Abstract
The hepatitis C virus (HCV) nonstructural (NS) protein 4B is known for protein–protein interactions with virus and host cell factors. Only little is known about the corresponding protein binding sites and underlying molecular mechanisms. Recently, we have predicted a putative basic leucine zipper (bZIP) motif within the aminoterminal part of NS4B. The aim of this study was to investigate the importance of this NS4B bZIP motif for specific protein–protein interactions. We applied in silico approaches for 3D-structure modeling of NS4B-homodimerization via the bZIP motif and identified crucial amino acid positions by multiple sequence analysis. The selected sites were used for site-directed mutagenesis within the NS4B bZIP motif and subsequent co-immunoprecipitation of wild-type and mutant NS4B molecules. Respective interaction energies were calculated for wild-type and mutant structural models. NS4B-homodimerization with a gradual alleviation of dimer interaction from wild-type towards the mutant-dimers was observed. The putative bZIP motif was confirmed by a co-immunoprecipitation assay and western blot analysis. NS4B-NS4B interaction depends on the integrity of the bZIP hydrophobic core and can be abolished due to changes of crucial residues within NS4B. In conclusion, our data indicate NS4B-homodimerization and that this interaction is facilitated by the aminoterminal part containing a bZIP motif.
Keywords: basic leucine zipper, bZIP, HCV, NS4B, homodimerization
Introduction
Chronic hepatitis C is a major cause of liver disease with progression to liver cirrhosis and its sequelae, including an elevated risk for the development of hepatocellular carcinoma.1,2 The hepatitis C virus (HCV) is a positive single-stranded RNA virus, belonging to the Flaviviridae family, genus hepacivirus.3 The genome comprises ∼9600 nucleotides coding for a polyprotein of about 3000 amino acids, which is processed by cellular and viral proteases in three structural and six nonstructural (NS) proteins.3–5
The HCV protein NS4B is a key player in the viral replication cycle.6–8 Adaptive amino acid mutations within NS4B have been shown to enhance replication efficacy in the HCV replicon system.6,8–11
NS4B is highly hydrophobic,12 which makes experimental structure determination a difficult task. Amongst others, bioinformatics was used to study protein structure and function as an alternative approach.12,13 From several experimental and bioinformatics' analyses, it is accepted that NS4B contains at least four transmembrane domains (TMDs) in its middle part, while the topology of a putative fifth TMD close to the aminoterminal part remains controversial.12–15 Furthermore, amphipathic α-helix structure elements were predicted within the aminoterminal part and recently reported to be potential targets of HCV replication inhibitors.7,16,17
The HCV NS4B protein integrates into the endoplasmatic reticulum (ER) with subsequent specific membrane alterations, described as membranous web.13,18,19 The subcellular structure is considered to harbour the HCV replication complex, since all nonstructural proteins as well as viral genomic RNA were found to co-localize within this ER alterations.19,20 The aminoterminal amphipathic α-helix (amino acid 1 to 26) has been described to mediate membrane association and correct localization of the replication complex within the ER.7 Recently, Gouttenoire et al. proposed the amphipathic α-helix to be located between amino acid 42 to 66.17 In this publication no membrane association of amino acid 1 to 29 or amino acid 1 to 40 was detected.17 Most likely, the HCV NS4B protein is anchored to the ER membrane by the four TMDs, initiating ER membrane alterations and recruiting other host and viral factors of the replication complex,20,21 while the aminoterminal region contains an amphipathic α-helix and mediates membrane association.7,17 Exact mechanisms how NS4B causes ER membrane alterations or interacts with other proteins are still unknown.
Polymerization of different NS4B molecules has been described previously.22 Thereby, the main polymerization determinants were mapped within the aminoterminal cytosolic domain of NS4B.22 Furthermore, the aminoterminal part of NS4B was shown to be involved in physical interaction with the cellular ER-stress response element CREB-RP/ATF6ß.23 However, neither the molecular basis for specific protein–protein interactions nor the biological relevance of this finding is known.
