytes. We also performed benzidine staining and observed more than 30% benzidine-positive cells upon a 24-hr treatment with 1 µM imatinib (Figure S1), which is consistent with previous findings 20, 21. The up-regulation of hemoglobin proteins, together with benzidine staining result, supported unambiguously the imatinib-induced erythroid differentiation of K562 cells.
Apart from the observation of the up-regulation of hemoglobins and γ-globin, we also found three important enzymes required for heme biosynthesis, i.e., porphobilinogen deaminase, coproporphyrinogen III oxidase and ferrochelatase, to be substantially up-regulated upon imatinib treatment (Table 1). Porphobilinogen deaminase (PBGD) converts porphobilinogen to hydroxymethylbilane, coproporphyrinogen III oxidase oxidizes coproporphyrinogen III to protoporphyrinogen IX, and ferrochelatase catalyzes the final step of heme biosynthesis, which converts protoporphyrin IX into heme (Figure S2). Thus, the hemoglobinization in K562 cells could stem, in part, from the enhanced heme biosynthesis induced by the over-expression of these three enzymes.
Imatinib induced histone hyperacetylation in K562 cells
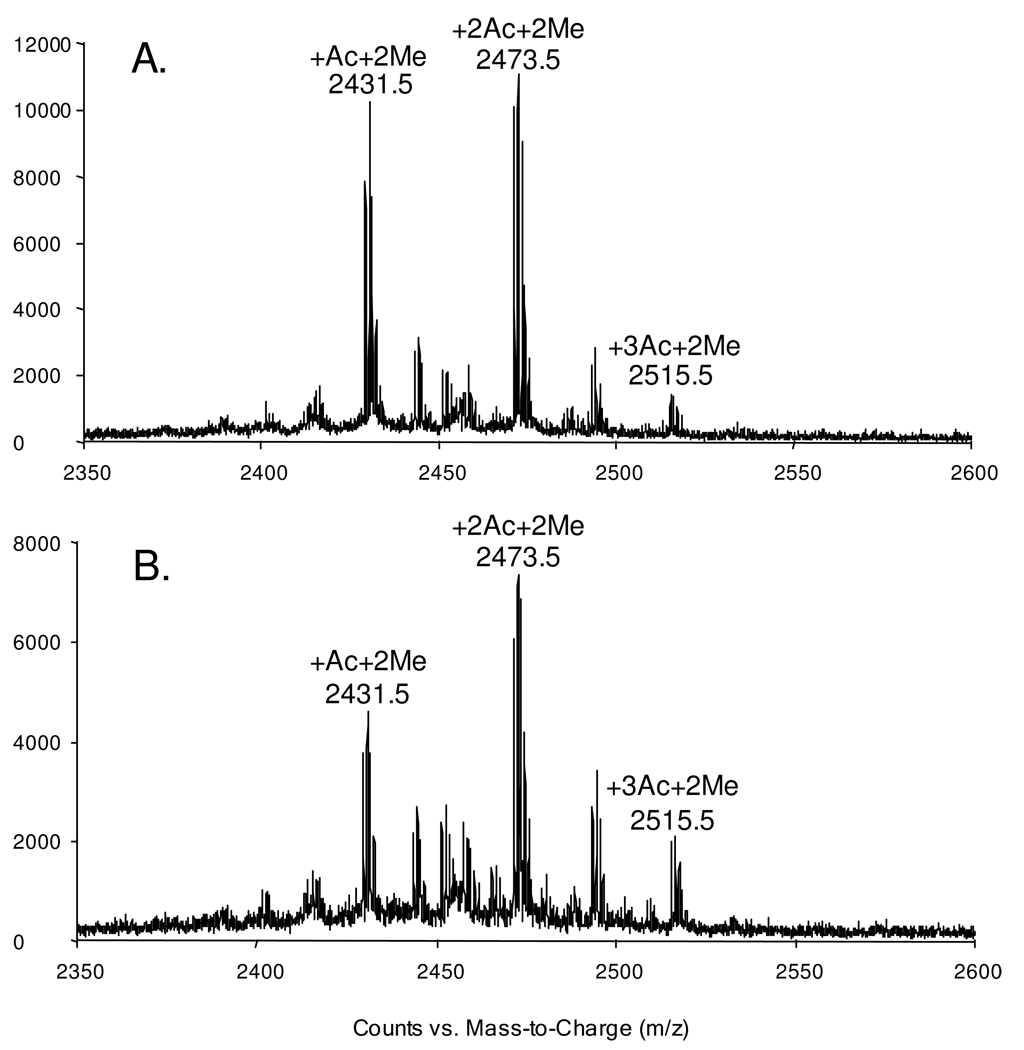
Viewing that many inducers of erythroid differentiation, which include sodium butyrate and valproic acid, were reported to act as histone deacetylase inhibitors and induce histone hyperacetylation in leukemia cells 22, 23, we reason that the imatinib-induced erythroid differentiation of K562 cells might also involve histone hyperacetylation. To test this, we extracted core histones from K562 cells that are treated with imatinib or mock-treated, and subjected the intact histone H4 and its constituent N-terminal peptide to MS analyses. It turned out that we indeed observed increased histone H4 acetylation based on the LC-MS analysis of the intact protein (Figure S3A & B). Likewise, we found a greater than 25% increase in the levels of di- and tri-acetylation of the N-terminal peptide upon treatment with imatinib (the MALDI-MS for the Asp-N-produced H4 N-terminal peptide 1SGRGKGGKGLGKGGAKRHRKVLR23 is shown in Figure 3). LC-MS/MS for the mono-, di-and tri-acetylated peptides revealed the acetylation of the N-terminus (for all three acetylated forms of the peptide), K16 (for both the di- and tri-acetylated peptide), and K8 or K12 (for the triacetylated peptide, Figure S4).
Figure 3.
MALDI-MS for the Asp-N-produced peptide 1SGRGKGGKGLGKGGAKRHRKVLR23 of histone H4 extracted from control (A) and imatinib-treated (B) K562 cells. The peptide with dimethylation and mono-, di-, tri-acetylation were labeled, while unmodified as well as mono- and tri-methylated peptides could also be observed as low-intensity peaks.
