Abstract
Analysis of the Coxiella burnetii RSA 493 (Nine Mile Phase I strain) genome revealed open reading frames with significant homology to the type IVB secretion system (T4BSS) of Legionella pneumophila. The T4BSS genes exist primarily at two loci, designated regions I (RI) and II. In C. burnetii, little is known about the T4BSS regions and the role they play in establishing and/or maintaining infection. C. burnetii T4BSS RI contains genes arranged in three linkage groups: (i) icmW→CBU1651→icmX, (ii) icmV→dotA→CBU1647, (iii) icmT→icmS→dotD→dotC→dotB→CBU1646. We used RT-PCR to demonstrate transcriptional linkage within the groups, and that icmX, icmV, and icmT are transcribed de novo by 8 hours post infection (hpi). We then examined transcript levels for icmX, icmW, icmV, dotA, dotB and icmT during the first 24 hours of an infection using RT-qPCR. Expression initially increased for each gene, followed by a decrease at 24 hpi. Subsequently, we analyzed IcmT protein levels during infection and determined that expression increases significantly from 8 to 24 hpi then remains relatively constant. These data demonstrate temporal changes in the RNA of several C. burnetii T4SS RI homologs and the IcmT protein. These changes correspond to early stages of the C. burnetii infectious cycle.
Keywords: Coxiella, Type four secretion system, gene expression, obligate intracellular
Introduction
Coxiella burnetii is an obligate intracellular pathogen that exhibits a bi-phasic life cycle that starts with the environmentally stable small cell variant (SCV) form and converts into the metabolically active and replicative large cell variant (LCV) form during the first 24 hours post infection (hpi) (McCaul & Williams, 1981, McCaul, 1991, Heinzen, et al., 1999). Upon infection of a host cell, C. burnetii is trafficked along the endocytic pathway and eventually resides within a parasitophorous vacuole (PV) retaining features of a mature phagolysosomes (Akporiaye, et al., 1983, Heinzen, et al., 1996, Ghigo, et al., 2002, Gutierrez, et al., 2005, Sauer, et al., 2005, Howe & Heinzen, 2006). The early trafficking, enlargement, and maintenance of the C. burnetii PV is dependent on C. burnetii protein synthesis (Howe, et al., 2003, Howe, et al., 2003). Infected cells treated with chloramphenicol early during infection contained small tightly bound LAMP-1 positive PVs containing single C. burnetii that were dispersed throughout the host cell (Howe, et al., 2003). However, with the removal of the chloramphenicol vacuolar fusion resumed, resulting in spacious PVs (SPV) containing multiple C. burnetii (Howe, et al., 2003). C. burnetii infected cells treated with carbenicillin or nalidixic acid were found to have mature SPVs containing multiple non-replicating C. burnetii, suggesting that vacuolar development requires metabolically active C. burnetii and is not dependant on bacterial density for complete PV maturation (Howe, et al., 2003, Howe, et al., 2003). These studies demonstrate that the expression of C. burnetii genes during the first 24 hpi of the PV niche establishment is crucial for the development of a productive infection. (Coleman, et al., 2004).
Interestingly, during PV establishment C. burnetii delays trafficking for approximately two hours (Howe & Mallavia, 2000) and the nascent PV appears to associate with the LC3 autophagy pathway marker (Beron, et al., 2002, Gutierrez, et al., 2005, Romano, et al., 2007). Moreover, C. burnetii actively mediates the inhibition of host cell apoptosis by activating Akt and Erk1/2 (Voth & Heinzen, 2009), allowing this relatively slow growing pathogen (10–12 h replication rate) the opportunity to replicate to high numbers prior to host cell lysis. These characteristics may be attributable to C. burnetii proteins containing the ankyrin repeat eukaryotic motifs, which have been shown to associate with the PV membrane, microtubules, and mitochondria when expressed ectopically within eukaryotic cells (Voth, et al., 2009). In addition, recent reports show a series of C. burnetii encoded ankyrin repeat domain containing proteins that are secreted into host cells by Legionella pneumophila in a type IVB secretion system (T4BSS) dependant manner (Pan, et al., 2008, Voth, et al., 2009), highlighting the versatility and importance of this secretion system.
Bacterial secretion systems specifically involved in virulence include the type IV secretion systems (T4SS). The T4SSs has been subdivided into two groups, the type IVA secretion system (T4ASS), encoded by the virB operon (Sexton & Vogel, 2002), and the T4BSS (Segal, et al., 1998, Vogel, et al., 1998). L. pneumophila’s T4BSS is essential for effector protein secretion, bacterial intracellular trafficking, and replication within macrophages as well as amoeba (Marra, et al., 1992, Berger & Isberg, 1993, Bruggemann, et al., 2006, Ninio & Roy, 2007, Shin & Roy, 2008). Analysis of the C. burnetii RSA 493 (Nine Mile phase I strain) genome sequence revealed loci with significant homology and gene organization to both Region I (RI) and Region II of the L. pneumophila T4BSS (Seshadri, et al., 2003). The genomic sequence, combined with studies using C. burnetii T4BSS analogs (IcmW, DotB, IcmS, and IcmT) to complement L. pneumophila mutants (Zamboni, et al., 2003, Zusman, et al., 2003), indicates that C. burnetii expresses a functional T4BSS during infection. Gene expression analysis of the C. burnetii T4BSS has been limited both in the number of homologs analyzed as well as the breadth of the temporal analysis.
In an effort to develop an understanding of the transcriptional and translational expression of the C. burnetii T4BSS with an emphasis on early stages of the infectious cycle, we analyzed the RNA expression profile of select RI genes. The C. burnetii T4BSS RI loci contains twelve genes (CBU1652 – CBU1641), nine of which are L. pneumophila T4BSS homologs (Seshadri, et al., 2003). Following a synchronous infection of host cells by C. burnetii SCVs, total RNA isolated during the initial stages of the infectious cycle was used to analyze the transcription of the C. burnetii T4BSS RI homologs. Here, we provide the first demonstration of transcriptional linkages between the C. burnetii T4BSS RI homologs, demonstrate de novo transcription by 8 hpi, analyze the relative RNA expression during the initial stages of the infectious cycle as well as the relative expression of the C. burnetii IcmT homolog throughout infection.
