Abstract
Thioesterases (TEs) are classified into EC 3.1.2.1 through EC 3.1.2.27 based on their activities on different substrates, with many remaining unclassified (EC 3.1.2.–). Analysis of primary and tertiary structures of known TEs casts a new light on this enzyme group. We used strong primary sequence conservation based on experimentally proved proteins as the main criterion, followed by verification with tertiary structure superpositions, mechanisms, and catalytic residue positions, to accurately define TE families. At present, TEs fall into 23 families almost completely unrelated to each other by primary structure. It is assumed that all members of the same family have essentially the same tertiary structure; however, TEs in different families can have markedly different folds and mechanisms. Conversely, the latter sometimes have very similar tertiary structures and catalytic mechanisms despite being only slightly or not at all related by primary structure, indicating that they have common distant ancestors and can be grouped into clans. At present, four clans encompass 12 TE families. The new constantly updated ThYme (Thioester-active enzYmes) database contains TE primary and tertiary structures, classified into families and clans that are different from those currently found in the literature or in other databases. We review all types of TEs, including those cleaving CoA, ACP, glutathione, and other protein molecules, and we discuss their structures, functions, and mechanisms.
Keywords: clan, primary structure, protein family, tertiary structure, thioesterases, ThYme
Introduction
The thioesterases (TEs), or thioester hydrolases, comprise a large enzyme group whose members hydrolyze the thioester bond between a carbonyl group and a sulfur atom. They are classified by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) into EC (enzyme commission) 3.1.2.1 to EC 3.1.2.27, as well as EC 3.1.2.– for unclassified TEs.1 Substrates of 15 of these 27 groupings contain coenzyme A (CoA), two contain acyl carrier proteins (ACPs), four have glutathione or its derivatives, one has ubiquitin, and two contain other moieties. In addition, three groupings have been deleted.
The EC classification system is based on enzyme function and substrate identity, and it was first formulated when very few amino acid sequences (primary structures) and three-dimensional (tertiary) structures of enzymes were available. Another way to classify enzymes is by primary structure into families and by tertiary structure into clans or superfamilies. Some databases are built this way: Pfam2 has a collection of protein families and domains, and SCOP3 classifies protein structures into classes, folds, families, and superfamilies. Other databases treat certain enzyme groups more specifically. For instance, MEROPS4 is a major database for peptidases, and CAZy5 covers carbohydrate-active enzymes.
It is common to observe that members of more than one EC grouping are found in one enzyme family based on similar amino acid sequences, implying that they have a common ancestor, mechanism, and tertiary structure. Conversely, members of a single EC grouping may be located in more than one enzyme family, being totally or almost totally unrelated in primary structure and potentially in mechanism and tertiary structure.
A further observation is that members of two different enzyme families may have very similar tertiary structures and mechanisms even though their primary structures are very different. This may imply that they are members of the same clan or superfamily, descended from a more distant common ancestor.
In this work, TE primary and tertiary structures will be analyzed to conclude how TEs are divided (and united) into families and clans. Structures, mechanisms, and catalytic residues are compared between families and clans. We compare our findings with existing databases such as Pfam and SCOP. Results also appear in a new continuously updated database, ThYme (Thioester-active enzYmes, http://www.enzyme.cbirc.iastate.edu) that includes families and clans of enzyme groups that are part of the fatty acid synthesis cycle, TEs among them.
Family identification
Family members must have strong (>15%, but typically >30%) sequence similarity and near-identical tertiary structures, and they must share general mechanisms as well as catalytic residues located in the same position.
In general, TE families were identified in the following way: (1) experimentally confirmed TE sequences were used as queries, (2) a series of successive Basic Local Alignment Search Tool (BLAST)6 searches and comparison among results reduced query sequences to a few representative ones, (3) the catalytic domains of representative query sequences were subjected to BLAST to populate the families, (4) experimentally confirmed TEs were surveyed to search for missing potential TE families, and (5) the uniqueness of the families was confirmed by multiple sequence alignments (MSAs), by tertiary structure superposition and comparison, and by catalytic residue positions. Methods are detailed in Supporting Information.
Clan identification
Two or more families are grouped into a clan if all the sequences within them show some (<15%) sequence similarity, if their structures are strongly similar (narrowing the search to families with the same fold), and if they share similar active sites and general mechanisms. To consider all aspects of clan classification criteria, several methods are used to combine sequence and structural analysis. In addition, catalytic mechanisms of members of each family were gathered from the literature, and positions of catalytic residues were determined to verify that they coincided. A more detailed description of these methods is found in the Supporting Information.
ThYme database
All the sequences in each family are displayed on the ThYme database website (http://www.enzyme.cbirc.iastate.edu). These sequences are taken, using a series of scripts, from the BLAST results of the catalytic domains of the representative query sequences. Matching accessions, taxonomical data, protein names, and EC numbers are taken from UniProt7 and GenBank8 databases. Each TE family is shown on a page where sequences are arranged into archaea, bacteria, and eukaryota, then alphabetically by species. In each row, a single sequence or group of sequences with 100% identical catalytic domains are shown with their protein name and UniProt and/or GenBank accession codes. EC numbers are shown only when they appear in a sequence's UniProt or GenBank annotation. If a crystal structure is known, the Protein Data Bank (PDB, http://www.rcsb.org) accession code also appears. ThYme will be continuously updated: the content of each family will grow as GenBank, UniProt, and PDB do. However, to create a new family, or to merge or delete existing ones, human judgment and manual changes will be necessary.
Thioesterase families
Use of BLAST with TE query sequences followed by construction of MSAs and superposition of tertiary structures yielded 23 families almost completely unrelated by primary structure (Table I).
Tabel I.
Thioesterase Families and Common Names of their Members
| Family | Producing organisms | Genes and/or other names of family members |
|---|---|---|
| TE1 | A, B, Ea | Ach1 |
| TE2 | A, B, E | Acot1–Acot6, BAAT thioesterase |
| TE3 | A, B | tesA, acyl-CoA thioesterase I, protease I, lysophospholipase L1 |
| TE4 | B, E | tesB, acyl-CoA thioesterase II, Acot8 |
| TE5 | B | tesC (ybaW), acyl-CoA thioesterase III |
| TE6 | A, B, E | Acot7 (BACH), Acot11 (BFIT, Them1), Acot12 (CACH), YciA |
| TE7 | B, E | Acot9, Acot10 |
| TE8 | A, B, E | Acot13 (Them2) |
| TE9 | B | YbgC |
| TE10 | B | 4HBT-I |
| TE11 | B | 4HBT-II, EntH (YbdB) |
| TE12 | B,E | DNHA-CoA hydrolase |
| TE13 | A, B | paaI, paaD |
| TE14 | B, E | FatA, FatB |
| TE15 | B | Thioesterase CalE7 |
| TE16 | A, B, E | TE domain of FAS (Thioesterase I), TE domain of PKS or NRP (type I thioesterase (TE I)) |
| TE17 | B | TE domain of PKS |
| TE18 | B,E | Thioesterase II, type II thioesterase (TE II) |
| TE19 | B | luxD |
| TE20 | E | ppt1, ppt2, palmitoyl-protein thioesterase |
| TE21 | A, B, E | apt1, apt2, acyl-protein thioesterase, phospholipase, carboxylesterase |
| TE22 | A, B, E | S-formylglutathione hydrolase, esterase A, esterase D |
| TE23 | A, B, E | Hydroxyglutathione hydrolase, glyoxalase II |
A, archaea; B, bacteria; E, eukaryota. Most prevalent producers bolded.
Enzymes in families TE1–TE13 hydrolyze substrates with various acyl moieties and CoA, those in TE14–TE19 attack bonds between acyl groups and ACP, and those in TE20 and TE21 cleave the bonds between acyl groups and proteins. Members of TE22 and TE23 break bonds between acyl groups and glutathione and its derivatives (Table II). The sulfur-carrying moiety in CoA and ACP is a pantethiene residue, whereas glutathione itself carries the sulfur moiety, and in non-ACP proteins, the sulfur-carrying moiety is built up mainly from a cysteine residue.
Tabel II.
Thioesterase Functions and Substrate Specificities
| Family | General function | EC number | Preferred substrate specificity (if known) |
|---|---|---|---|
| TE1 | Acyl-CoA hydrolase | 3.1.2.1, 2.8.3.– | Acetyl-CoA |
| TE2 | Acyl-CoA hydrolase | 3.1.2.–, 3.1.2.2, 2.3.1.65 | Palmitoyl-CoA, bile-acid-CoA |
| TE3 | Acyl-CoA hydrolase | 3.1.2.–, 3.1.2.20, 3.1.1.2, 3.1.1.5 | Medium- to long-chain acyl-CoA |
| TE4 | Acyl-CoA hydrolase | 3.1.2.–, 3.1.2.2, 3.1.2.27 | Short- to long-chain acyl-CoA, palmitoyl-CoA, choloyl-CoA |
| TE5 | Acyl-CoA hydrolase | 3.1.2.– | Long-chain acyl-CoA, 3,5-tetradecadienoyl-CoA |
| TE6 | Acyl-CoA hydrolase | 3.1.2.–, 3.1.2.1, 3.1.2.2, 3.1.2.18, 3.1.2.19, 3.1.2.20 | Short- to long-chain acyl-CoA, C4–C18 |
| TE7 | Acyl-CoA hydrolase | 3.1.2.–, 3.1.2.1, 3.1.2.2, 3.1.2.20 | Short- to long-chain acyl-CoA |
| TE8 | Acyl-CoA hydrolase | 3.1.2.– | Short- to long-chain acyl-CoA, C6–C18 |
| TE9 | Acyl-CoA hydrolase | 3.1.2.–, 3.1.2.18 | Short- to long-chain acyl-CoA, 4-hydroxybenzoyl-CoA |
| TE10 | Acyl-CoA hydrolase | 3.1.2.23 | 4-Hydroxybenzoyl-CoA |
| TE11 | Acyl-CoA hydrolase | 3.1.2.– | 4-Hydroxybenzoyl-CoA |
| TE12 | Acyl-CoA hydrolase | 3.1.2.– | 1,4-Dihydroxy-2-napthoyl-CoA |
| TE13 | Acyl-CoA hydrolase | 3.1.2.– | Short and medium-chain acyl-CoA, several hydroxyphenylacetyl-CoA substrates |
| TE14 | Acyl-ACP hydrolase | 3.1.2.–, 3.1.2.14 | Short- to long-chain acyl-ACP, C8–C18 |
| TE15 | Acyl-ACP hydrolase | — | — |
| TE16 | Acyl-ACP hydrolase | 3.1.2.14a | Long-chain acyl-ACP, various polyketides and non-ribosomal peptides |
| TE17 | Acyl-ACP hydrolase | 3.1.2.14b | Several polyketides |
| TE18 | Acyl-ACP hydrolase | 3.1.2.–, 3.1.2.14 | Medium-chain acyl-ACP, various polyketides and nonribosomal peptides |
| TE19 | Acyl-ACP hydrolase | 2.3.1.– | Myristoyl-ACP |
| TE20 | Protein-palmitoyl hydrolase | 3.1.2.–, 3.1.2.22 | Palmitoyl-protein |
| TE21 | Protein-acyl hydrolase | 3.1.2.–, 3.1.1.1 | — |
| TE22 | Glutathione hydrolase | 3.1.2.12, 3.1.1.1, 3.1.1.6 | S-Formylglutathione |
| TE23 | Glutathione hydrolase | 3.1.2.6 | d-Lactoylglutathione |
TE domain. FASs, PKSs, and NRPs can have several EC numbers such as 2.3.1.85, 2.3.1.94, 2.3.1.–, 2.7.7.–, and 5.1.1.–.