Imatinib affects the expression of histones and proteins involved in translation
Aside from the considerable upregulation of hemoglobins and related proteins, imatinib treatment also led to significant down-regulation of some important proteins associated with translational machinery, including ribosomal proteins, translation initiation factors and nuclear ribonucleoproteins (Table 1). These results are in accordance with the previous findings 24, 25 and with the imatinib-induced growth inhibition.
All four core histones and linker histone H1 were markedly up-regulated in imatinib-treated K562 cells (Table 1). The substantially increased expression of histones might reflect the considerable change in chromatin structure induced by imatinib treatment. In this regard, we chose to use only those peptides that do not bear any known post-translational modifications (PTMs) for the quantification 26. This avoids the inaccurate quantification emanating from imatinib-induced perturbation in histone PTMs (e.g. H4 hyperacetylation).
Imatinib induced the alteration in expression of other important enzymes
Imatinib treatment also gave rise to considerable changes in the expression levels of some other important enzymes, including thymidylate synthase (TS), S-adenosylmethionine synthetase (AdoMetS), mitochondrial glycerol-3-phosphate dehydrogenase (GPD2), NAD(P) transhydrogenase, poly(ADP-ribose) polymerase 1, etc. (Table 1). These proteins play pivotal roles in different cellular pathways, and we would like to discuss some of them in detail.
TS is a folate-dependent enzyme that catalyzes the reductive methylation of 2′-deoxyuridine-5′-monophosphate to generate thymidine-5′-monophosphate and the latter is subsequently phosphorylated to thymidine-5′-triphosphate, which is an essential precursor for DNA synthesis 27. We observed that TS was down-regulated by approximately 2 fold upon imatinib treatment. The inhibition of TS results in a lack of thymidine-5′-triphosphate and an accumulation of 2′-deoxyuridine-5′-monophosphate, which may be subsequently converted to 2′-deoxyuridine-5′-triphosphate and incorporated into DNA and induces DNA strand breaks and cell death. In this vein, Lee and coworkers 28 reported that a histone deacetylase inhibitor trichostatin A enhanced 5-fluorouracil cytotoxicity by down-regulating TS in human cancer cells. The diminished expression of TS might be attributed to the imatinib-induced histone hyperacetylation.