Materials and Methods
Bacterial Cultivation and Purification
C. burnetii NMII was propagated in African green monkey kidney (Vero) cells in RPMI-1640 medium with 5% fetal bovine serum (FBS) and the SCV form of the organism was isolated as previously described (Coleman, et al., 2004). Following differential centrifugation, SCV preparations were resuspended in SPG buffer (0.7 M sucrose, 3.7 mM KH2PO4, 6.0 mM K2HPO4, 0.15 M KCl, 5.0 mM glutamic acid, pH 7.4) and stored at −80°C. Organisms were enumerated by genome equivalents using qPCR (Brennan & Samuel, 2003).
Cell culture and infection
Uninfected Vero cells were propagated in RPMI-1640 media containing 5% FBS with gentamicin (20 µg mL−1) at 37°C and 5% CO2. The culture medium was exchanged for medium without antibiotics two hours prior to bacterial infections. Vero cells were infected with C. burnetii NMII at a genome equivalent multiplicity of infection (MOI) of 100, resulting in 40% infection. After two hours (designated as time-zero), inoculums were removed, cells washed three times with RPMI, then incubated in RPMI with 5% FBS at 37°C and 5% CO2. To determine de novo synthesis of C. burnetii RNA upon infection of Vero cells, parallel cultures were either treated with the RNA synthesis inhibitor rifampin (+Rif) at 20 µg mL−1 in the culture media or mock treated (−Rif).
RNA isolation and quality control
Total RNA was harvested at 0, 8, 16, and 24 hpi using Tri Reagent (Ambion, Austin, TX). In some cases, enriched C. burnetii RNA was isolated using a modification of the digitonin based bacterial isolation method (Cockrell, et al., 2008). Briefly, GeneLock™ (Sierra Molecular) was added to 20% in SP buffer (250mM sucrose, 12.8mM KH2PO4, 72.6mM NaCl, 53.9mM Na2HPO4 at pH 7.4). SPD-GL buffer (SP buffer containing digitonin at 0.2 mg ml−1 and GeneLock™ solution) was added to infected culture flasks. Flasks were incubated on ice for 30min. with moderate rocking during which time cell lysis occurs (Cockrell, et al., 2008). Cell lysates were then collected and centrifuged at 1,200 × g for 15min. (4°C) to pellet host cellular debris. Supernatants were then transferred to new tubes and centrifuged for 10min. at 13,000 × g (4°C) to pellet the released C. burnetii. The C. burnetii pellets were solubilized in TRI Reagent® Solution (Ambion), and processed according to the manufacturer’s instruction. This process was found to protect the integrity of the RNA during bacterial enrichment while substantially enriching the relative amount of C. burnetii specific RNA in a given sample (J. K. Morgan & E. I. Shaw, unpublished data). To remove contaminating DNA, all RNA samples were treated with RQ1 DNase (Promega, Madison, WI). Removal of contaminating DNA was confirmed using PCR.
RNA analysis
RT-PCR analysis was carried out using the Access Quick RT-PCR Kit (Promega) following the manufacturer’s instructions. All oligonucleotide primers used in this study (Integrated DNA Technologies, Coralville, IA) are shown in Table 1. Forward [f] and reverse [r] primer pairs: CB58 [f] and CB57 [r] (icmW – CBU1651 – icmX), CB59 [f] and CB60 [r] (icmV – dotA), CB718 [f] and CB716 [r] (dotA – CBU1647), CB603 [f] and CB602 [r] (dotB – CBU1646), CB63 [f] and CB64 [r] (dotD – dotC – dotB), and CB62 [f] and CB61 [r] (icmT – icmS – dotD); were used to demonstrate transcriptional linkage. RT-PCR analysis of icmT, icmV, and icmW ORFs was performed using CB78 [f] and CB79 [r], CB70 [f] and CB71 [r], and CB40 [f] and CB41 [r], respectively.
TABLE 1.
Oligonucleotide primers used in this study.
| Gene Target | RefSeq ID | Designation | Gene Primer Sequence (5’ → 3’) | Purpose |
|---|---|---|---|---|
| icmT | CBU 1641 | CB62f | GCAAAATCGCCATAGCATGGTG | RT-PCR |
| CB578f | GGGATGGCAAAACAGCCTTT | RT-qPCR | ||
| CB579r | CCGTCACCGCTACGATGAG | RT-qPCR | ||
| CB78f | †CACCATGAAATCTCTCGATGAGG | PCR, RT-PCR | ||
| CB79r | ‡TTAGTTATCCCACCATGCTATGG | PCR, RT-PCR | ||
| dotD | CBU 1643 | CB63f | TCCAGACGGATCATTGAGC | RT-PCR |
| CB61r | CAACGCCAGAAAGAGGGGCAGC | RT-PCR | ||
| dotB | CBU 1645 | CB64r | CAATGTGTTTGGGTTCGAAGCG | RT-PCR |
| CB603f | ACTCGACAGTGATCCCGAAC | RT-PCR | ||
| CB586f | CCACGGGTTCGGGTAAAAG | RT-qPCR | ||
| CB587r | GCGCTTCGGCCAATTCT | RT-qPCR | ||
|
glycine betaine transporter§ |
CBU 1646 | CB602r | TTACCCAGCGGCGTTAATAC | RT-PCR |
|
hypothetical ORF |
CBU 1647 | CB716r | ACGCTAACGGCGTTAATACG | RT-PCR |
| dotA | CBU 1648 | CB60r | CGATAATGCCTTCATTGAGC | RT-PCR |
| CB718f | TAGTGGCAGTATCCTTCACG | RT-PCR | ||
| CB654f | ACAATCAATCCCCGTTGAAA | RT-qPCR | ||
| CB655r | AGCTATCATCGCCTGGCTTA | RT-qPCR | ||
| icmV | CBU 1649 | CB59f | TTCCAAATGAAGCAACGC | RT-PCR |
| CB592f | TGGCGGGTTACACGGTTT | RT-qPCR | ||
| CB593r | CGCAGACGAAAGCCGATAA | RT-qPCR | ||
| CB70f | †CACCATGATTCTTTTGGAGTCTTCC | RT-PCR | ||
| CB71r | ‡TTATTGTTTGGACCCCTTAAAGGTG | RT-PCR | ||
| icmW | CBU 1650 | CB58f | CAAACTCTTGAGGAAGG | RT-PCR |
| CB594f | CGCCGCTGCGAAAGTG | RT-qPCR | ||
| CB595r | ACCGGCGGTGTCTATTTCC | RT-qPCR | ||
| CB40f | †CACCATGCCAGATCTGTCGC | RT-PCR | ||
| CB41r | ‡TTATAAACCACCTTCCTCAAGAG | RT-PCR | ||
| icmX | CBU 1652 | CB57r | GAAGCAATACCAAGAACACG | PCR, RT-PCR |
| CB658f | CCGCTTATAATTCGGACCAA | RT-qPCR | ||
| CB659r | TTGATAAGCGGGATTGTTCA | RT-qPCR | ||
CACC, non-C. burnetii sequence, engineered directional cloning sequence for pET200/D-TOPO plasmid.