TE domain of PKSs.
All tertiary structures within each family have almost identical cores and very strong overall resemblance (Table III) shown by RMSDave values of <1.8 Å and Pave values of >75% (see Supporting Information for definitions), with two exceptions. TE4 has a Pave value of 33.3% because it has only two crystal structures, of which one monomer (1C8U) is a double HotDog, whereas another monomer (1TBU) is incomplete with only a single HotDog. Similarly, in TE16 the Pave value is 65.8% because the TE domain of one structure (2VSQ) is smaller than the rest.
Tabel III.
Thioesterase Folds
| Family | Fold | RMSDave (Å) | Pave (%) | PDB files |
|---|---|---|---|---|
| TE1 | NagB | 1.25 | 96.4 | 2G39, 2NVV |
| TE2 | α/β-Hydrolase | 1.00 | 96.6 | 3HLK, 3K2I |
| TE3 | Flavodoxin-like | 0.58 | 96.6 | 1IVN, 1J00, 1JRL, 1U8U, 1V2G, 3HP4 |
| TE4 | HotDog | 0.90 | 33.3 | 1C8U, 1TBU |
| TE5 | HotDog | — | — | 1NJK |
| TE6 | HotDog | 1.39 | 75.9 | 3B7K, 2Q2B, 2V1O, 2QQ2, 1YLI, 3BJK, 3D6L |
| TE7 | HotDog | — | — | |
| TE8 | HotDog | 0.58 | 88.3 | 2H4U, 3F5O, 2F0X, 2CY9 |
| TE9 | HotDog | 1.19 | 88.8 | 2PZH, 1S5U, 3HM0, 1Z54 |
| TE10 | HotDog | 0.67 | 97.1 | 1BVQ, 1LO7, 1LO8, 1LO9 |
| TE11 | HotDog | 0.87 | 93.9 | 1Q4S, 1Q4T, 1Q4U, 1VH9, 2B6E, 1SC0, 3LZ7 |
| TE12 | HotDog | — | — | 2VEU |
| TE13 | HotDog | 0.43 | 94.6 | 2FS2, 1PSU, 2DSL0, 1J1Y, 1WLU, 1WLV, 1WM6, 1WN3 |
| TE14 | HotDog | 1.65 | 87.7 | 2OWN, 2ESS |
| TE15 | HotDog | — | — | 2W3X |
| TE16 | α/β-Hydrolase | 1.51 | 66.9 | 2VZ8,a 2VZ9,a 2PX6, 1XKT, 2ROQ,b 2CB9, 2CBG, 2VSQ, 1JMK |
| TE17 | α/β-Hydrolase | 1.67 | 82.4 | 1MO2, 1KEZ, 1MN6, 2H7X, 2H7Y, 2HFK, 2HFJ, 1MNA, 1MNQ |
| TE18 | α/β-Hydrolase | 0.83 | 97.2 | 3FLA, 3FLB, 2RON,b 2K2Qb |
| TE19 | α/β-Hydrolase | — | — | 1THT |
| TE20 | α/β-Hydrolase | 1.41 | 91.2 | 1EH5, 3GRO, 1EI9, 1EXW, 1PJA |
| TE21 | α/β-Hydrolase | 0.82 | 96.7 | 1FJ2, 1AUO, 1AUR, 3CN7, 3CN9 |
| TE22 | α/β-Hydrolase | 1.69 | 78.9 | 3FCX, 3C6B, 2UZ0, 1PV1, 3I6Y, 3E4D, 3LS2 |
| TE23 | Lactamase | 1.67 | 78.5 | 2QED, 1XM8, 2P18, 2GCU, 2Q42, 1QH3, 1QH5, 2P1E |
2VZ8 and 2VZ9 have TE domains in their FASTA format. Therefore, these were picked up by BLAST, but their PDB files do not include the TE domain, and they were not included in the RMSD calculation.
NMR-resolved structures not included in RMSD calculation.
Of the families whose members hydrolyze acyl-CoAs, all have HotDog9,10 folds (Table III, Figs. 1 and 2) except for TE1, TE2, and TE3. TE1 enzymes have NagB folds, and they have acetyl-CoA hydrolase (EC 3.1.2.1) activity as well as acetate or succinate-CoA transferase (EC 2.8.3.–) activity. They are found mainly in bacteria and fungi, although they are also present in archaea. Enzymes coded by the acetyl-CoA hydrolase ACH1 gene from Saccharomyces cerevisiae are present in TE1.11 Fungal enzymes in this family are involved with acetate levels and CoA transfer in mitochondria.12
Figure 1.
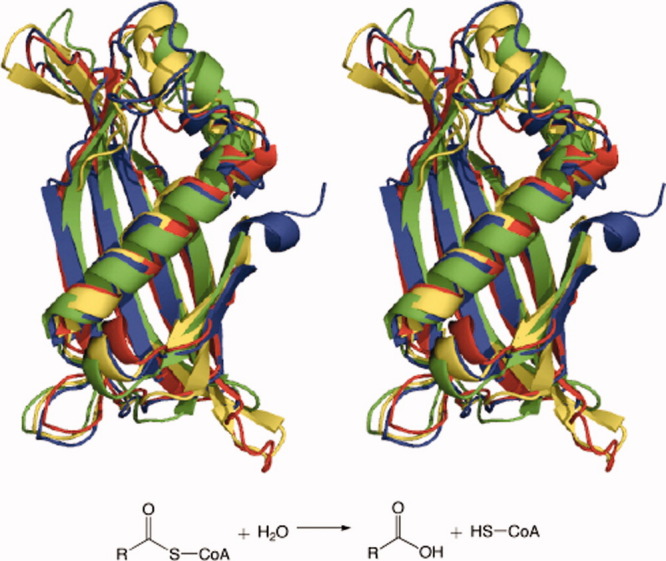
Superimposed tertiary structures of single representatives of each TE family in a clan: TE-A acyl-CoA hydrolases from Escherichia coli (TE5) (green), Helicobacter pylori (TE9) (red), Pseudomonas sp. (TE10) (yellow), and Prochlorococcus marinus (TE12) (blue).
Figure 2.
Superimposed tertiary structures of single representatives of each TE family in a clan: TE-B acyl-CoA hydrolases from Homo sapiens (TE8) (blue), Arthrobacter sp. (TE11) (red), and E. coli (TE13) (yellow).
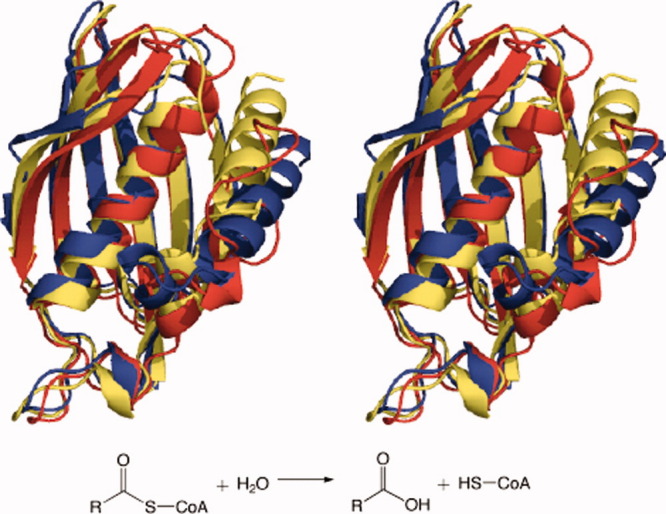
TE2 enzymes have α/β-hydrolase13 folds (Figs. 3 and 4). They are mainly found in eukaryotes (animals), but they are also present in bacteria. They have mostly palmitoyl (EC 3.1.2.2) and bile acid-CoA:amino acid N-acyl transferase (BAT) (EC 2.3.1.65) activities. The acyl-CoA TE (Acot) enzymes ACOT1, ACOT2, ACOT4, and ACOT6 from Homo sapiens are present in this family, as well as the Acot1 through Acot6 enzymes from Mus musculus, Rattus norvegicus, and similar species.14 Also in TE2 are the BAAT TEs that transfer bile acid from bile acid-CoA to amino acids in the liver; these conjugates later solvate fatty acids in the gastrointestinal tract.15
Figure 3.