We have previously used a bioinformatics approach to predict a putative basic leucine zipper (bZIP) motif,12 in part overlapping with the aminoterminal amphipathic α-helical element. A leucine zipper is a common supersecondary (coiled-coil) structural element involved in protein–protein interactions via parallel α-helices with similar sequence motifs.24 Until now, existence of the NS4B bZIP has not been tested and confirmed on the basis of experimental data. Furthermore, its role as a potential protein–protein interaction site within NS4B is unknown. Therefore in the present study, we investigated the existence of the bZIP motif and its relevance in mediating specific protein–protein interactions, particularly NS4B-homodimerization by co-expression and co-immunoprecipitation studies, as well as by bioinformatics' analysis.
Results
Multiple sequence analysis and homology structure modeling
The aminoterminal part of HCV NS4B contains a putative basic leucine zipper (bZIP) motif ranging from amino acid position 22 to 49 according to the HCVJ prototype sequence of NS4B12,25 (Fig. 1). Protein–protein interactions mediated by bZIP motifs are characterized by the formation of a non-covalent hydrophobic core between the bZIP motifs with hydrophobic amino acids at specific positions within the motif and leucine residues for stacking interactions, as well as amino acids at given positions outside the core for putative ionic interactions (salt bridges). We used multiple sequence alignments to determine amino acid positions of functional importance within the putative NS4B bZIP motif. The four heptad repeats of the prototype sequence HCVJ (genotype 1b) possess 7 over 8 apolar amino acids, A (alanine), L (leucine), V (valine), and W (tryptophane) at positions “a” and “d,” important for the formation of the bZIP hydrophobic core, and only 2 over 4 possible L (leucine) residues for stacking interactions at position d. Furthermore, HCVJ possesses 5 over 8 polar amino acids, E (glutamate), K (lysine), Q (glutamine), and R (arginine) at positions “e” and “g” (Fig. 2).
Figure 1.

Sequence alignment of the bZIP motif for the HCVJ prototype sequence (wild-type) and the corresponding modified sequence upon site-directed mutagenesis (modified sites in bold and framed by grey shaded boxes). The alignment is annotated with the corresponding heptad positions “a” to “g” for each heptad repeat and numbered according to the NS4B protein of prototype strain HCVJ. Note that a genotype 1b sequence was used within the present study, while Gouttenoire et al. used a HCV genotype 1a sequence.17
Figure 2.
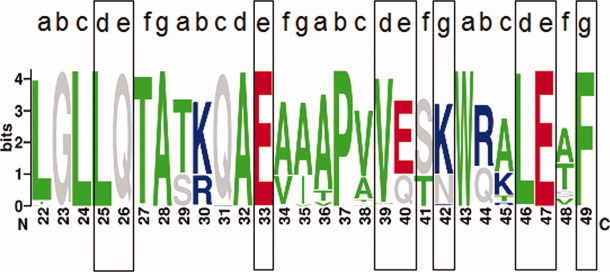
Sequence logo of HCV NS4B in genotype 1 (487 sequences) showing the suspected bZIP motif region (amino acid positions, X-axis) and the positions for site-directed mutagenesis in black boxes. Amino acids are colored as follows, according to their physico-chemical properties: hydrophobic (green) – ACFILMPTVWY; charged, positive (blue) – HKR; charged, negative (red) – DE; rest (grey) – GNQS. The overall height of each stack indicates the sequence conservation at a position (measured in bits, Y-axis), whereas the height of each symbol within a stack reflects the relative frequency of the corresponding amino acid at a given position.
Natural polymorphisms within the putative bZIP motif of HCV genotype 1 were investigated to determine relevance and importance of the hydrophobic core and potential ionic interactions for protein–protein interactions. For that purpose, we used sequence alignments and a sequence logo of the bZIP motif within HCV genotype 1 (Fig. 2 and Table I). Amino acid patterns at heptad positions a and d as well as e and g are given in Table I. The hydrophobic core (heptad positions a and d) seems to be better conserved than residues for putative ionic interactions (heptad positions e and g). We found 6 over 9 polymorphisms to be hydrophobic at heptad position a, and 5 over 5 hydrophobic polymorphisms at position d. The 5 polymorphisms found at heptad position d comprised 2 leucine residues. On the other hand, we identified only 3 over 5 charged polymorphisms, negative or positive, at position e and even solely 1 over 7 at position g. None of the heptad repeats differed markedly from the other heptads in term of the frequency of their polymorphisms. The hydrophobic core seems to be mostly conserved and therefore an important site for protein–protein interactions via the bZIP motif of HCV NS4B in genotype 1 (Table I). Change-to-alanine mutations were performed at six positions putatively involved in the formation of salt bridges (position e, n = 4; g, n = 2) and at three positions considered to be involved in the formation of the zipper hydrophobic core and stacking interactions (position d).12 Because alanine is compatible with α-helix structure elements, these amino acid exchanges should only impact the bZIP function while preserving the α-helix.