TTA, non-C. burnetii sequence, engineered stop codon.
glycine betaine transporter (Beare, et al., 2009).
designate Forward and Reverse primers respectively.
Oligonucleotide primers (Table 1) for RT-qPCR analysis of icmX, icmW, icmV, dotA, dotB, and icmT were designed using Primer3Plus (Andreas Untergasser, 2007). The primer efficiency of all primer sets were within the efficiency window for the 2−ΔCT calculation method (Livak & Schmittgen, 2001, Schmittgen & Livak, 2008). Single step RT-qPCR analysis using SuperScript III (Invitrogen) reverse transcriptase and the SYBR Green Master Mix Kit (Applied Biosystems) was performed on an ABI 7500 cycler. Each reaction contained 15 µL total volume and 20 ng total RNA. To calculate relative temporal RNA expression fold changes over the time course, we employed 2−ΔCT to each individual gene respectively. For each gene we employed their respective mean time-zero cycle threshold (CT) value as the baseline reference point for all other (respective) mean time point CT values over the evaluation period. Therefore, for each individual gene their relative fold expression at each time point is internally referenced to time zero RNA levels. Each time-zero point has been referenced to themselves resulting in a calculated fold value of 1 (Fig. 3). Statistical significance between the time points was evaluated by single factor ANOVA with a 95% confidence interval using MS Excel 2007 (Microsoft). A p value of <0.05 was considered significant.
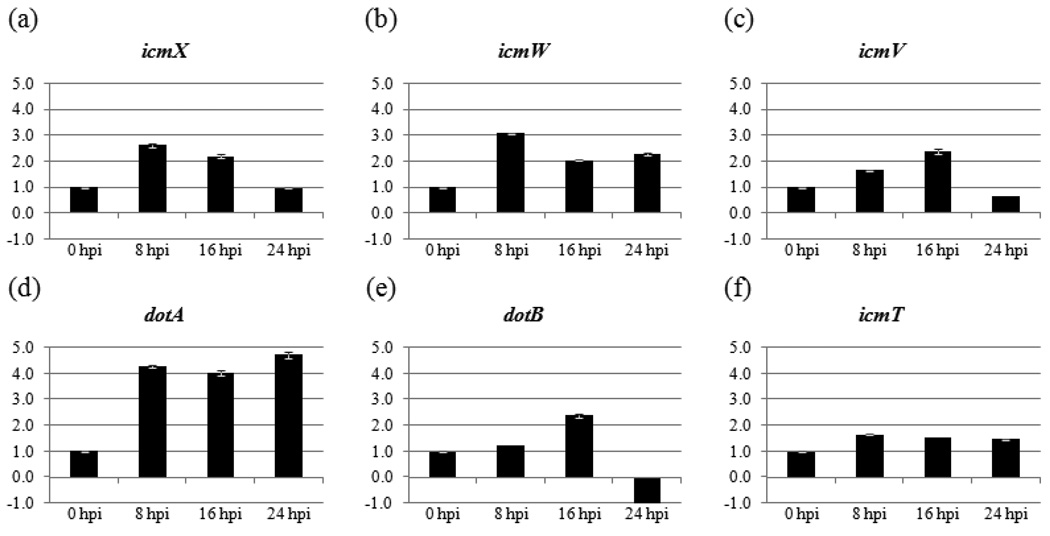
Fig. 3. C. burnetii icmX, icmW, icmV, dotA, dotB, and icmT transcript levels during the early stages of infection.
(a–f) Fold changes in mRNA levels relative to 0 hpi. An equal amount of total RNA from each sample was analyzed by RT-qPCR. The time (in hpi) when total RNA was harvested is indicated below the X-axis. Results represent the mean of three biological samples with no fewer than three technical replicates of each sample. Standard error bars represent the combined standard error of the mean (S.E.M.) per time point.
C. burnetii IcmT expression analysis
Recombinant C. burnetii IcmT and protein specific antibody were the same as previously described (Morgan, et al., 2010). Briefly, to ensure specificity the rabbit sera against recombinant C. burnetii IcmT was absorbed against Vero cell lysates as well as the E. coli DH5α expression strain to remove any cross reactive antibodies. This antibody was designated RαIcmT. For immunoblot analysis, purified C. burnetii NMII was pelleted and resuspended in protein lysis/running buffer (Tris-HCl pH 6.8 62.5mM, SDS 2%, Glycerol 25%, Bromophenol Blue 0.01%, 2-mercaptoethanol 5 % added prior to loading). Protein representing 108 C. burnetii genome equivalents and 104 Vero cells, respectively, was separated by 16% SDS PAGE and transferred to nitrocellulose membrane (Whatman, Dassel, Germany) along with a protein ladder (Bio-Rad, Hercules, CA). Immunoblot analysis was carried out using a Pico Western Chemilluominescent Kit (Pierce, Rockford, IL) following the manufacturer’s directions using the RαIcmT primary antibody at a 1:1000 dilution in hybridization buffer.
Vero cells were seeded on 12 mm glass coverslips in 24 well plates and allowed to attach overnight. C. burnetii NMII infections were initiated as described and fixed at 0, 8, 16, 24, 48, 96, and 168 hpi with 4% paraformaldehyde, 0.05% Tween-20 in PBS for 15 minutes at room temperature. Indirect immunofluorescent antibody (IFA) analysis was performed by dual staining as previously described (Morgan, et al., 2010). Micrograph images were captured via a Nikon DS FI1 camera on a Nikon Eclipse TE 2000-S microscope at 400× magnification, with NIS-Elements F 3.00 software. Magnification of 400× was used as opposed to 600×, as used previously (Morgan, et al., 2010). Micrograph capture settings were uniform for all images. Using a modification of a method previously used in C. burnetii studies which employs relative pixel ratios in sample quantitation (Zamboni, et al., 2001), each micrograph image was analyzed using ImageJ version 1.42n (Wayne Rasband, NIH) software. Five fields of view from each time sampled (three biological samples of each) were digitally captured. The matching Alexa Fluor® 555 and Alexa Fluor® 488 images were stacked (paired) and converted to grey scale (8 bit, see Fig. 4a inset). No fewer than five regions of interest (ROI) were blindly selected from each field of view of the 555 wavelength grey scale images. This provided at least 75 ROIs for each time point analyzed. Saturated regions of an image were not selected and ROI size varied depending on PV size. The pixel density within the identical ROIs from each stacked image were then measured as previously published (Collins, 2007). The mean pixel densities were then compared to obtain the 488:555 ratio for each ROI. These individual ratios were then averaged (≥75 individual ratios/time point) to determine the relative expression of IcmT to whole C. burnetii NMII. The final 488:555 (IcmT:C. burnetii) ratio for each time point was then divided into the 0 hpi ratio to obtain the final IcmT relative expression levels. Statistical significance between each time point was evaluated using single factor ANOVA with a 95% confidence interval with MS Excel 2007 (Microsoft).