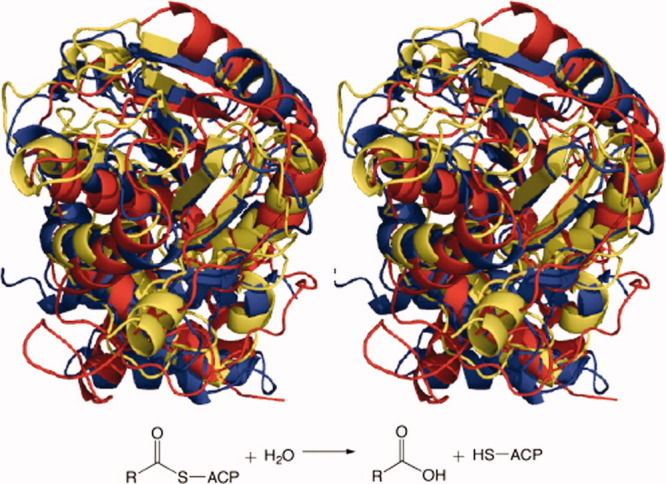
Superimposed tertiary structures of single representatives of each TE family in a clan: TE-C acyl-ACP hydrolases from Homo sapiens (TE16) (blue), Saccharopolyspora erythraea (TE17) (red), and Amycolatopsis mediterranei (TE18) (yellow).
Figure 4.
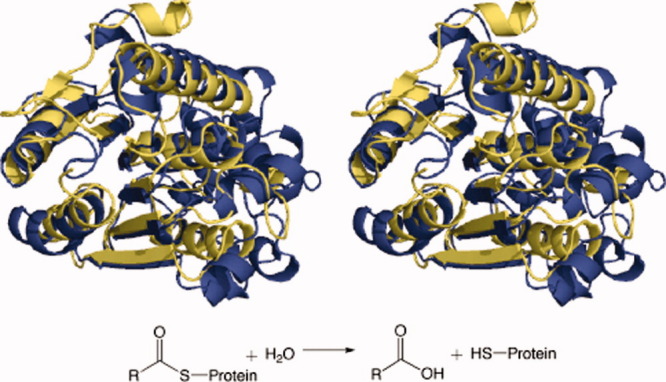
Superimposed tertiary structures of single representatives of each TE family in a clan: TE-D protein-acyl hydrolases from Bos taurus (TE20) (blue) and Homo sapiens (TE21) (yellow).
Enzymes in TE3 are part of the SGNH hydrolase superfamily with a flavodoxin-like fold. They are mainly found in bacteria and have acyl-CoA hydrolase (EC 3.1.2.20), arylesterase (EC 3.1.1.2), and lysophospholipase (EC 3.1.1.5) activities. Some TE3 enzymes come from the tesA gene, and they are located in the periplasm and are involved in fatty acid synthesis.16 TE3 enzymes are also called acyl-CoA thioesterase I, protease I, and lysophospholipase L1, and the genes that code for them, tesA, apeA, and pldC, respectively, are nearly identical.17
The rest of the acyl-CoA hydrolase families have HotDog folds. TE4 enzymes, present in bacteria and eukaryotes, are acyl-CoA hydrolases as well as palmitoyl-CoA (EC 3.1.2.2) and choloyl-CoA (EC 3.1.2.27) hydrolases. The Acot8 gene encodes for peroxisomal TEs,18 which are found in TE4. Also in this family are acyl-CoA thioesterase II enzymes, encoded by the tesB gene, that can hydrolyze a broad range of medium- to long-chain acyl-CoA thioesters, but whose physiological function is not known.19
TE5 acyl-CoA enzymes, also known as thioesterase IIIs, are present in bacteria. They are encoded by the tesC (or ybaW) gene and are long-chain acyl-CoA TEs preferring 3,5-tetradecadienoyl-CoA as a substrate.20
TE6 members, present in eukaryotes, bacteria, and archaea, hav e acyl-CoA hydrolase activities with various specificities. Acot enzymes 7, 11, and 12, present in eukaryotes, are found in TE6. Acot7 enzymes (also known as BACH: brain acyl-CoA hydrolases) are expressed mainly in brain tissue and preferentially attack C8–C18 acyl-CoA chains.21 Acot11 (also known as BFIT: brown fat inducible TE, or Them1: TE superfamily member 1) enzymes are specific toward medium- and long-chain acyl-CoA molecules, and they may be involved with obesity in humans.22 Acot12 (also known as CACH: cytoplasmic acyl-CoA hydrolase) enzymes in humans hydrolyze acetyl-CoA.23 Many bacterial TE6 sequences are YciA TEs that hydrolyze a wide range of acyl-CoA thioesters and may help to form membranes.24 They preferentially attack butyryl, hexanoyl, lauroyl, and palmitoyl-CoA substrates.25
TE7 enzymes are acyl-CoA TEs found in eukaryota and bacteria. In this family are the Acot9 and Acot10 enzymes (previously known as MT-ACT48), which are expressed in the mitochondria and have short- to long-chain acyl-CoA TE activity, showing preference for C14 chains.26
Most TE8 members, mainly present in eukaryota but also in bacteria, are acyl-CoA thioesterase 13 (Acot13) enzymes, also known as TE superfamily member 2 (Them2). Enzymes in this family hydrolyze short-to-long acyl-CoA (C4–C18) chains, preferring the latter.27
TE9 members are found only in bacteria, and they have acyl-CoA hydrolase activity, mostly unclassified (3.1.2.–), but ADP-dependent short-chain acyl-CoA hydrolases (EC 3.1.2.18), and 4-hydroxybenzoyl-CoA hydrolases (EC 3.1.2.23) are also found. The YbgC TEs are found in this family; some hydrolyze primarily short-chain acyl-CoA thioesters,28 whereas others prefer long-chain acyl-CoA thioesters.29 Also, the TE domain of methylketone synthase, MKS2, recently discovered in tomato, is found in TE9.30
The enzymes in TE10 and TE11 are found only in bacteria, and most have 4-hydroxybenzoyl-CoA hydrolase (EC 3.1.2.23) activity. They, along with other enzymes, convert 4-chlorobenzoate to 4-hydroxybenzoate in soil-dwelling bacteria.31 Also in TE11 are the EntH (YbdB) TEs, involved with enterobactin (an iron chelator) biosynthesis in Escherichia coli.32 This is a unique example of a HotDog-fold enzyme involved in nonribosomal peptide biosynthesis.
Most TE12 enzymes are 1,4-dihydroxy-2-napthoyl (DNHA)-CoA hydrolases, involved in vitamin K1 biosynthesis,33 and they are found mostly in bacteria. TE13 enzymes occur in archaea and bacteria. Most are either PaaI or PaaD enzymes in the phenylacetic acid degradation pathway, and they are part of the paa gene cluster.34
TE14–TE19 enzymes hydrolyze acyl-ACP thioesters, with those in TE14 and TE15 having HotDog folds, whereas the rest have α/β-hydrolase folds. TE14 enzymes are found in bacteria and plants; they have acyl-ACP hydrolase (EC 3.1.2.14) activity. Many plant enzymes in this family have been experimentally characterized: they contain FatA and FatB enzymes and can hydrolyze C8–C18 acyl-ACP thioesters.35 All TE14 bacterial sequences come from genomic or structural genomic studies.
TE15 is a small family whose enzymes are present mainly in bacteria. Among them is the TE CalE7 involved with enediyne biosynthesis. After substrate-ACP hydrolysis, these enzymes decarboxylate the product before release.36 Enzymes in this family are the few TEs with HotDog domains involved with polyketide biosynthesis.
TE16 enzymes occur in both eukaryotes and bacteria, and they have oleoyl-ACP hydrolase (EC 3.1.2.14) activity. They include the TE domains of fatty acid synthases (FASs), also known as Thioesterase I, that terminate fatty acid synthesis,37 and the TE domain of polyketide synthases (PKSs) and nonribosomal peptide synthases (NRPs), also known as Type I thioesterases (TE I), that terminate polyketide biosynthesis,38 or nonribosomal peptide biosynthesis.39 In the case of NRPs, instead of an ACP as the carrier molecule, a polypeptide carrier protein (PCP) is used.
TE17 enzymes are only found in bacteria, mainly in Streptomyces. They are the TE domains of various PKSs. FASs, PKSs, and NRPs are large multimodular enzymes with many domains having different functions. Only the TE domains were used to identify these family members.
Enzymes in TE18 are present in eukaryotes and bacteria and mainly have oleoyl-ACP hydrolase (EC 3.1.2.14) activity. Some enzymes in this family are S-acyl fatty acid synthetases/thioester hydrolases (Thioesterase II).40 They work with FASs to produce medium-chain (C8–C12) fatty acids in milk.41 The Type II thioesterases (TE IIs) are found in TE18; these enzymes play an important role in polyketide and nonribosomal peptide biosynthesis by removing aberrant acyl chains from multimodular polyketide synthases and nonribosomal peptide synthases.42,43 TE18 enzymes are independent TEs, not integrated to the multimodular FASs, PKSs, or NRPs.
TE19 enzymes are classified as acyltransferases (EC 2.3.1.–), but they hydrolyze acyl-ACP molecules, mainly myristoyl-ACP.44 These enzymes divert fatty acids to the luminescent system in certain bacteria.
TE20 members, found only in eukaryotes, are palmitoyl-protein TEs (EC 3.1.2.22) encoded by PPT genes. They hydrolyze the thioester bond between a palmitoyl group and a cysteine residue in proteins.45 Mutations in PPT enzymes have been linked to neuronal ceroid lipofuscinosis, a genetic neurodegenerative disorder.46
TE21 enzymes were originally identified as lysophospholipases,47 but they are also acyl-protein APT1 TEs.48 They hydrolyze thioester bonds between acyl chains and cysteine residues on proteins. Many proteins in this family also have carboxyesterase (EC 3.1.1.1) activity.
Among TE22 enzymes are S-formylglutathione hydrolases (EC 3.1.2.12) catalyzing formaldehyde detoxification; they hydrolyze S-formylglutathione into formate and glutathione.49 Also in TE22 are enzymes with acetyl esterase (EC 3.1.1.6) and carboxyesterase (EC 3.1.1.1) activity.