Poly(ADP-ribosyl)ation (PAR) catalyzed by poly(ADP-ribose) polymerase (PARP) is a major mediator of cell death after exposure to a variety of DNA damaging agents 29. PARP1 is rapidly activated by DNA strand breaks and this enzyme catalyzes the transfer of the ADP-ribose moiety from the co-enzyme NAD+ to a number of nuclear acceptor proteins 29. Imatinib treatment was found to lead to a rapid increase in PAR which preceded the loss of integrity of mitochondrial membrane and DNA fragmentation; inhibition of PAR in imatinib-treated cells partially prevented cell death 30. Therefore, the imatinib-induced up-regulation of PARP1 may contribute to the growth inhibition and apoptosis induction in K562 cells through stimulating PAR.
Conclusions
Imatinib mesylate is considered the most effective and relatively safe drug for the treatment of chronic phase of CML. In this study, we employed SILAC, together with LC-MS/MS, and assessed quantitatively the perturbation of protein expression in K562 human CML cells by imatinib. Although a modest number of proteins were quantified in the present study, which is mainly due to the limitation in instrument sensitivity, the expression of many important proteins was found to be perturbed by imatinib. Our results revealed that the imatinib treatment led to the up-regulation of all the hemoglobinization-related proteins and the perturbation in the expression of many important enzymes.
Upon imatinib treatment, all the quantified hemoglobins and γ-globin were markedly up-regulated, and benzidine staining provided direct evidence supporting the imatinib-induced hemoglobinization and erythroid differentiation of K562 cells. More importantly, three key enzymes for heme biosynthesis were found up-regulated by 1.5–2 folds, demonstrating the enhanced endogenous hemoglobin biosynthesis. Viewing that many well-known histone deacetylase inhibitors could lead to differentiation of leukemia cells 22, 23, our quantitative proteomic results led us to predict and confirm that imatinib treatment gave rise to histone hyperacetylation in K562 cells. Although the imatinib-induced hemoglobinization has been reported previously 20, 21, the results from the present study paints the most complete picture for this pathway, by providing quantitative measurements of many proteins including hemoglobins and enzymes involved in heme biosynthesis, and demonstrating the ability of imatinib to induce histone hyperacetylation. The latter perturbation in histone acetylation epigenetic mark might be important in the imatinib-induced differentiation of CML cells.
Aside from proteins involved with erythroid differentiation, we found that many other important proteins were up- or down-regulated upon imatinib treatment. Future studies about the biological implications of the perturbation in expression of these proteins may also facilitate the discovery of additional molecular pathways that are altered by imatinib. This may contribute to an improved understanding of the imatinib-induced cytotoxicity and the development of resistance toward this drug. Taken together, the pharmacoproteomic profiling could constitute a valuable tool for the identification of drug-responsive biomarkers and for the establishment of a molecular basis for developing novel and more effective approaches for the therapeutic intervention of human CML.
Supplementary Material
Acknowledgment
This work was supported by the National Institutes of Health (R01 CA 116522).