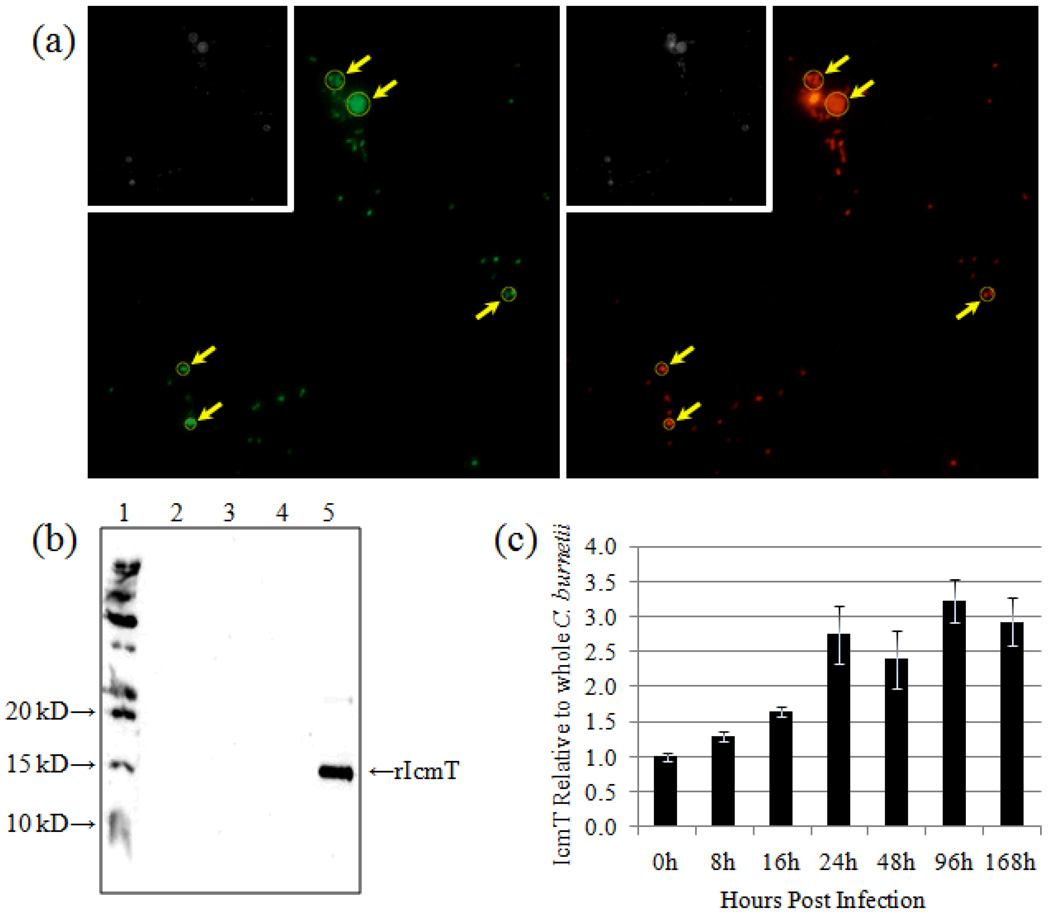
Fig. 4. Relative IcmT expression over a time course of infection.
(a) Representative dual channel IFA micrograph (400× magnification) images of C. burnetii infected Vero cells 24 hpi. Left panel - Alexa® 488 indicates RαIcmT antibody binding. Right panel - Alexa® 555 indicates Guinea pig anti-whole C. burnetii antibody. Inset images are 8 bit (gray scale) conversions of the color images. Representative Regions of Interest (ROI) are shown (arrows). (b) Immunoblot using RαIcmT to probe total protein from purified C. burnetii (lane 2), Vero cells (lane 3), blank lane (lane 4), and purified recombinant IcmT (rIcmT, lane 5). Lane 1 contains the protein size ladder. (c) Pixel density ratios of IcmT to whole C. burnetii (488:555) from digital images captured at 0, 8, 16, 24, 48, 96, and 168 hpi were converted to quotient values relative to 0 hpi. Error bars represent the S.E.M. for each respective quotient value.