TE23 members are hydroxyglutathione hydrolases (EC 3.1.2.6), also known as glyoxalase II enzymes, that hydrolyze S-d-lactoyl-glutathione to glutathione and lactic acid in methylglyoxal detoxification.50 TE23 enzymes occur in archaea, bacteria, and eukaryotes and have a metallo-β-lactamase fold.51
Correspondence to EC groupings
These TE families bear rather limited resemblance to EC numbers representing TEs. For instance, acetyl-CoA hydrolases (3.1.2.1) occur in TE1, TE6, and TE7; palmitoyl-CoA hydrolases (EC 3.1.2.2) are found in TE2, TE4, TE6, and TE7; oleoyl-ACP hydrolases (EC 3.1.2.14) occur in TE14 and TE16–TE18, and acyl-CoA hydrolases (EC 3.1.2.20) are found in TE3, TE6, and TE7. Conversely, of the 24 EC numbers remaining after three deletions, only 11 of them (EC 3.1.2.1, 3.1.2.2, 3.1.2.6, 3.1.2.12, 3.1.2.14, 3.2.1.18, 3.1.2.19, 3.1.2.20, 3.1.2.22, 3.1.2.23, and 3.1.2.27, along with unclassified TEs (EC 3.1.2.–)) occur in significant numbers among the 23 TE families. Of course, further EC numbers characteristic of TEs will likely appear as more TEs are sequenced and characterized.
Other thioesterases
Ubiquitin carboxyl-terminal hydrolases (EC 3.1.2.15) cleave a wide variety of products from the C-terminal glycine residue of ubiquitin. They were first identified as thiolesterases because they cleave dithiothreitol from ubiquitin, and they were thought to also hydrolyze ubiquitin-glutathione and other ubiquitin thiolesters.52 It was later shown that they hydrolyze amides and other groups from ubiquitin.53 These enzymes belong to a larger class of peptidases called deubiquitinating enzymes that hydrolyze lysine-glycine amide bonds in ubiquitinated proteins.54 Several families of these enzymes can be found in MEROPS, the peptidase database. We identified 11 ubiquitin thiolesterase families by the methods described above, but we have not included them here or in the ThYme database, as peptidase activity is their main function, and they can be found in MEROPS.
Certain acyl transferases (EC 2.3.1.–), for example, 2.3.1.9, 2.3.1.16, 2.3.1.38, and 2.3.1.39 among others, can hydrolyze acyl-CoA or acyl-ACP substrates and later join the liberated acyl group to another acyl-CoA or acyl-ACP molecule. Although they hydrolyze thioesters, this is not their main function, and therefore, we also decided not to include these enzymes here.
Thioesterase clans
TE families 4–6 and 8–15, all with members having HotDog crystal structures, were subjected to the methods described above and two clans were found: TE-A comprising families TE5, TE9, TE10, and TE12; and TE-B with TE8, TE11, and TE13.
PSI–BLAST6 analysis suggested that TE5, TE9, TE10, and TE12 should be grouped into one clan and TE8, TE11, and TE13 into another, because slight sequence similarities among these families were found. Secondary structure element analysis of the structures pointed to TE5, TE6, TE10, TE12, and TE15 (having five β-strands) being placed in one clan and TE8, TE11, and TE13 (having six β-strands) being placed in another (Table IV); visual inspection suggested the same two groupings, with the first also including TE9. All crystal structures in candidate families of both possible clans were tested with superpositions and RMSD analysis (Figs. 1 and 2, Table V). These different tests led to the two clans being defined. Members of TE-A are all acyl-CoA hydrolases active on many substrates including short, long, branched, and aromatic acyl chains. Catalytic residues (see below) in TE6 are placed differently than those of other TE-A families, and therefore, TE6 was not included in this clan. The different substrate specificities, catalytic residues, and mechanism (see below) of TE15 members suggested that it also be excluded from TE-A. TE-B enzymes are also acyl-CoA hydrolases, except for the YbdB TEs in TE11 involved with enterobactin biosynthesis. TE4, TE7 (which has no known tertiary structure), and TE14 enzymes are sufficiently different from members of TE-A and TE-B that they were not considered for placement in either clan; the first 2 are acyl-CoA hydrolases, whereas the third is an acyl-ACP hydrolase.
Tabel IV.
Thioesterase Core Secondary Structure Elements
| Clan | Family | Secondary structural element |
|---|---|---|
| HotDog | ||
| TE-A | TE5 | β-α-β-β-β-βa |
| TE-A | TE9 | α-β-β-β-β |
| TE-A | TE10 | β-α-β-β-β-β |
| TE-A | TE12 | β-α-β-β-β-β |
| TE-B | TE8 | β-β-α-β-β-β-β |
| TE-B | TE11 | β-β-α-β-β-β-β |
| TE-B | TE13 | β-β-α-β-β-β-β |
| — | TE4 | α-β-β-β-β-β-β-α-β-β-β-β |
| — | TE6 | β-α-β-β-β-β |
| — | TE14 | β-α-β-β-β-β-β-α-β-β-β-β |
| — | TE15 | β-α-β-β-β-β |
| α/β-Hydrolase | ||
| TE-C | TE16 | β-α-β-α-β-α-β-β-α-β-α |
| TE-C | TE17 | β-α-β-α-β-α-β-β-α-β-α |
| TE-C | TE18 | β-β-α-β-α-β-α-β-β-α-β-α |
| TE-D | TE20 | β-α-β-α-α-β-α-β-α-β-α-β-α |
| TE-D | TE21 | β-α-β-β-α-β-α-β-β-α-β-α |
| — | TE2 | β-β-β-α-β-α-β-α-β-β-α-β-α |
| — | TE19 | β-β-β-α-β-α-β-α-β-α-β-α-β-α-α |
| — | TE22 | β-β-β-β-α-β-α-β-α-β-α-β-α-β-α |
α, α-helix; β, β-strand.
Tabel V.
RMSD Analysis of TE Clan Members
| Clan | RMSDmin (Å) | RMSDave (Å) | RMSDmax (Å) | Pmin (%) | Pave (%) | Pmax (%) | Cutoff (Å) |
|---|---|---|---|---|---|---|---|
| TE-A | 1.14 | 1.33 | 1.53 | 77.5 | 87.1 | 90.9 | 3.81 |
| TE-B | 0.11 | 0.97 | 2.02 | 72.3 | 86.8 | 100.0 | 3.80 |
| TE-C | 1.81 | 1.94 | 2.13 | 52.6 | 58.3 | 75.2 | 3.82 |
| TE-D | 0.44 | 1.45 | 2.00 | 67.0 | 80.9 | 100.0 | 3.79 |
TE families 2 and 16 through 22, whose members all have α/β-hydrolase crystal structures, belong to two clans: TE-C comprising TE16, TE17, and TE18, and TE-D with TE20 and TE21.
Both sequence analysis and secondary structure element arrangement suggested only one clan of TE16, TE17, and TE18 (Table IV). Visual inspection suggested the two clans described above, and they were confirmed by superpositions, RMSD analysis, and the position of catalytic residues (Figs. 3 and 4, Table V). Families in TE-C contain acyl-ACP hydrolases present in multidomain FASs, PKSs, and NRPs, as well as independent acyl-ACP TEs involved in those pathways. TE-D enzymes hydrolyze palmitoyl and other acyl groups from protein surfaces. TE2, an acyl-CoA hydrolase, TE19, a myristoyl-ACP hydrolase, and TE22, active on glutathione-activated molecules, are not part of either clan.
TE tertiary structures, catalytic residues, and mechanisms
Catalytic mechanisms and residues of each TE family were gathered from crystal structure articles. The PDB files, proposed catalytic residues, and producing organisms of the relevant TEs are listed in Table VI.
Tabel VI.