Tabel I.
Natural Amino Acid Polymorphisms Within the bZIP Motif of HCV Genotype 1 at Positions Important for the bZIP Functionality
| bZIP Heptad position | Amino acid position within NS4Ba | Natural amino acid polymorphismsb | Hydrophobic or charged residues versus all polymorphisms |
|---|---|---|---|
| a | 22 | FIL | 6/9 (hydrophobic) |
| 29 | ST | ||
| 36 | ATV | ||
| 43 | W | ||
| d | 25 | L | |
| 32 | A | 5/5 (hydrophobic) | |
| 39 | MV | 2/5 (leucine) | |
| 46 | L | ||
| e | 26 | Q | 3/5 (charged) |
| 33 | E | ||
| 40 | EQ | ||
| 47 | E | ||
| g | 28 | A | 1/7 (charged) |
| 35 | AIV | ||
| 42 | KN | ||
| 49 | F |
Numbering according to the NS4B protein within the prototype strain HCVJ.
Natural polymorphisms in alphabetical order with hydrophobic or charged residues in italic/bold.
We computed sequence logos for HCV genotypes 1 to 6 based on multiple sequence alignments (comprising a total of 614 HCV NS4B sequences) to compare the bZIP region among the different genotypes (Supporting Information Fig. 1). Here, we found the bZIP heptad repeat best conserved within HCV genotype 1, whereas genotypes 4, 5, and 6 have been the most diverse. Residues of the hydrophobic core (heptad positions a and d) seem to be better conserved among different genotypes than residues for putative ionic interactions (heptad positions e and g), with a highest degree of conservation found in HCV genotype 1.
The transcription factor c-Jun was used to model the 3D-structure of the bZIP interaction within an NS4B-homodimer. The 3D model shows a highly symmetrical dimer interface with conserved hydrophobic residues in dense packaging buried within a hydrophobic core (Fig. 3). We also modeled structures for the heterodimer and the mutant-homodimer. The wild-type dimer [Fig. 4(A)] showed the largest interaction interface and densest side chain packaging in comparison with the hetero- and mutant-homodimer investigated by co-immunoprecipitation experiments [Fig. 4(B,C)].
Figure 3.
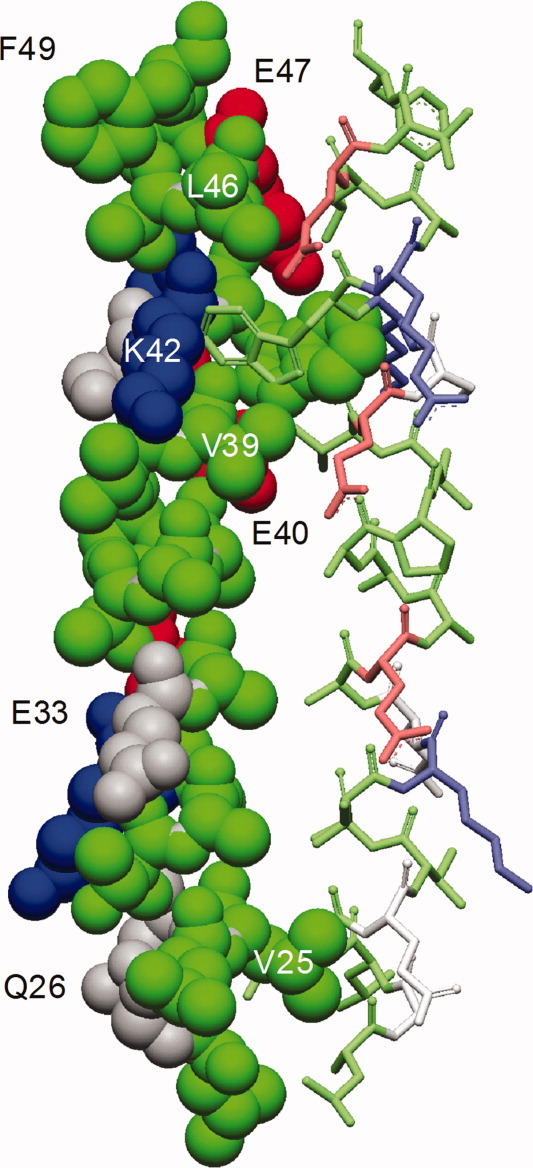
Side view of the bZIP homology model for the proposed NS4B homodimer structure. Alpha-helical element in CPK mode on the left with corresponding α-helical element as stick model on the right, colored according to the sequence logo in Figure 1 with residues labeled for site-directed mutagenesis.