Results and Discussion
Transcriptional linkage within the C. burnetii T4BSS Region I
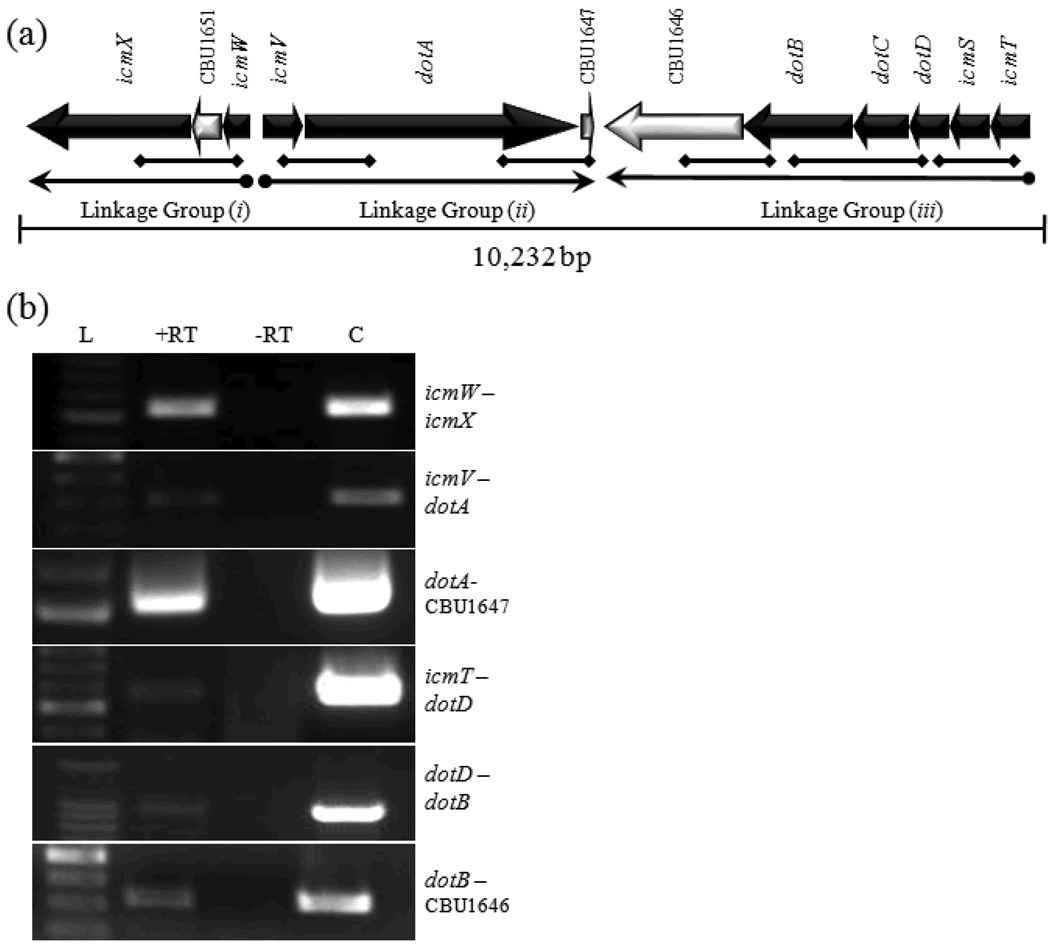
The C. burnetii T4BSS RI gene linkage map suggests three groups of transcriptionally linked genes exist (see Fig. 1a). These include: (i) icmX←CBU1651←icmW, (ii) icmV→dotA→CBU1647, and (iii) CBU1646←dotB←dotC←dotD←icmS←icmT. To demonstrate transcriptional linkage between the genes within each group, RT-PCR analysis was performed using oligonucleotide 200 primers (see Table 1) designed to span intergenic sequences and/or adjoining ORFs. The diamond-ended lines in Fig.1a indicate the position of primers and DNA products that would result from RT-PCR amplification. Using total RNA harvested from Vero cells infected with C. burnetii NMII as template, amplification products were observed (Fig. 1b) for each linkage region: (i) icmW – icmX, (ii) icmV – dotA, dotA – CBU1647, and (iii) icmT – dotD, dotD – dotB, dotB – CBU1646 (Fig. 1b). Taken together, the data indicate that the C. burnetii T4BSS RI is expressed as three operons. This does not preclude the possibility that additional transcriptional regulation exists within the operons.
Fig. 1. Coxiella burnetii T4BSS Region I gene linkage.
(a) Physical map of C. burnetii T4BSS RI. Solid arrows represent T4BSS homologs. Open arrows represent non-T4BSS ORFs. Gene designations are indicated above each ORF. Diamond-ended lines indicate the position of primers and DNA products that would result from RT-PCR amplification. (b) Agarose gel image(s) showing RT-PCR products corresponding to co-transcribed genes from Linkage Group i, ii, and iii (correlating to the diamond tipped lines in (a). L, 100 bp DNA ladder (size designated on left). +RT, with reverse transcriptase. −RT, without reverse transcriptase. C, DNA control.
Sequence data from the C. burnetii genome indicate that the T4BSS ORFs within each linkage group have little non-coding intervening sequences (Seshadri, et al., 2003). Only icmW, icmV, icmT, dotD, icmQ, and dotP have more than 90bp of non-coding sequence upstream and none have more than 262bp. The compact nature of the C. burnetii T4BSS contrasts with that of the L. pneumophila system in that the L. pneumophila T4BSS has non-coding sequences upstream of transcriptional units that range from 91–400bp (Gal-Mor, et al., 2002).
De novo synthesis of C. burnetii T4BSS genes
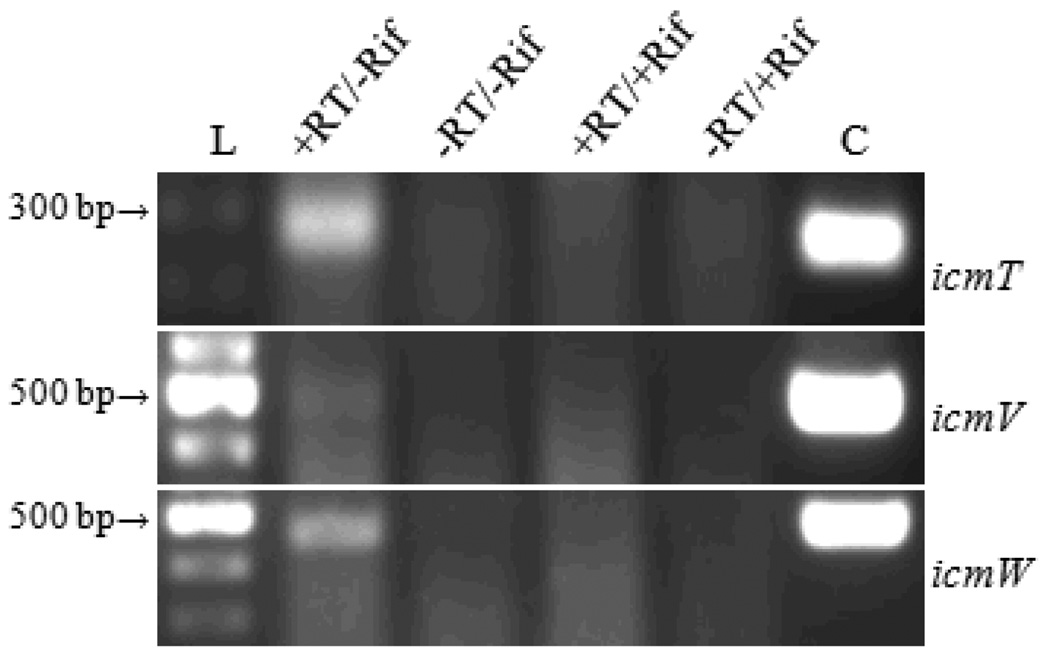
The utility of the mRNA carried within SCVs from one host cell infection to the next is unknown. To determine when de novo synthesis of mRNA for C. burnetii T4BSS genes begins post infection, RT-PCR analysis was used on total RNA samples that were enriched for the C. burnetii RNA fraction (J. K. Morgan & E. I. Shaw, unpublished data) from infected Vero cells. Vero cells were inoculated with C. burnetii NMII (see methods) and RNA samples were collected at 8 hpi from +Rif and −Rif samples. Using RNA from −Rif samples as template, RT-PCR produces amplicons representing full-length mRNA for icmT, icmV, and icmW by 8 hpi (Fig. 2). In contrast, amplification products are not produced from the +Rif RNA samples (Fig. 2). Together, these data indicate that by 8 hpi the transcripts carried into the cell by SCVs had degraded and that de novo transcription was occurring for the three genes assayed.