TE Family and Clan Mechanisms
| Clan | Family | Catalytic residuesa | PDB file | Producing organism |
|---|---|---|---|---|
| — | TE1 | — | — | — |
| — | TE2 | Ser294, His422, Asp388 | 3HLK | Homo sapiens |
| Ser232,b His360,b Asp326b | 3K2I | H. sapiens | ||
| — | TE3 | Ser10, His157, Asp154 | 1IVN, 1JRL, 1J00, 1U8U, 1V2G | Escherichia coli |
| — | TE4 | Asp204, Thr228, Gln278 | 1C8U | E. coli |
| TE-A | TE5 | Asp13c | 1NJK | E. coli |
| — | TE6 | Asn24, Asp213 | 2Q2B, 2V1O | Mus musculus |
| Asp44 | 1YLI, 3BJK | Haemophilus influenzae | ||
| Asp34 | 3D6L | Campylobacter jejuni | ||
| Asp36b | 3B7K | H. sapiens | ||
| Asp245b | 2QQ2 | H. sapiens | ||
| — | TE7 | — | — | — |
| TE-B | TE8 | Asn50, Asp65, Ser83, Gly57 | 2F0X, 3F5O, 2H4Ub | H. sapiens |
| Asn50,b Asp65,b Ser83,b Gly57b | 2CY9 | M. musculus | ||
| TE-A | TE9 | Tyr7, Asp11, His18 | 2PZH | Helicobacter pylori |
| Tyr14,b Asp18,b His25b | 1S5U | E. coli | ||
| Tyr23,b Asp28,b His35b | 3HM0 | Bartonella henselae | ||
| Tyr11,b Asp15,b His22b | 1Z54 | Thermus thermophilus | ||
| TE-A | TE10 | Asp17 | 1BVQ, 1LO7, 1LO8, 1LO9 | Pseudomonas sp. |
| TE-B | TE11 | Gly65, Glu73 | 1Q4S, 1Q4T, 1Q4U | Arthrobacter sp. |
| Gly55,b Glu63b | 1VH9 | E. coli | ||
| Gly55,b Glu63b | 2B6E, 1SC0, 3LZ7 | H. influenzae | ||
| TE-A | TE12 | Asp19c | 2HX5 | Prochlorococcus marinus |
| TE-B | TE13 | Gly40,b Asp48 | 1WLU, 1J1Y, 1WM6, 1WLV, 1WN3, 2DSLb | T. thermophilius |
| Gly53, Asp61 | 2FS2, 1PSUb | E. coli | ||
| — | TE14 | Cys264, His229, Asn227 | Arabidopsis thaliana | |
| — | TE15 | Asn19, Tyr29, Arg37 | 2W3X | Micromonospora echinospora |
| TE-C | TE16 | Ser2308, His2481, Asp2338 | 1XKT, 2PX6 | H. sapiens |
| Ser80, His207, Asp107 | 1JMK | Bacillus subtilis | ||
| Ser84, His201, Asp111 | 2CB9, 2CBG | B. subtilis | ||
| TE-C | TE17 | Ser142, His259, Asp169 | 1KEZ, 1MO2 | Saccharopolyspora erythraea |
| Ser148, His268, Asp176 | 1MN6, 1MNA, 1MNQ | S. erythraea | ||
| Ser148, His268, Asp176 | 2HFK, 2HFJ, 2H7X, 2H7Y | Streptomyces venezuelae | ||
| TE-C | TE18 | Ser94, His228, Asp200 | 3FLA, 3FLB | Amycolatopsis mediterranei |
| — | TE19 | Ser114, His241, Asp211 | 1THT | Vibrio harveyi |
| TE-D | TE20 | Ser115, His289, Asp233 | 1EI9, 1EH5, 1EXW | Bos taurus |
| Ser115, His289, Asp233 | 3GRO | H. sapiens | ||
| Ser111, His283, Asp228 | 1PJA | H. sapiens | ||
| TE-D | TE21 | Ser114, His199, Asp168 | 1AUO, 1AUR | Pseudomonas fluorescens |
| Ser114, His203, Asp169 | 1FJ2 | H. sapiens | ||
| Ser113,b His197,b Asp166b | 3CN7, 3CN9 | Pseudomonas aeruginosa | ||
| — | TE22 | Ser120, His231, Asp202 | 2UZ0 | Streptococcus pneumoniae |
| Ser161, His276, Asp241 | 1PV1, 2C6B | S. cerevisae | ||
| Ser149, His260, Asp226 | 3FCX | H. sapiens | ||
| Ser147, His256, Asp223 | 3E4D | Agrobacterium tumefaciens | ||
| Ser147, His258, Asp255 | 3LS2 | Pseudoalteromonas haloplanktis | ||
| Ser148,b His257,b Asp224b | 3I6Y | Oleispira antartica | ||
| — | TE23 | Various His and Asp residues, along with Zn, Fe, or Mn ions | 1QH3 | H. sapiens |
As proposed by the authors in the literature cited, except as noted.
Predicted by the authors of this article based upon the position of catalytic residues in superimposed structures within families.
Predicted by the authors of this article based upon the position of catalytic residues in superimposed structures within clan TE-A.
HotDog-fold enzymes lack defined nonsolvated binding pockets and conserved catalytic residues,24 thus a variety of catalytic residues and mechanisms exist.
In TE-A, only TE9 and TE10 can be further analyzed, as TE5 and TE12 at present have only one crystal structure each with no corresponding refereed article. In TE9, the YbgC structure 2PZH is a tetramer of two dimers. After comparing this structure to 1LO9 in TE10 and other YbgC crystals, the authors proposed that His18, Tyr7, and Asp11 play important roles in catalysis.29
TE10 4-hydroxybenzoyl-CoA TEs have homotetrameric quaternary structures. It was suggested from structures 1LO7, 1LO8, and 1LO9 that hydrogen bonds and the positive end of a helix dipole moment make the thioester carbonyl group more susceptible to a nucleophilic attack by Asp17 through an acyl-enzyme intermediate.55
TE-B families include TE8, TE11, and TE13. Members of TE8 are tetramers composed of two HotDog dimers. Based on a crystal structure of a human Them2 enzyme (3F5O), it was proposed that Gly57 and Asn50 bind and polarize the thioester carbonyl group, whereas Asp65 and Ser85 orient and activate the water nucleophile.56
In TE11, Arthrobacter sp. strain SU 4-hydroxybenzoyl-CoA TE crystal structures reveal a tetrameric enzyme with a dimer of dimers. Structures 1Q4S, 1Q4T, and 1Q4U led to the proposal that Gly65 polarizes the carbonyl group for a nucleophilic attack carried out by Glu73.57
Both TE10 and TE11 are 4-hydroxybenzoyl-CoA TEs of similar substrate specificities and metabolic functions; however, their tertiary and quaternary structures are different and they use different active-site regions and residues for catalysis. This supports placing these two families in two different clans.
TE13 PaaI TE from Thermusthermophilus HB8 yielded homotetrameric quaternary structures 1WLU, 1J1Y, 1WM6, 1WLV, and 1WN3. From those structures, a study proposed that these enzymes use an induced-fit mechanism to hydrolyze the substrate via an Asp48-activated water nucleophile.58 Comparison of the structure of another PaaI, from E. coli (2FS2) with the Arthrobacter TE11 structures, as well as site-directed mutagenesis, pointed to a mechanism similar to that in TE11: Gly53 prepares the thioester for a nucleophilic attack from Asp61.59 4-Hydrozybenzoyl-CoA enzymes from TE11 and the PaaI enzymes from TE13 catalyze two different reactions in different organisms, and their primary sequences are not related; yet their tertiary structures, catalytic residues, and mechanisms are similar, supporting the conclusion that both TE11 and TE13 are part of TE-B.
Of TE families with HotDog structures not placed in clans, TE4 catalytic residues and mechanisms have been identified based on structure 1C8U and site-directed mutagenesis: an Asp204–Gln278–Thr228 triad orients a water molecule for nucleophilic attack on the substrate.60 The double HotDog structure of 1C8U and its catalytic residues and mechanism differ from other HotDog enzymes, supporting the exclusion of TE4 from TE-A and TE-B.
In TE6, Acot7 structure 2Q2B is a trimer of HotDog dimers; both domains are required for activity and Asn24 from the N-domain and Asp213 from the C-domain have been identified as catalytic residues through site-directed mutagenesis.61 Also in TE6, YciA structures 1YLI and 3D6L have a trimer of dimers with two binding sites across the dimer interface. A D44A mutation (3BJK) of 1YLI eliminated activity.62
The two known tertiary structures in TE14 come from structural genomic studies and do not have supporting literature. However, based on a bioinformatics-guided site-directed mutagenesis study on a FatB enzyme from Arabidopsis, a study proposed that Cys264, His229, and Asn227 make up a papain-like catalytic triad.63
Structure 2W3X of TE15 CalE7 has no acidic residues in the catalytic region. Based on site-directed mutagenesis, a mechanism different from other HotDogs was proposed: Asn19 and Arg37 anchor the substrate with hydrogen bonds, whereas a water molecule or hydroxide anion acts as a nucleophile, attacking the substrate carbonyl group. Arg37 also acts as an oxyanion hole to stabilize the intermediate, and Tyr29 facilitates decarboxylation following hydrolysis.36
Unlike HotDogs, α/β hydrolases have very conserved catalytic residues: a nucleophile–histidine–acid triad.13 The nucleophile can be serine, cysteine, or aspartate; the histidine is always conserved, and the third residue is always acidic. Fold architecture, substrate specificities, and binding sites vary within this fold.64 Generally, the acid stabilizes a histidine residue that acts as a base, accepting a proton from the nucleophile that forms an intermediate with the substrate, which is then attacked by water. In some PKSs and NRPs that make cyclic products, a hydroxyl group from the substrate chain is used for lactonization instead of a water molecule.
TE-C encompasses TE16, TE17, and TE18. Members of all three families have the same Ser–His–Asp triad. The position of the acid residue differs from what is usually found in α/β hydrolases, as was seen in human FAS TE domain 1XKT.65 In TE17, a substrate channel was found in the TE domain of 6-deoxyerythronolide B synthase (DEBS-TE) from Saccharopolyspora erythraea (1KEZ), unique among TE α/β hydrolases.66 The substrate channel depends on pH and supports the prediction that all macrocycle-forming TEs from PKSs have a similar catalytic mechanism. Many macrocycle-forming TE domains from PKSs were found in TE17, again supporting this prediction.
Members of the two families, TE20 and TE21, in TE-D also have Ser–His–Asp catalytic triads. TE21 structure 1AUO confirmed that some bacterial carboxylesterases with broad substrate specificity have an α/β hydrolase fold.67 These enzyme structures are very similar to the mainly eukaryotic acyl-protein hydrolases, even though they have very different specificities.
TE2, TE19, and TE22 are other α/β hydrolases that are not part of any clan, but they still maintain the characteristic catalytic triad.
TEs are found in NagB (TE1), SGNH (TE3), and lactamase (TE23) folds. TE1 has two structures with no refereed literature, so their mechanism and catalytic residues remain unknown.
From TE3 structures 1IVN, 1JRL, and 1J00, the existence of a Ser–His–Asp catalytic triad similar to those in the TE α/β hydrolases was proposed.68 The later crystal structures 1U8U and 1V2G suggested that a conformational change, described as a switch loop movement, occurs during catalysis.69
Because TE23 hydroxyglutathione hydrolases (glyoxalase IIs) have metallo-β-lactamase folds, their mechanisms are very different from the rest of TEs that do not have catalytic metal ions. Two crystal structures, 1QH3 and 1QH5, have seven His and Asp residues and a water molecule interacting with two zinc ions. Based on this, a study proposed that a hydroxide ion bonded with both ions attacks the carbonyl carbon atom of the glutathione thioester substrate, forming a tetrahedral intermediate, followed by breakage of the C–S bond.70
Predicted catalytic residues
Some TE tertiary structures, including all those in TE1, TE5, TE12, and TE14, are not supplemented by refereed literature. Furthermore, TE7 does not have a known tertiary structure. Therefore, the catalytic residues and mechanisms for TEs in these families are not securely known.
Because the positions of catalytic residues must be conserved within families and clans, unknown identities of catalytic residues in TEs can be predicted by viewing the positions of the catalytic residues in all superimposed structures in a family or clan.