Figure 4.
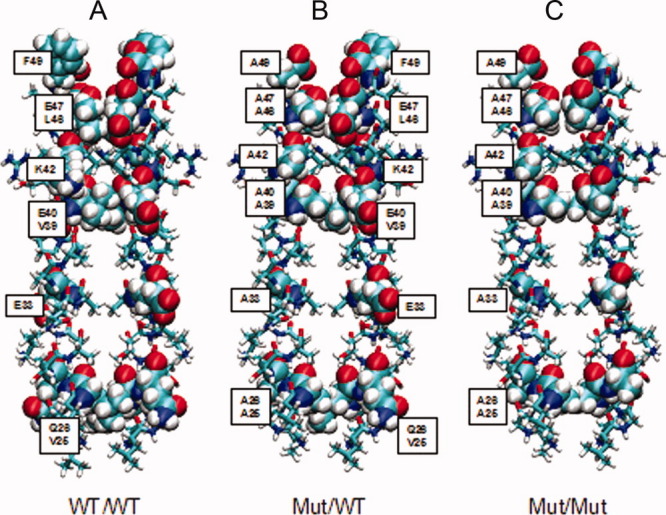
Energy minimized bZIP homology structures of the wild-type homodimer (3A), the heterodimer (3B), and the mutant-dimer (3C). The residues mutated for the mutant structures are labelled and schematically represented by van-der-Waals spheres to illustrate the side chain packaging effects. The VMD software26 was used for the figure construction.
Interaction energies for the bZIP wild-type and the two modified NS4B dimer structures
For calculation of differences in the chain-chain interaction energies of the whole chains of the dimers and of all residues, structures of the energy minimized homology models of the wild-type NS4B homodimer structure [Fig. 4(A)] and of the two mutated structures, heterodimer [Fig. 4(B)] and mutant-homodimer [Fig. 4(C)] were used. The energy differences between the mutated structures and the wild-type structure were all positive, indicating reduced interaction energy for mutated structures (Table II). Comparing the chain-chain interaction energies of all three structural models, the structure of the wild-type homodimer showed the highest interaction energy, whereas the mutant-homodimer showed the lowest interaction energy, and the heterodimer had an intermediate interaction energy value (Table II). For a more detailed, quantitative analysis of the chain-chain interactions and their dynamics, molecular dynamics simulations could be performed. This, however, would go beyond the scope of the theoretical investigations in this manuscript, which were performed with the goal of providing a qualitative, structural interpretation of the experimental results, which was achieved with the studies performed.
Tabel II.
Differences of Interaction Energies [kJ/mol] Between the Whole Chains (Chain/Chain) and the Mutated Residues for the Three Modeled bZIP Structures (Wild-Type Dimer, Heterodimer, and Mutant Homodimer)
| Heterodimer versus wild-type dimer | Mutant homodimer versus wild-type dimer | Mutant homodimer versus heterodimer | |
|---|---|---|---|
| Chain/chain | 302.76 | 596.01 | 293.25 |
| Mutated residues | 813.21 | 1606.47 | 793.26 |
Immunoblotting of wild-type and modified NS4B expression plasmids
To investigate the role of the bZIP motif in NS4B-homodimerization, HEK 293T cells were co-transfected with equal amounts of different pairs of expression plasmids (NS4B-bZIP+/MYC and NS4B-bZIP+/FLAG; NS4B-bZIP+/MYC and NS4B-bZIP-/FLAG; NS4B-bZIP-/MYC and NS4B-bZIP+/FLAG; NS4B-bZIP-/MYC and NS4B-bZIP-/FLAG). Expression of each plasmid in the presence of the respective other expression plasmid was shown by immunoblotting of the lysates with either FLAG- or MYC-specific antibodies in sequential experimental steps [Fig. 5(A)]. Thereby, we found no evidence for perturbance in expression of plasmids in any combination. Moreover, immunoprecipitation of wild-type and modified NS4B proteins with consecutive immunoblotting was performed. These results also showed that expression of wild-type and modified plasmids were not affected (data not shown).