Fig. 2. RT-PCR detection of C. burnetii T4BSS transcripts, icmT, icmV, and icmW.
RNA template was isolated at 8 hpi from rifampicin-treated (+Rif) and mock-treated (−Rif) cells. L, 100 bp DNA ladder (size designated on left). +RT/−Rif, with reverse transcriptase and mock-treated. −RT/−Rif, without reverse transcriptase and mock-treated. +RT/+Rif, with reverse transcriptase and rifampin-treated. −RT/+Rif, without reverse transcriptase and rifampin-treated. C, DNA control.
Use of a bacterial RNA synthesis inhibitor to demonstrate de novo RNA synthesis suggests that previous studies where C. burnetii T4BSS transcripts were detected by RT-PCR post infection (Shaw, 2003, Zamboni, et al., 2003, Zusman, et al., 2003, Coleman, et al., 2004, Shaw, 2004) were likely detecting de novo synthesized mRNA. De novo synthesis of C. burnetii T4BSS dotA transcript by 8 hpi was previously implied using RT-qPCR (Coleman, et al., 2004). Predictably, comparisons of +Rif and −Rif samples harvested later during infection demonstrated that de novo synthesis of RNA continued when RT-PCR assays were performed (data not shown). Therefore, it is unlikely that carryover RNA within an SCV makes a substantial contribution in the translation of proteins during the early stages of C. burnetii infection of a host cell.
Relative transcript levels of icmX, icmW, icmV, dotA, dotB, and icmT during infection
To determine the temporal expression of C. burnetii T4BSS RI genes during the first 24 hpi we used RT-qPCR to determine the relative amounts of icmX, icmW, icmV, dotA, dotB, and icmT mRNA at 0, 8, 16, and 24 hpi. Fig. 3 shows a graphical representation of the relative abundance of these transcripts as a function of time. These data points represent the relative fold ratio as calculated by the 2−ΔCT method (Livak & Schmittgen, 2001, Schmittgen & Livak, 2008) in which each gene transcript was normalized to itself at 0 hpi.
RT-qPCR analysis (Fig. 3) of icmX indicates a statistically significant increase in its expression from 0 to 8 hpi followed by decreases in expression from 8 to 16 hpi, and 16 to 24 hpi. Expression of icmW is similar in showing an increase between 0 and 8 hpi followed by a significant decrease from 8 to 16 hpi. This was followed by insignificant change from 16 to 24 hpi. The C. burnetii icmV transcripts increased significantly from 0 to 8, and 8 to 16 hpi followed by a significant decrease from 16 to 24hpi. However, for dotA the initial significant increase in expression from 0 to 8 hpi was followed by relatively constant RNA levels. Early expression changes of both dotB and icmT were subtle (Figure 3). After no significant change in dotB RNA from 0 to 8 hpi, a significant increase from 8 to 16 hpi was followed by a decrease from 16 to 24 hpi at which time the dotB RNA, while present, was less than the 0 hpi. The expression of icmT increased significantly from 0 to 8 hpi, after which little change occurred thru 24 hpi.
Our analysis indicates that for the icmW→icmX and icmT→dotB linkage groups, the relative expression of the 3´ gene declines at 24 hpi while the 5´ gene remains relatively constant. The mechanism for this decrease is not readily apparent in the primary sequence, although partial transcription termination and/or RNase degradation of transcripts could account for the relative decline in the 3´ gene RNA. The icmV→dotA linkage group demonstrates a different profile in that the relative amount of RNA for the 3´ gene (dotA) remains elevated at 24 hpi while RNA for the 5´ gene (icmV) declines. This may be a case where an additional promoter of transcription exists for dotA, and this promoter region is activated or increased at 24 hpi while the promoter upstream of icmV decreases. In each of these cases, the differential expression patterns are observed at 24 hpi. This time point during infection is at the end of the lag phase (Coleman, et al., 2004) and may indicate that the need for the different T4BSS homologs changes as C. burnetii transitions into the log growth phase of the infectious cycle. The genome sequence of C. burnetii Nine Mile Phase I strain indicated that the bacteria possesses three RNA polymerase sigma subunits (rpoD, rpoS and rpoH, (Seshadri, et al., 2003)). The rpoS subunit has been shown to be increased in C. burnetii LCVs relative to SCVs (Seshadri & Samuel, 2001) indicating a role in log growth of the organism. However, a conserved nucleotide sequence binding site has not been established in C. burnetii (Melnicakova, et al., 2003), making searches of the C. bunetii T4BSS RI primary sequence a challenge. In addition, a conserved rpoH binding sequence is poorly defined. Searches of the sequence upstream of each open reading frame did not reveal any apparent or consensus (rpoD) −10 or −35 binding sequences for the sigma subunits. Nevertheless, it may be that the alternate sigma factors act in conjunction with transcription factors and/or partial termination to regulate the expression of the C. burnetii T4BSS during the transition from SCVs to LCVs.
Samples harvested at 0, 8, 16, and 24 hpi were used to analyze the expression of the C. burnetii T4BSS as it relates to early events of infection such as bacterial trafficking and SCV to LCV conversion. While the changes in mRNA are relatively subtle, the fact that it is compared to the mRNA present within SCVs at the time of infection (0 hpi), and that this SCV RNA appears to degrade within the first 8 hpi (see Fig. 2), makes the mRNA concentration increase observed at 8 hpi for the C. burnetii T4BSS genes crucial for ongoing T4BSS production. However, it is likely that T4BSS expression may begin even earlier during the infectious process. Electron microscopy evidence showing SCV to LCV conversion by 8 hpi (Coleman, et al., 2004), prior to replication, supports this assumption.