Footnotes
Supporting Information Available: Tables for quantification results and LC-MS data for monitoring histone H4 acetylation. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Rowley JD. New consistent chromosomal abnormality in chronic myelogenous leukemia identified by quinacrine fluorescence and giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- 2.Sherbenou DW, Druker BJ. Applying the discovery of the Philadelphia chromosome. J. Clin. Invest. 2007;117:2067–2074. doi: 10.1172/JCI31988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chu S, Holtz M, Gupta M, Bhatia R. BCR/ABL kinase inhibition by imatinib mesylate enhances MAP kinase activity in chronic myelogenous leukemia CD34+ cells. Blood. 2004;103:3167–3174. doi: 10.1182/blood-2003-04-1271. [DOI] [PubMed] [Google Scholar]
- 4.Horita M, Andreu EJ, Benito A, Arbona C, Sanz C, Benet I, Fernandez-Luna JL. Blockade of the Bcr-Abl kinase activity induces apoptosis of chronic myelogenous leukemia cells by supressing STAT5-dependent expression of Bcl-xL. J. Exp. Hematol. 2000;191 doi: 10.1084/jem.191.6.977. 977-784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Savage DG, Antman KH. Drug therapy: Imatinib mesylate - A new oral targeted therapy. N. Eng. J. Med. 2002;346:683–693. doi: 10.1056/NEJMra013339. [DOI] [PubMed] [Google Scholar]
- 6.Balabanov S, Gontarewicz A, Ziegler P, Hartmann U, Kammer W, Copland M, Brassat U, Priemer M, Hauber I, Wilhelm T, Schwarz G, Kanz L, Bokemeyer C, Hauber J, Holyoake TL, Nordheim A, Brummendorf TH. Hypusination of eukaryotic imtiation factor 5A (eIF5A): a novel therapeutic target in BCR-ABL-positive leukemias identified by a proteomics approach. Blood. 2007;109:1701–1711. doi: 10.1182/blood-2005-03-037648. [DOI] [PubMed] [Google Scholar]
- 7.Smith DL, Evans CA, Pierce A, Gaskell SJ, Whetton AD. Changes in the proteome associated with the action of Bcr-Abl tyrosine kinase are not related to transcriptional regulation. Mol. Cell. Proteomics. 2002;1:876–884. doi: 10.1074/mcp.m200035-mcp200. [DOI] [PubMed] [Google Scholar]
- 8.Unwin RD, Sternberg DW, Lu YN, Pierce A, Gilliland DG, Whetton AD. Global effects of BCR/ABL and TEL/PDGFR beta expression on the proteome and phosphoproteome - Identification of the rho pathway as a target of BCR/ABL. J. Biol. Chem. 2005;280:6316–6326. doi: 10.1074/jbc.M410598200. [DOI] [PubMed] [Google Scholar]
- 9.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 10.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Liang XQ, Hajivandi M, Veach D, Wisniewski D, Clarkson B, Resh MD, Pope RM. Quantification of change in phosphorylation of BCR-ABL kinase and its substrates in response to imatinib treatment in human chronic myelogenous leukemia cells. Proteomics. 2006;6:4554–4564. doi: 10.1002/pmic.200600109. [DOI] [PubMed] [Google Scholar]
- 12.Xiong L, Ping LY, Yuan BF, Wang YS. Methyl group migration during the fragmentation of singly charged ions of trimethyllysine-containing peptides: Precaution of using MS/MS of singly charged ions for interrogating peptide methylation. J. Am. Soc. Mass Spectrom. 2009;20:1172–1181. doi: 10.1016/j.jasms.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SK, Venable JD, Xu T, Yates JR. A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods. 2008;5:319–322. doi: 10.1038/nmeth.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McClatchy DB, Liao LJ, Park SK, Venable JD, Yates JR. Quantification of the synaptosomal proteome of the rat cerebellum during post-natal development. Genome Res. 2007;17:1378–1388. doi: 10.1101/gr.6375007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tabb DL, McDonald WH, Yates JR. DTASelect and contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uitto PM, Lance BK, Wood GR, Sherman J, Baker MS, Molloy MP. Comparing SILAC and two-dimensional gel electrophoresis image analysis for profiling urokinase plasminogen activator signaling in ovarian cancer cells. J. Proteome Res. 2007;6:2105–2112. doi: 10.1021/pr060638v. [DOI] [PubMed] [Google Scholar]