Figure 5.
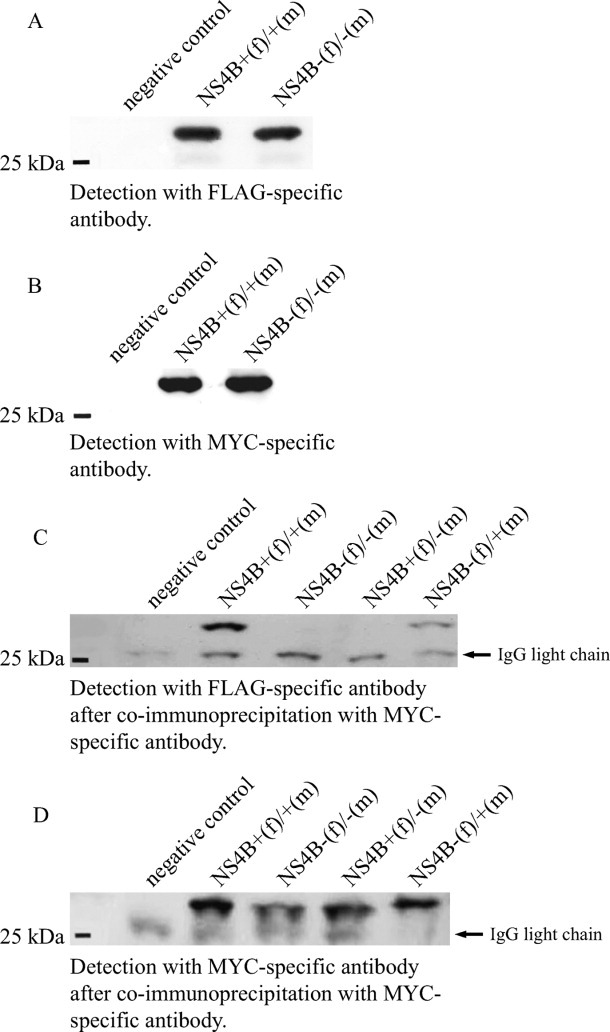
Western-blot analyses after diverse co-transfection experiments showing undisturbed co-expression of wild-type and mutant NS4B full-length protein (A, B) and interaction of NS4B proteins (C, D). (A, B) Co-expression of wild-type NS4B as well as mutant NS4B with (A) FLAG- and (B) MYC-tag-specific antibodies, demonstrating that neither insertion of FLAG-tag or MYC-tag, nor site-directed mutations within the bZIP region affected co-expression of NS4B full-length plasmids. HEK 293T-cells were co-transfected with GFP (negative control) or NS4B full-length expression plasmids containing both the wild-type (+) bZIP region and either FLAG-tag (f) or MYC-tag (m) or NS4B full-length expression plasmids containing both the modified (-) bZIP region and either FLAG-tag (f) or MYC-tag (m). Immunoblotting of the lysates was performed, and NS4B proteins were detected with FLAG-specific (A) or MYC-specific antibodies (B), subsequently visualized by enhanced chemiluminescence after binding of an anti-mouse, horseradish peroxidase-conjugated antibody. (C, D) Co-immunoprecipitation with a MYC-specific antibody was performed with lysates of HEK 293T-cells either transfected with GFP (negative control) or co-transfected with different combinations of NS4B full-length expression plasmids containing either the wild-type (+) or the mutant (-) bZIP motif, respectively. Plasmids were labelled either with FLAG-tag (f) or MYC-tag (m). Consecutive immunoblotting with a FLAG-specific antibody (C) showed interaction of different NS4B molecules when both interaction partners contained the wild-type bZIP motif, whereas interaction was undetectable when both partners contained the mutant bZIP motif, and weakly detectable when one plasmid contained the mutated and the other plasmid contained the wild-type bZIP motif (left). Control staining (D) of co-immunoprecipitation was performed with MYC-specific antibodies. Note the band below the NS4B band (all columns) as an artefact, most likely corresponding to the IgG light chain.