IcmT expression during infection
To determine the relative expression of a C. burnetii T4BSS RI protein, IcmT expression was analyzed over the course of an infectious cycle. We hypothesized that individual C. burnetii T4BSS proteins might be present in low quantities relative to total protein, making temporal analysis by immunoblot challenging, especially early during infection when bacterial numbers are low. In addition, we have previously used RαIcmT for IFA analysis and observed adequate fluorescent signal and polar localization at 600× magnification (Morgan, et al., 2010). To demonstrate specificity and determine whether RαIcmT could be used for immunoblot analysis, total protein from Vero cells, purified C. burnetii, and recombinant IcmT was probed with RαIcmT (Fig 4b). Our previous study and Fig. 4b indicates that while the antibody is very sensitive when used in IFA analysis of C. burnetii infected cells, it is unable to detect native IcmT (10.15 kD predicted size) in protein lysates from 108 purified C. burnetii. Reactivity of the antibody against a relatively high concentration (200 ng) of the recombinant IcmT protein control (Fig 4b, lane 5, 13.3 kD predicted size), and the lack of reactivity with either Vero (Fig 4b, lane 3) or purified C. burnetii (Fig 4b, lane 2) whole protein suggests that the antibody, (i) is specific for C. burnetii IcmT, (ii) has a higher affinity for fixed antigen presented on an intact C. burnetii cell, and (iii) the IcmT protein is present at levels below the level of detection by immunoblot analysis with this antibody, restricting our ability to use immunoblot analysis for temporal protein studies. As such, Guinea pig antibodies against whole cell C. burnetii NMII and RαIcmT, previously employed for C. burnetii T4BSS analysis (Morgan, et al., 2010), were used in IFA microscopy assays using dual fluorescence and relative signal intensity.
Infected Vero cells were fixed at 0, 8, 16, 24, 48, 96, and 168 hpi. Fig. 4a, shows a representative color micrograph image from a 24 hpi sample using 400× magnification. Although 400× magnification and split fluorescent channels does not provide the resolving power to differentiate the polar expression we have reported at 600× magnification with merged fluorescent channels (Morgan, et al., 2010), it provides a broader view of fixed samples, aiding in comparative fluorescent channel intensity calculations. As would be expected, the Guinea pig antibody against whole C. burnetii produced a strong fluorescent signal. Changes in IcmT levels were measured against this standard. Fluorescence was observed in both channels for each time point sampled (data not shown). Fig. 4c is a graphical representation of this data relative to 0 hpi. Analysis revealed that from 0 to 24 hpi, a significant increase (p < 0.05) in the amount of IcmT relative to whole C. burnetii occurred between each time point measured. From 24 to 168 hpi, a statistically significant change was not detected. Although subtle, these data demonstrate that IcmT expression increases early during infection, and then remains relatively unchanged for the duration of the infectious cycle. Whether these changes during this crucial time in PV development are required for C. burnetii survival is yet to be determined. However, the need for secreted effector proteins to control the development and trafficking of the PV early during the infectious cycle would likely be central to C. burnetii’s survival.
Whether the IcmT detected at 0 hpi is part of a functional T4BSS structure poised to secrete effector proteins upon host cell contact, or whether a functional T4BSS structure is assembled once the bacteria enters the host cell remains to be elucidated. Combined with our RT-qPCR analysis, these data suggest that C. burnetii T4BSS IcmT expression closely follows the increase in icmT transcript early during infection (0 to 24 hpi) and becomes relatively uniform for the duration of the infection (24 to 168 hpi). A comparison of Fig. 3 (icmT) and Fig. 4c indicates that an increase in RNA expression early during infection is followed by a rise in IcmT protein levels from a low at 0 hpi. Although the increase in RNA is modest, the relationship between the RNA and corresponding IcmT protein expression indicate that temporal regulation of IcmT expression exists during the C. burnetii infectious cycle.
In summary, we have shown that the C. burnetii T4BSS RI is expressed as a set of three operons and that de novo transcription and translation of C. burnetii T4BSS genes is present as early as 8 hpi. In addition, we have shown that an increase in transcription is accompanied by an increase in at least one protein, IcmT, in the first 24 hpi. Protein levels for the C. burnetii T4BSS RI IcmT homolog appear relatively constant at later stages of an infection (48 to 168 hpi). These data provide a substantial increase in our understanding of the temporal regulation of the C. burnetii T4BSS early during infection and indicate that this bacterial virulence mechanism is maintained throughout infection.
Acknowledgments
We would like to thank William Picking, Wendy Picking, Saugata Mahapatra, and Brandon Luedtke for critical review of this manuscript. This research was supported by National Institutes of Health grant A1072710 (E.I.S.).
References
- Akporiaye ET, Rowatt JD, Aragon AA, Baca OG. Lysosomal response of a murine macrophage-like cell line persistently infected with Coxiella burnetii. Infect Immun. 1983;40:1155–1162. doi: 10.1128/iai.40.3.1155-1162.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreas Untergasser HN, Rao Xiangyu, Bisseling Ton, Geurts René, Leunissen Jack AM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Research 2007. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. doi:10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beare PA, Unsworth N, Andoh M, et al. Comparative genomics reveal extensive transposon-mediated genomic plasticity and diversity among potential effector proteins within the genus Coxiella. Infect Immun. 2009;77:642–656. doi: 10.1128/IAI.01141-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun. 2002;70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan RE, Samuel JE. Evaluation of Coxiella burnetii antibiotic susceptibilities by real-time PCR assay. J Clin Microbiol. 2003;41:1869–1874. doi: 10.1128/JCM.41.5.1869-1874.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggemann H, Cazalet C, Buchrieser C. Adaptation of Legionella pneumophila to the host environment: role of protein secretion, effectors and eukaryotic-like proteins. Curr Opin Microbiol. 2006;9:86–94. doi: 10.1016/j.mib.2005.12.009. [DOI] [PubMed] [Google Scholar]