In TE2, we predict that in the human ACOT4 structure 3K2I the catalytic triad is Ser232–His360–Asp326, based on the human ACOT2 structure 3HLK. In TE6, based on structures from mouse Acot7 (2Q2B), Haemophilus influenzae YciA (3BJK), and Campylobacter jejuni (3D6L), Asp245 in human ACOT7, and Asp36 in human ACOT12 appear to be catalytic residues. In TE8, study of the structures of human Them2 (3F5O and 2F0X) leads to the prediction that human TE structure 2H4U and mouse TE structure 2CY9 have the same catalytic residues: Asn50, Asp65, Ser83, and Gly57. In TE9, based on Helicobacter pylori YbgC enzyme 2PZH, we predict that, in E. coli 1S5U, the catalytic residues are Tyr14, Asp18, and His25, in Bartonella henselae 3HM0, they are Tyr23, Asp28, and His35, and, in T. thermophilus, they are 1Z54 Tyr11, Asp15, and His22. All three residues in the three structures superimposed very closely and in the same position except for Tyr35 in 3HM0, where the ring is facing in another direction and the residue is one amino acid position displaced. This might lead to a difference in enzyme substrate specificity. In TE11, based on TE structures 1Q4S, 1Q4T, and 1Q4U from Arthrobacter sp., we predict that the catalytic residues are Gly55 and Glu63 in both structure 1VH9 of the putative TE from E. coli and in structures 2B6E, 1SC0, and 3LZ7 of a hypothetical H. influenzae TE. In TE13, E. coli structure 1PSU shares the same Gly53 and Asp61 catalytic residues with E. coli PaaI 2FS2, and T. thermophilus 2DSL shares the same Gly40 and Asp48 catalytic residues with the other T. thermophilus PaaI structures 1J1Y, 1WLU, 1WLV, 1WM6, 1WN3.
Comparing TE20 bovine PPT1 (1EI9) and human PPT2 (1PJA) suggests that human PPT1 structure 3GRO has the catalytic triad Ser115–His289–Asp233, the same residues as the bovine PPT1. In TE21, Pseudomonas fluorescens carboxylesterases 1AUO and 1AUR, and human acyl-protein TE 1FJ2 lead to predicting that the catalytic residues in Pseudomonas aeruginosa structures 3CN7 and 3CN9 should be Ser113, His197, and Asp166. In TE22, esterase A 2UZ0, S-formylglutathione hydrolase 1PV1, human esterase D 3FCX, and a putative enzyme (3E4D), suggest that, in Oleispira antartica esterase 3I6Y, the catalytic triad is Ser148–His257–Asp224.
Within TE-A, two families, TE5 and TE12, only have one crystal structure each (1NJK and 2HX5, respectively) with no supporting refereed article. Superimposition of structures within TE-A lead us to predict that the catalytic residue in TE5 YbaW probable enzyme is Asp13, whereas, in TE12 putative TEs (2HX5), the catalytic residue is Asp19.
Convergent and divergent evolution
These results show the effects of both convergent and divergent evolution. The former is exemplified by the fact that members in TE families from different clans and/or folds, descended from different ancestors, are active on substrates, many of them the same and all containing the thioester group, attacking the bond between its carbonyl carbon atom and its adjacent sulfur atom. Divergent evolution is shown by the presence in many TE families of enzymes with separate names and EC numbers (beyond the undifferentiated number EC 3.1.2.–), signifying activities on different substrates, even though they have similar primary and tertiary structures and mechanisms. A more profound indication of divergent evolution is the presence of clans containing families with very different primary structures but common tertiary structures and mechanisms.
Thioesterases in existing databases and previous classifications
Some TEs have been previously classified by sequence or structure in different databases. The subjective boundaries between subfamilies, families, and clans or superfamilies, as well as the use of different methodologies, give rise to differences among databases. A summary of these differences can be found on Table VII.
Tabel VII.
Comparison of Family and Clan Nomenclature
| ThYme clan | ThYme family | Ref. 10 subfamily | ESTHER/Lipase Engineering Database | Pfam-A family | SCOP database family |
|---|---|---|---|---|---|
| TE1 | — | — | AcetylCoA_hydro | CoA transferase α subunit-like | |
| TE2 | — | — | BAAT_C | — | |
| TE3 | — | — | Lipase_GDSL | TAP-like | |
| TE4 | TesB-like | — | Acyl_CoA_thio | — | |
| TE-A | TE5 | YbaW | — | 4HBT | 4HBT-like |
| TE6 | Acyl-CoA | — | 4HBT | 4HBT-like | |
| thioesterase | |||||
| TE7 | — | — | 4HBT | — | |
| TE-B | TE8 | — | — | 4HBT | PaaI/YdiI-like |
| TE-A | TE9 | YbgC-like | — | 4HBT | 4HBT-like |
| TE-A | TE10 | 4HBT-I | — | 4HBT | 4HBT-like |
| TE-B | TE11 | 4HBT-II | — | 4HBT | PaaI/YdiI-like |
| TE-A | TE12 | — | — | 4HBT | 4HBT-like |
| TE-B | TE13 | PaaI | — | 4HBT | PaaI/YdiI-like |
| TE14 | Fat subfamily | — | Acyl-ACP_TE | Acyl-ACP thioesterase-like | |
| TE15 | — | — | 4HBT | — | |
| TE-C | TE16 | — | Thioesterasea | Thioesterase | Thioesterase domain of polypeptide, polyketide, and fatty acid synthases |
| TE-C | TE17 | — | — | Thioesterase | Thioesterase domain of polypeptide, polyketide, and fatty acid synthases |
| TE-C | TE18 | — | — | Thioesterase | — |
| TE19 | — | Thioesterase_ acyltransferasea | Acyl_transf_2 | Thioesterases | |
| Acyl transferasesb | |||||
| TE-D | TE20 | — | Palmitoyl-protein thioesterasea | Palm_thioest | Thioesterases |
| Thioesterasesb | |||||
| TE-D | TE21 | — | Lysophospholipase_ carboxylesterasea | Abhydrolase_2 | Carboxylesterase/thioesterase 1 |
| Lysophospholipaseb | |||||
| TE22 | — | — | Esterase | Hypothetical esterase YLLJ068C | |
| TE23 | — | — | Lactamase B | Glyoxalase II |
ESTHER
Lipase Engineering Database.
The SCOP classification system3 is based on protein structure, and the SCOP database was assembled mainly by visual inspection and comparison, with some automation. We used it to search for the structures of TE family members. Our classes and folds correspond with SCOP; however, some differences arise at the superfamily and family levels. For example, structures in TE2, TE4, TE15, and TE18 are not classified, nor are those in TE7, which does not have a known structure. Some of our clans correspond to SCOP families: TE-B corresponds to the PaaI/YdiI-like SCOP family, and TE-A is somewhat similar to the 4HBT-like SCOP family.
Pfam has identified and classified many protein families. Pfam-A is manually curated and its families are identified with hidden Markov model profiles built from carefully chosen seed multiple alignments.71 The main difference in methodology is that in Pfam, sequences that fit a profile from a seed alignment are part of a family, whereas in ThYme sequences strongly similar to a query sequence that has TE function and evidence at protein level are part of a TE family. This leads to differences in the families: Pfam families are more inclusive and cover a wider range of sequences, whereas ThYme families are smaller, with all sequences having a very strong sequence similarity. For example, Pfam-A family 4HBT includes our families TE5–TE13 and TE15. We have shown that these families have varied structures and overall functions, as well as different mechanisms and catalytic residues. Furthermore, some of these ThYme families appear in two clans, TE-A and TE-B, and some are not part of any clan.
Dillon and Bateman10 defined subfamilies for the HotDog fold. Among HotDog fold enzymes are TEs, and some of the subfamilies presented correspond with our families. For instance, TE4 corresponds to the TesB-like subfamily, TE5 to the YbaW subfamily, TE6 to the Acyl-CoA thioesterase subfamily, TE9 to the YbgC-like subfamily, TE10 and TE11 to the 4HBT-I and 4HBT-II subfamilies, TE13 to the PaaI subfamily, and TE14 to the Fat subfamily. However, the exact content, sequences, and structures of these subfamilies might not correspond to the corresponding families in ThYme.
Two other more specific databases, ESTHER,72 a database for the α/β hydrolase fold, and the Lipase Engineering Database,73 contain some TEs. Both have families that resemble TE19, TE20, and TE21, and ESTHER also has one family similar to TE16. Again, the exact content, sequences, and structures in these families might not correspond to those in ThYme.
ThYme has detailed structural and biological annotation for TEs and other thioester-active enzymes. It does not compete with but rather supplements existing databases such as Pfam, as it is focused on thioester-active enzymes in the same way that CAZy is focused on carbohydrate-active enzymes and MEROPS is focused on peptidases. It can accurately point out differences and similarities in structure, mechanisms, and catalytic residues. The differences in methodology and purpose lead to differences with other databases. We follow the SCOP classification system, but differences in the databases arise because SCOP's purpose is to present a tertiary structure classification, whereas ThYme classifies TEs and other thioester-active enzymes mainly by primary structure. Pfam has a wide scope, and it is an extremely useful tool to identify domains within sequences, but it is not specific to a particular enzyme group. Dillon and Bateman's article,10 as well as ESTHER and the Lipase Engineering Database, have different focuses, and their overall contents do not correspond to that in ThYme.
Concluding Remarks
Analysis of primary and tertiary structures has led to classifying all TEs into families and clans. Within families, all enzymes share very similar primary and tertiary structures, active sites, catalytic residues, and mechanisms. Within clans, enzymes share some sequence similarity and similar structures, active sites, catalytic residues, and mechanisms. This classification system can help to predict an individual enzyme's structure, function, or mechanism. It also provides a standardized nomenclature.
Acknowledgments
The authors thank Matthew Lemons of the Iowa State University Web Development Services office for his help in developing ThYme. They also thank Dr. Luis Petersen for helping to write the scripts to automate BLAST, and Professor Basil Nikolau and his group for helpful advice.