Co-immunoprecipitation of wild-type and modified NS4B expression plasmids
After co-transfection of HEK 293T cells with two full-length NS4B expression plasmids of the bZIP wild-type, which differed only by their implemented tag (FLAG-tag, MYC-tag), co-immunoprecipitation (co-IP) of the lysate was performed with either FLAG- or MYC-specific antibodies and immunoblotting was carried out with the accordant complementary antibody. Independently of the antibodies used for co-IP and immunoblotting, interaction between different NS4B molecules was shown by immunoblotting of the binding partner with the complementary tag [Fig. 5(B)]. The use of the MYC-specific antibody for co-IP in combination with the FLAG-specific antibody for immunoblotting showed best results, and MYC-specific antibodies for co-IP with FLAG-specific antibodies for immunoblotting were used for the following experiments. We tested different combinations of full-length NS4B expression plasmids containing either wild-type or modified bZIP regions. In comparison with combinations of two NS4B molecules with the wild-type bZIP motif, the interaction of two NS4B molecules containing the modified bZIP region was considerably attenuated or even undetectable by co-IP and immunoblotting [Fig. 5(B,C)]. When only one NS4B molecule contained the mutated bZIP region, whereas the interaction partner possesses the wild-type bZIP motif, a weaker protein–protein interaction compared with the wild-type/wild-type combination was found [Fig. 5(C)].
Discussion
The hepatitis C virus (HCV) nonstructural (NS) protein 4B is a key player in the viral replication cycle, probably acting as a protein hub recruiting host and viral determinants important for viral replication.19,20,27–29 It is largely unknown how NS4B interacts with other proteins. Recently, we have predicted a basic leucine zipper (bZIP) motif within the aminoterminal part of HCV NS4B.12 A bZIP is a common structural motif, known for a multitude of protein–protein interactions and thus important for diverse cellular functions and biochemical pathways.12 The bZIP motif in HCV NS4B is located in the aminoterminal part of the protein, which was reported to mediate membrane association.7,17 Interestingly, amino acid substitutions within the predicted bZIP motif (Q26H, L46I) have been described to influence viral replication efficacy in the replicon model.11,25,30 The aim of the present study was to investigate the capability of NS4B to interact with each other due to specific interactions via this bZIP motif.
In this study, a homology-modeling approach was used to propose a 3D-structural model for the putative NS4B-NS4B interaction via the aminoterminal bZIP function. NS4B-homodimerization most likely depends on the integrity of the hydrophobic core of the bZIP structure as the major determinant for protein–protein interaction and dimer stability in this case. The hydrophobic core of the bZIP motif appeared to be better conserved than residues outside the core and therefore crucial for protein–protein interactions via the bZIP motif of HCV NS4B. Amino acid positions outside the core are assumed to be involved in putative ionic interactions (salt bridges). Overall, the bZIP motif seems to be best conserved within HCV genotype 1, whereas genotypes 4 to 6 are least conserved. Our structural model of the genotype 1b bZIP homodimer exhibited two characteristic leucine residues for stacking interactions with the parallel α-helical element. Based on the structure modeling and a comprehensive in silico sequence analysis, sites for subsequent mutagenesis were selected to disturb the bZIP functionality, while maintaining the α-helical structure and amphipathic properties of this region.
Subsequently, interaction energies for the different dimer structural models of wild-type and mutant NS4B molecules were calculated. Positive energy differences were observed for the mutant-dimers in comparison to the wild-type homodimer. Here, the NS4B bZIP wild-type homodimer represented the most favourable 3D-structural model with the lowest interaction energy, whereas the mutant-homodimer was the least stable model. The heterodimer of a wild-type and a mutant NS4B bZIP motif showed an intermediate energy value. When investigating the different 3D-structural models visually, it was observed that the side-chain packaging within the bZIP hydrophobic core was the densest in the wild-type homodimer with also the largest size of its interaction surface, whereas both quantities, the side-chain packaging and the interaction surfaces, became gradually smaller for the heterodimer towards the mutant-homodimer, confirming the energetic observations. Thus, we proposed the bZIP motif of NS4B to be crucial for protein–protein interactions and homodimerization.