- 17.Romijn EP, Christis C, Wieffer M, Gouw JW, Fullaondo A, van der Sluijs P, Braakman I, Heck AJR. Expression clustering reveals detailed coexpression patterns of functionally related proteins during B cell differentiation - A proteomic study using a combination of one-dimensional gel electrophoresis, LC-MS/MS, and stable isotope labeling by amino acids in cell culture (SILAC) Mol. Cell. Proteomics. 2005;4:1297–1310. doi: 10.1074/mcp.M500123-MCP200. [DOI] [PubMed] [Google Scholar]
- 18.Horikoshi A, Takei K, Sawada S. Relationship between daily dose of imatinib per square meter and its plasma concentration in patients with chronic-phase chronic myeloid leukemia (CML) Leuk. Res. 2007;31:574–575. doi: 10.1016/j.leukres.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 19.Tsutsumi Y, Kanamori H, Yamato H, Ehira N, Miura T, Kawamura T, Obara S, Masauzi N, Tanaka J, Imamura M, Asaka M. Monitoring of plasma imatinib concentration for the effective treatment of CML patients. Leuk. Res. 2004;28:1117–1118. doi: 10.1016/j.leukres.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 20.Fang G, Kim CN, Perkins CL, Ramadevi N, Winton E, Wittmann S, Bhalla KN. CGP57148B (STI-571) induces differentiation and apoptosis and sensitizes Bcr-Abl-positive human leukemia cells to apoptosis due to antileukemic drugs. Blood. 2000;96:2246–2253. [PubMed] [Google Scholar]
- 21.Jacquel A, Herrant M, Legros L, Belhacene N, Luciano F, Pages G, Hofman P, Auberger P. Imatinib induces mitochondria-dependent apoptosis of the Bcr-Abl positive K562 cell line and its differentiation towards the erythroid lineage. FASEB J. 2003;17:2160–2162. doi: 10.1096/fj.03-0322. [DOI] [PubMed] [Google Scholar]
- 22.Gurvich N, Tsygankova OM, Meinkoth JL, Klein PS. Histone deacetylase is a target of valproic acid-mediated cellular differentiation. Cancer Res. 2004;64:1079–1086. doi: 10.1158/0008-5472.can-03-0799. [DOI] [PubMed] [Google Scholar]
- 23.Wei GH, Zhao GW, Song W, Hao DL, Lv X, Liu DP, Liang CC. Mechanisms of human gamma-globin transcriptional induction by apicidin involves p38 signaling to chromatin. Biochem. Biophys. Res. Commun. 2007;363:889–894. doi: 10.1016/j.bbrc.2007.06.191. [DOI] [PubMed] [Google Scholar]
- 24.Prabhu S, Saadat D, Zhang M, Halbur L, Fruehauf JP, Ong ST. A novel mechanism for Bcr-Abl action: Bcr-Abl-mediated induction of the eIF4F translation initiation complex and mRNA translation. Oncogene. 2007;26:1188–1200. doi: 10.1038/sj.onc.1209901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ly C, Arechiga AF, Melo JV, Walsh CM, Ong ST. Bcr-Abl kinase modulates the translation regulators ribosomal protein S6 and 4E-BP1 in chronic myelogenous leukemia cells via the mammalian target of rapamycin. Cancer Res. 2003;63:5716–5722. [PubMed] [Google Scholar]
- 26.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 27.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Ann. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 28.Lee JH, Park JH, Jung Y, Kim JH, Jong HS, Kim TY, Bang YJ. Histone deacetylase inhibitor enhances 5-fluorouracil cytotoxicity by down-regulating thymidylate synthase in human cancer cells. Mol. Cancer Ther. 2006;5:3085–3095. doi: 10.1158/1535-7163.MCT-06-0419. [DOI] [PubMed] [Google Scholar]
- 29.D'Amours D, Desnoyers S, D'Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem. J. 1999;342:249–268. [PMC free article] [PubMed] [Google Scholar]
- 30.Moehring A, Wohlbold L, Aulitzky WE, van der Kuip H. Role of poly(ADP-ribose) polymerase activity in imatinib mesylate-induced cell death. Cell Death Differ. 2005;12:627–636. doi: 10.1038/sj.cdd.4401608. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.