Supplementary material
References
- 1.Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC–IUBMB) Enzyme nomenclature. San Diego, CA: Academic Press; 1992. http://www.chem.qmul.ac.uk/iubmb/enzyme/Available at. [Google Scholar]
- 2.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund C, Holm L, Sonnhammer ELL, Eddy SR, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. Available at http://pfam.sanger.ac.uk/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. http://scop.mrc-lmb.cam.ac.uk/scop/ Available at. [DOI] [PubMed] [Google Scholar]
- 4.Rawlings ND, Morton FR, Kok CY, Kong J, Barrett AJ. MEROPS: the peptidase database. Nucleic Acids Res. 2008;36:D320–D325. doi: 10.1093/nar/gkm954. http://merops.sanger.ac.uk/Available at: Accessed on Jun 7 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 2009;37:D233–2388. doi: 10.1093/nar/gkn663. http://www.cazy.orgAvailable at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–2402. doi: 10.1093/nar/25.17.3389. http://blast.ncbi.nlm.nih.gov/Blast.cgiAvailable at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.UniProt Consortium. The universal protein resource (UniProt) Nucleic Acids Res. 2008;36:D190–D195. doi: 10.1093/nar/gkm895. http://www.uniprot.orgAvailable at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. http://www.ncbi.nlm.nih.gov/genbank/Available at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leesong M, Henderson BS, Gillig JR, Schwab JM, Smith JL. Structure of a dehydratase–isomerase from the bacterial pathway for biosynthesis of unsaturated fatty acids: two catalytic activities in one active site. Structure. 1996;4:253–264. doi: 10.1016/s0969-2126(96)00030-5. [DOI] [PubMed] [Google Scholar]
- 10.Dillon SC, Bateman A. The hotdog fold: wrapping up a superfamily of thioesterases and dehydratases. BMC Bioinformatics. 2004;5:109–123. doi: 10.1186/1471-2105-5-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee F-JS, Lin L-W, Smith JA. A glucose-represible gene encodes acetyl-CoA hydrolase from Saccharomyces cerevisiae. J Biol Chem. 1990;265:7413–7418. [PubMed] [Google Scholar]
- 12.Fleck CB, Brock M. Re-characterisation of Saccharomyces cerevisiae Ach1p: fungal CoA-transferases are involved in acetic acid detoxification. Fungal Genet Biol. 2009;46:473–485. doi: 10.1016/j.fgb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Marel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A. The α/β hydrolase fold. Protein Eng. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 14.Hunt MC, Rautenen A, Westin MAK, Svensson LT, Alexson SEH. Analysis of the mouse and human acyl-CoA thioesterase (ACOT) gene clusters show that convergent, functional evolution results in a reduced number of human peroxisomal ACOTs. FASEB J. 2006;20:1855–1864. doi: 10.1096/fj.06-6042com. [DOI] [PubMed] [Google Scholar]
- 15.Johnson MR, Barnes S, Kwakye JB, Diasio RB. Purification and characterization of bile acid-CoA:amino acid N-acyltransferase from human liver. J Biol Chem. 1991;266:10227–10233. [PubMed] [Google Scholar]
- 16.Cho H, Cronan JE., Jr Defective export of a periplasmic enzyme disrupts regulation of fatty acid synthesis. J Biol Chem. 1995;270:4216–4219. doi: 10.1074/jbc.270.9.4216. [DOI] [PubMed] [Google Scholar]
- 17.Karasawa K, Yokoyama K, Setaka M, Nojima S. The Escherichia coli pldC gene encoding lysophospholipase L1 is identical to the apeA and tesA genes encoding protease I and thioesterase I respectively. J Biochem. 1999;126:445–448. doi: 10.1093/oxfordjournals.jbchem.a022470. [DOI] [PubMed] [Google Scholar]
- 18.Jones JM, Nau K, Geraghty MT, Erdmann R, Gould SJ. Identification of peroxisomal acyl-CoA thioesterases in yeast and humans. J Biol Chem. 1999;274:9216–9223. doi: 10.1074/jbc.274.14.9216. [DOI] [PubMed] [Google Scholar]
- 19.Naggert J, Narasimhan ML, Deveaux L, Cho H, Randhawa ZI, Cronan JE, Jr, Green BN, Smith S. Cloning, sequencing, and characterization of Escherichia coli thioesterase II. J Biol Chem. 1991;266:11044–11050. [PubMed] [Google Scholar]
- 20.Nie L, Ren Y, Schulz H. Identification and characterization of Escherichia coli thioesterase III that functions in fatty acid β-oxidation. Biochemistry. 2008;47:7744–7751. doi: 10.1021/bi800595f. [DOI] [PubMed] [Google Scholar]
- 21.Yamada J, Furihata T, Tamura H, Watanabe T, Suga T. Long-chain acyl-CoA hydrolase from rat brain cytosol: purification, characterization, and immunohistochemical localization. Arch Biochem Biophys. 1996;326:106–114. doi: 10.1006/abbi.1996.0053. [DOI] [PubMed] [Google Scholar]
- 22.Adams SH, Chui C, Schilbach SL, Yu XX, Goddard AD, Grimaldi JC, Lee J, Dowd P, Colman S, Lewin DA. BFIT, a unique acyl-CoA thioesterase induced in thermogenic brown adipose tissue: cloning, organization of the human gene and assessment of a potential link to obesity. Biochem J. 2001;360:135–142. doi: 10.1042/0264-6021:3600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suematsu N, Isohahshi F. Molecular cloning and functional expression of human cytosolic acetyl-CoA hydrolase. Acta Biochim Pol. 2006;53:553–561. [PubMed] [Google Scholar]
- 24.Zhuang Z, Song F, Zhao H, Li L, Cao J, Eisenstein E, Herzberg O, Dunaway-Mariano D. Divergence of function in the hotdog fold enzyme superfamily: the bacterial thioesterase YciA. Biochemistry. 2008;47:2789–2796. doi: 10.1021/bi702334h. [DOI] [PubMed] [Google Scholar]
- 25.Yokoyama T, Choi K-J, Bosch AM, Yeo H-J. Structure and function of a Campylobacter jejuni thioesterase Cj0915, a hexameric hot dog fold enzyme. Biochim Biophys Acta. 2009;1794:1073–1081. doi: 10.1016/j.bbapap.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poupon V, Begue B, Gagnon J, Dautry-Varsat A, Cerf-Bensussan N, Benmerah A. Molecular cloning and characterization of MT-ACT48, a novel mitochondrial acyl-CoA thioesterase. J Biol Chem. 1999;274:19188–19194. doi: 10.1074/jbc.274.27.19188. [DOI] [PubMed] [Google Scholar]
- 27.Wei J, Kang HW, Cohen DE. Thioesterase superfamily member 2 (Them2)/acyl-CoA thioesterase 13 (Acot13): a homotetrameric hotdog fold thioesterase with selectivity for long-chain fatty acyl-CoAs. Biochem J. 2009;421:311–322. doi: 10.1042/BJ20090039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhuang Z, Song F, Martin BM, Dunaway-Mariano D. The YbgC protein encoded by the ygbC gene of the tol-pal cluster of Haemophilus influenzae catalyzes acyl-coenzyme A thioester hydrolysis. FEBS Lett. 2002;516:161–163. doi: 10.1016/s0014-5793(02)02533-4. [DOI] [PubMed] [Google Scholar]
- 29.Angelini A, Cendron L, Goncalves S, Zanotti G, Terradot L. Structural and enzymatic characterization of HP0496, a YbgC thioesterase from Helicobacter pylori. Proteins: Struct Funct Bioinformatics. 2008;72:1212–1221. doi: 10.1002/prot.22014. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Israel I, Yu G, Austin MB, Bhuiyan N, Auldridge M, Nguyen T, Schauvinhold I, Noel JP, Pichersky E, Fridman E. Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol. 2009;151:1952–1964. doi: 10.1104/pp.109.146415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benning MM, Wesenberg G, Liu R, Taylor KL, Dunaway-Mariano D, Holden HM. The three-dimensional structure of 4-hydroxybenzoyl-CoA thioesterase from Pseudomonas sp. strain CBS-3. J Biol Chem. 1998;273:33572–33579. doi: 10.1074/jbc.273.50.33572. [DOI] [PubMed] [Google Scholar]
- 32.Leduc D, Battesti A, Bouveret E. The hotdog thioesterase EntH (YbdB) plays a role in vivo in optimal enterobactin biosynthesis by interacting with the ArCP domain of EntB. J Bacteriol. 2007;189:7112–7126. doi: 10.1128/JB.00755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Widhalm JR, van Oostende C, Furt F, Basset GJ. A dedicated thioesterase of the hotdog-fold family is required for the biosynthesis of the naphthoquinone ring of vitamin K1. Proc Natl Acad Sci USA. 2009;106:5599–5603. doi: 10.1073/pnas.0900738106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrández A, Miñambres B, García B, Olivera ER, Luengo JM, García JL, Díaz E. Catabolism of phenylacetic acid in Escherichia coli. J Biol Chem. 1998;273:25974–25986. doi: 10.1074/jbc.273.40.25974. [DOI] [PubMed] [Google Scholar]
- 35.Jones A, Davies HM, Voelker TA. Palmitoyl-acyl carrier protein (ACP) thioesterase and evolutionary origin of plant acyl-ACP thioesterases. Plant Cell. 1995;7:359–371. doi: 10.1105/tpc.7.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kotaka M, Kong R, Qureshi I, Ho QS, Sun H, Liew CW, Goh LP, Cheung P, Mu Y, Lescar J, Liang Z-X. Structure and catalytic mechanism of the thioesterase CalE7 in enediyne biosynthesis. J Biol Chem. 2009;284:15739–15749. doi: 10.1074/jbc.M809669200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakil SJ. Fatty acid synthase, a proficient multifunctional enzyme. Biochemistry. 1989;28:4523–4530. doi: 10.1021/bi00437a001. [DOI] [PubMed] [Google Scholar]