- Cockrell DC, Beare PA, Fischer ER, Howe D, Heinzen RA. A method for purifying obligate intracellular Coxiella burnetii that employs digitonin lysis of host cells. J Microbiol Methods. 2008;72:321–325. doi: 10.1016/j.mimet.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman SA, Fischer ER, Howe D, Mead DJ, Heinzen RA. Temporal analysis of Coxiella burnetii morphological differentiation. J Bacteriol. 2004;186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- Gal-Mor O, Zusman T, Segal G. Analysis of DNA regulatory elements required for expression of the Legionella pneumophila icm and dot virulence genes. J Bacteriol. 2002;184:3823–3833. doi: 10.1128/JB.184.14.3823-3833.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigo E, Capo C, Tung CH, Raoult D, Gorvel JP, Mege JL. Coxiella burnetii survival in THP-1 monocytes involves the impairment of phagosome maturation: IFN-gamma mediates its restoration and bacterial killing. J Immunol. 2002;169:4488–4495. doi: 10.4049/jimmunol.169.8.4488. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol. 2005;7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- Heinzen RA, Hackstadt T, Samuel JE. Developmental biology of Coxiella burnetii. Trends Microbiol. 1999;7:149–154. doi: 10.1016/s0966-842x(99)01475-4. [DOI] [PubMed] [Google Scholar]
- Heinzen RA, Scidmore MA, Rockey DD, Hackstadt T. Differential interaction with endocytic and exocytic pathways distinguish parasitophorous vacuoles of Coxiella burnetii and Chlamydia trachomatis. Infect Immun. 1996;64:796–809. doi: 10.1128/iai.64.3.796-809.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D, Mallavia LP. Coxiella burnetii exhibits morphological change and delays phagolysosomal fusion after internalization by J774A.1 cells. Infect Immun. 2000;68:3815–3821. doi: 10.1128/iai.68.7.3815-3821.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe D, Heinzen RA. Coxiella burnetii inhabits a cholesterol-rich vacuole and influences cellular cholesterol metabolism. Cell Microbiol. 2006;8:496–507. doi: 10.1111/j.1462-5822.2005.00641.x. [DOI] [PubMed] [Google Scholar]
- Howe D, Melnicakova J, Barak I, Heinzen RA. Fusogenicity of the Coxiella burnetii parasitophorous vacuole. Ann N Y Acad Sci. 2003;990:556–562. doi: 10.1111/j.1749-6632.2003.tb07426.x. [DOI] [PubMed] [Google Scholar]
- Howe D, Melnicakova J, Barak I, Heinzen RA. Maturation of the Coxiella burnetii parasitophorous vacuole requires bacterial protein synthesis but not replication. Cell Microbiol. 2003;5:469–480. doi: 10.1046/j.1462-5822.2003.00293.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Marra A, Blander SJ, Horwitz MA, Shuman HA. Identification of a Legionella pneumophila locus required for intracellular multiplication in human macrophages. Proc Natl Acad Sci U S A. 1992;89:9607–9611. doi: 10.1073/pnas.89.20.9607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaul TF. The developmental cycle of Coxiella burnetii. In: Williams JC, Thompson HA, editors. Q Fever: The Biology of Coxiella burnetii. Boca Raton, Fl: CRC Press; 1991. pp. 223–258. [Google Scholar]
- McCaul TF, Williams JC. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J Bacteriol. 1981;147:1063–1076. doi: 10.1128/jb.147.3.1063-1076.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnicakova J, Lukacova M, Howe D, Heinzen RA, Barak I. Identification of Coxiella burnetii RpoS-dependent promoters. Ann N Y Acad Sci. 2003;990:591–595. doi: 10.1111/j.1749-6632.2003.tb07432.x. [DOI] [PubMed] [Google Scholar]
- Morgan JK, Luedtke BE, Shaw EI. Polar localization of the Coxiella burnetii Type IVB Secretion System. FEMS Microbiol Lett. 2010;305:177–183. doi: 10.1111/j.1574-6968.2010.01926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio S, Roy CR. Effector proteins translocated by Legionella pneumophila: strength in numbers. Trends Microbiol. 2007;15:372–380. doi: 10.1016/j.tim.2007.06.006. [DOI] [PubMed] [Google Scholar]
- Pan X, Luhrmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin Repeat Proteins Comprise a Diverse Family of Bacterial Type IV Effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol. 2007;9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- Sauer JD, Shannon JG, Howe D, Hayes SF, Swanson MS, Heinzen RA. Specificity of Legionella pneumophila and Coxiella burnetii vacuoles and versatility of Legionella pneumophila revealed by coinfection. Infect Immun. 2005;73:4494–4504. doi: 10.1128/IAI.73.8.4494-4504.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Segal G, Purcell M, Shuman HA. Host cell killing and bacterial conjugation require overlapping sets of genes within a 22-kb region of the Legionella pneumophila genome. Proc Natl Acad Sci U S A. 1998;95:1669–1674. doi: 10.1073/pnas.95.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Samuel JE. Characterization of a stress-induced alternate sigma factor, RpoS, of Coxiella burnetii and its expression during the development cycle. Infect Immun. 2001;69:4874–4883. doi: 10.1128/IAI.69.8.4874-4883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshadri R, Paulsen IT, Eisen JA, et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc Natl Acad Sci U S A. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton JA, Vogel JP. Type IVB secretion by intracellular pathogens. Traffic. 2002;3:178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- Shaw EI, Thompson HA. Coxiella burnetii. Maryland: Rocky Gap; 2003. The Expression of Select Type IV Secretion Homologs. eds.), pp. [Google Scholar]
- Shaw EI, Thompson HA. RNA Expression of the Coxiella burnetii Type IV Secretion System Region I During the Course of Infection. New Orleans, LA: 2004. [Google Scholar]
- Shin S, Roy CR. Host cell processes that influence the intracellular survival of Legionella pneumophila. Cell Microbiol. 2008;10:1209–1220. doi: 10.1111/j.1462-5822.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- Vogel JP, Andrews HL, Wong SK, Isberg RR. Conjugative transfer by the virulence system of Legionella pneumophila. Science. 1998;279:873–876. doi: 10.1126/science.279.5352.873. [DOI] [PubMed] [Google Scholar]
- Voth DE, Heinzen RA. Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect Immun. 2009;77:205–213. doi: 10.1128/IAI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voth DE, Howe D, Beare PA, Vogel JP, Unsworth N, Samuel JE, Heinzen RA. The Coxiella burnetii ankyrin repeat domain-containing protein family is heterogeneous, with C-terminal truncations that influence Dot/Icm-mediated secretion. J Bacteriol. 2009;191:4232–4242. doi: 10.1128/JB.01656-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamboni DS, Mortara RA, Rabinovitch M. Infection of Vero cells with Coxiella burnetii phase II: relative intracellular bacterial load and distribution estimated by confocal laser scanning microscopy and morphometry. J Microbiol Methods. 2001;43:223–232. doi: 10.1016/s0167-7012(00)00223-2. [DOI] [PubMed] [Google Scholar]
- Zamboni DS, McGrath S, Rabinovitch M, Roy CR. Coxiella burnetii express type IV secretion system proteins that function similarly to components of the Legionella pneumophila Dot/Icm system. Mol Microbiol. 2003;49:965–976. doi: 10.1046/j.1365-2958.2003.03626.x. [DOI] [PubMed] [Google Scholar]
- Zusman T, Yerushalmi G, Segal G. Functional similarities between the icm/dot pathogenesis systems of Coxiella burnetii and Legionella pneumophila. Infect Immun. 2003;71:3714–3723. doi: 10.1128/IAI.71.7.3714-3723.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]