- 38.Gokhale RS, Hunziker D, Cane DE, Khosla C. Mechanism and specificity of the terminal thioesterase domain from the erythromycin polyketide synthase. Chem Biol. 1999;6:117–125. doi: 10.1016/S1074-5521(99)80008-8. [DOI] [PubMed] [Google Scholar]
- 39.Kohli RM, Takagi J, Walsh CT. The thioesterase domain from a nonribosomal peptide synthetase as a cyclization catalyst for integrin binding peptides. Proc Natl Acad Sci USA. 2002;99:1247–1252. doi: 10.1073/pnas.251668398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Libertini LJ, Smith S. Purification and properties of a thioesterase from lactating rat mammary gland which modifies the product specificity of fatty acid synthetase. J Biol Chem. 1977;253:1393–1401. [PubMed] [Google Scholar]
- 41.Mikkelsen J, Witkowski A, Smith S. Interaction of rat mammary gland thioesterase II with fatty acid synthase is dependent on the presence of acyl chains on the synthase. J Biol Chem. 1987;262:1570–1574. [PubMed] [Google Scholar]
- 42.Heathcote ML, Staunton J, Leadlay PF. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem Biol. 2001;8:207–220. doi: 10.1016/s1074-5521(01)00002-3. [DOI] [PubMed] [Google Scholar]
- 43.Claxton HB, Akey DL, Silver MK, Admiraal SJ, Smith JL. Structure and functional analysis of RifR, the type II thioesterase from the rifamycin biosynthetic pathway. J Biol Chem. 2009;284:5021–5029. doi: 10.1074/jbc.M808604200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferri SR, Meighen EA. A Lux-specific myristoyl transferase in luminescent bacteria related to eukaryotic serine esterases. J Biol Chem. 1991;266:12852–12857. [PubMed] [Google Scholar]
- 45.Camp LA, Hofmann SL. Purification and properties of a palmitoyl-protein thioesterase that cleaves palmitate from H-Ras. J Biol Chem. 1993;268:22566–22574. [PubMed] [Google Scholar]
- 46.Vesa J, Hellsten E, Verkruyse LA, Camp LA, Rapola J, Santavuorl P, Hofmann SL, Peltonen L. Mutations in the palmitoyl protein thioesterase gene causing infantile neuronal ceroid lipofuscinosis. Nature. 1995;376:584–587. doi: 10.1038/376584a0. [DOI] [PubMed] [Google Scholar]
- 47.Sugimoto H, Hayashi H, Yamashita S. Purification, cDNA cloning, and regulation of lysophospholipase from rat liver. J Biol Chem. 1996;271:7705–7711. doi: 10.1074/jbc.271.13.7705. [DOI] [PubMed] [Google Scholar]
- 48.Duncan JA, Gilman AG. A cytoplasmic acyl-protein thioesterase that removes palmitate from G protein α subunits and p21RAS. J Biol Chem. 1998;273:15830–15837. doi: 10.1074/jbc.273.25.15830. [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez CF, Proudfoot M, Brown G, Korniyenko Y, Mori H, Savchenko AV, Yakunin AF. Characterization of two S-formylglutathione hydrolases from Escherichia coli, FrmB and YeiG. J Biol Chem. 2006;281:14514–14522. doi: 10.1074/jbc.M600996200. [DOI] [PubMed] [Google Scholar]
- 50.Vander Jagt DL. Glyoxalase II: molecular characteristics, kinetics and mechanism. Biochem Soc Trans. 1993;21:522–527. doi: 10.1042/bst0210522. [DOI] [PubMed] [Google Scholar]
- 51.Carfi A, Duee E, Galleni M, Duez C, Frere JM, Dideberg O. The 3-D structure of a zinc metallo-β-lactamase from Bacillus cereus reveals a new type of protein fold. EMBO J. 1995;14:4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rose IA, Warms JVB. An enzyme with ubiquitin carboxyl-terminal esterase activity from reticulocytes. Biochemistry. 1983;22:4234–4237. doi: 10.1021/bi00287a012. [DOI] [PubMed] [Google Scholar]
- 53.Pickart CM, Rose IA. Ubiquitin carboxyl-terminal hydrolase acts on ubiquitin carboxyl-terminal amides. J Biol Chem. 1985;260:7903–7910. [PubMed] [Google Scholar]
- 54.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 55.Thoden JB, Holden HM, Zhuang Z, Dunaway-Mariano D. X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase. J Biol Chem. 2002;277:27468–27476. doi: 10.1074/jbc.M203904200. [DOI] [PubMed] [Google Scholar]
- 56.Cao J, Xu H, Zhao H, Gong W, Dunaway-Mariano D. The mechanisms of human hotdog-fold thioesterase 2 (hTHEM2) substrate recognition and catalysis illuminated by a structure and function based analysis. Biochemistry. 2009;48:1293–1304. doi: 10.1021/bi801879z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thoden JB, Zhuang Z, Dunaway-Mariano D, Holden HM. The structure of 4-hydroxybenzoyl-CoA thioesterase from Arthrobacter sp. strain SU. J Biol Chem. 2003;278:43709–43716. doi: 10.1074/jbc.M308198200. [DOI] [PubMed] [Google Scholar]
- 58.Kunishima N, Asada Y, Sugahara M, Ishijima J, Nodake Y, Sugahara M, Miyano M, Kuramitsu S, Yokoyama S, Sugahara M. A novel induced-fit reaction mechanism of asymmetric hot dog thioesterase PaaI. J Mol Biol. 2005;352:212–228. doi: 10.1016/j.jmb.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 59.Song F, Zhuang Z, Finci L, Dunaway-Mariano D, Kniewel R, Buglino JA, Solorzano V, Wu J, Lima CD. Structure, function, and mechanism of the phenylacetate pathway hot dog-fold thioesterase PaaI. J Biol Chem. 2006;28:11028–11038. doi: 10.1074/jbc.M513896200. [DOI] [PubMed] [Google Scholar]
- 60.Li J, Derewenda U, Dauter Z, Smith S, Derewenda ZS. Crystal structure of the Escherichia coli thioesterase II, a homolog of the human Nef binding enzyme. Nat Struct Biol. 2000;7:555–559. doi: 10.1038/76776. [DOI] [PubMed] [Google Scholar]
- 61.Forwood JK, Thakur AS, Guncar G, Marfori M, Mouradov D, Meng W, Robinson J, Huber T, Kellie S, Martin JL, Hume DA, Kobe B. Structural basis for recruitment of tandem hotdog domains in acyl-CoA thioesterase 7 and its role in inflammation. Proc Natl Acad Sci USA. 2007;104:10382–10387. doi: 10.1073/pnas.0700974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willis MA, Zhuang Z, Song F, Howard A, Dunaway-Mariano D, Herzberg O. Structure of YciA from Haemophilus influenzae (UI0827), a hexameric broas specificity acyl-coenzyme A thioesterase. Biochemistry. 2008;47:2797–2805. doi: 10.1021/bi702336d. [DOI] [PubMed] [Google Scholar]
- 63.Mayer KM, Shanklin J. Identification of amino acid residues involved in substrate specificity of plant acyl-ACP thioesterases using a bioinformatics-guided approach. BMC Plant Biol. 2007;7:1. doi: 10.1186/1471-2229-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nardini M, Dijkstra BW. α/β Hydrolase fold enzymes: the family keeps growing. Curr Opin Struct Biol. 1999;9:732–737. doi: 10.1016/s0959-440x(99)00037-8. [DOI] [PubMed] [Google Scholar]
- 65.Chakravarty B, Gu Z, Chirala SS, Wakil SJ, Quiocho FA. Human fatty acid synthase: structure and substrate selectivity of the thioesterase domain. Proc Natl Acad Sci USA. 2004;101:15567–15572. doi: 10.1073/pnas.0406901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsai S-C, Lu H, Cane DE, Khosla C, Stroud RM. Insights into channel architecture and substrate specificity from crystal structures of two macrocycle-forming thioesterases of modular polyketide synthases. Biochemistry. 2002;41:12598–12606. doi: 10.1021/bi0260177. [DOI] [PubMed] [Google Scholar]
- 67.Kim KK, Song HK, Shin DH, Hwang KY, Choe S, Yoo OJ, Suh SW. Crystal structure of carboxylesterase from Pseudomonas fluorescens, an α/β hydrolase with broad substrate specificity. Structure. 1997;5:1571–1584. doi: 10.1016/s0969-2126(97)00306-7. [DOI] [PubMed] [Google Scholar]
- 68.Lo Y-C, Lin S-C, Shaw J-F, Liaw Y-C. Crystal structure of Escherichia coli thioesterase I/proteaseI/lysophospholipase L1: consensus sequence blocks constitute the catalytic center of SGNH-hydrolases through a conserved hydrogen bond network. J Mol Biol. 2003;330:539–551. doi: 10.1016/s0022-2836(03)00637-5. [DOI] [PubMed] [Google Scholar]
- 69.Lo Y-C, Lin S-C, Shaw J-F, Liaw Y-C. Substrate specificities of Escherichia coli thioesteraseI/protease I/lysophosphollipase L1 are governed by its switch loop movement. Biochemistry. 2005;44:1971–1979. doi: 10.1021/bi048109x. [DOI] [PubMed] [Google Scholar]
- 70.Cameron AD, Ridderstrom M, Olin B, Mannervik B. Crystal structure of human glyoxalase II and its complex with a glutathione thiolester substrate analogue. Structure. 1999;7:1067–1078. doi: 10.1016/s0969-2126(99)80174-9. [DOI] [PubMed] [Google Scholar]
- 71.Sonnhammer ELL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 72.Hotelier T, Renault L, Cousin X, Negre V, Marchot P, Chatonnet A. ESTHER, the database of the α/β-hydrolase fold superfamily of proteins. Nucleic Acids Res. 2004;32:D145–D147. doi: 10.1093/nar/gkh141. http://bioweb.ensam.inra.fr/ESTHER/general?what=indexAvailable at: Accessed on Jun 7 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fischer M, Pleiss J. The lipase engineering database: a navigation and analysis tool for protein families. Nucleic Acids Res. 2003;31:319–321. doi: 10.1093/nar/gkg015. http://www.led.uni-stuttgart.de/Available at: Accessed on Jun 7 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