Subsequently, an in vitro approach was applied to verify our in silico data. Protein–protein interaction of NS4B molecules among each other was shown upon co-expression and co-immunoprecipitation for plasmids containing the bZIP wild-type sequence. Furthermore, the question was addressed whether this interaction could be altered by selective modification of the aminoterminal bZIP functionality by side-directed mutagenesis. Non-conservative amino acid substitutions, for example, helix-braking amino acids, were considered to likely disturb the aminoterminal α-helical structure. Therefore, we used only conservative amino acid substitutions at different sites, each with a minor effect on physico-chemical properties at a given position. All substitutions were chosen to ensure the integrity of the α-helical structure as only change-to-alanine substitutions were performed. It seemed unlikely that a single change-to-alanine substitution would impair the bZIP functionality. Thus, several amino acid substitutions together have been performed to alter the bZIP functionality as suggested by the previous calculation of interaction energies. For further investigation of this issue, successive co-immunoprecipitations of NS4B proteins containing the wild-type or the mutated bZIP motif were performed. A protein–protein interaction of HCV NS4B was shown by co-immunoprecipitation experiments and apparently attenuated upon combining NS4B wild-type with mutant molecules or largely abolished when only mutant NS4B bZIP motifs were present in the co-immunoprecipitation assay. The bZIP motif of HCV NS4B is located in a region with an amphipathic α-helix, mediating membrane association.7,17 Gouttenoire et al. proposed transmembrane orientation of the amphipathic α-helix between amino acid 42 and 66, likely associated with oligomerization. They suggested this segment as a potential multifunctional site.17 These findings are in accordance with our results. Furthermore, it has been reported that the aminoterminal part of HCV-NS4B, containing the first 33 amino acids, has the ability to translocate at least partially towards the ER lumen.13,15 Thus, bZIP-mediated NS4B protein interactions with other host and viral factors may depend on different localization of NS4B during the viral life cycle. Nevertheless, it must be discussed whether dimerization of NS4B is mediated solely by the bZIP motif or if other structural elements are involved. The presence of an amphipathic α-helix and a bZIP motif within the same protein region is no discrepancy. In the current study, site-directed mutagenesis experiments were chosen to maintain the α-helix structure of this region and to specifically abolish the bZIP function. Therefore, based on our experimental data, we concluded NS4B dimerization to be specifically mediated by its bZIP motif.
The relevance of the NS4B bZIP motif for viral replication efficiency and the formation of the ER membranous web should be further investigated using replicon and cell culture systems, as well as biochemical und fluorescence microscopy techniques. Within additional in vitro mutagenesis experiments at specific sites within the bZIP region subsequent in silico analysis of the molecular effects using molecular dynamics simulations of the bZIP dimer should be performed.
In summary, we identified a specific protein–protein interaction of HCV-NS4B molecules via an aminoterminal bZIP motif. The existence of such a motif within HCV-NS4B was formerly only predicted using a bioinformatics approach. Our in silico and in vitro data are in agreement with previous publications, reporting polymerization of different NS4B molecules.22 It cannot be excluded yet that NS4B molecules may also form multimers. Furthermore, there may be uncertainty as to exclude a yet unidentified additional binding partner, mediating the NS4B-NS4B interaction observed in this study. Nevertheless, we provide the first experimental evidence for the protein–protein interaction facilitating bZIP motif within HCV NS4B. This protein–protein interaction largely depends on the integrity of the hydrophobic core within the leucine zipper and can be abolished due to an exchange of crucial residues at specific sites within the structural motif. We could show that NS4B-homodimerization can be gradually alleviated from the wild-type towards the mutant NS4B-dimer. A multitude of different protein–protein interactions is known to be mediated by bZIP motifs in general. Therefore, we suspect that the bZIP motif maybe enables HCV NS4B to act as a protein hub for interactions with other host and viral factors. Screening for bZIP motifs might be an appropriate approach to identify proteins important for the HCV replication cycle.
Material and Methods
Multiple sequence analysis of the NS4B bZIP motif
We used the sequence of a previously predicted bZIP motif for further analysis.12 We retrieved 614 NS4B sequences from public HCV databases, UniProtKB (http://au.expasy.org/uniprot/)31 and euHCVdb (http://euhcvdb.ibcp.fr/euHCVdb/),32 487 of HCV genotype 1, 48 of HCV genotype 2, 10 of HCV genotype 3, and 69 of HCV genotypes 4, 5 and 6. Sequence alignments were computed using CLUSTAL W33 and MUSCLE,34 and subsequently improved by minor manual modifications using the SEAVIEW alignment editor.35 WebLogo (http://weblogo.berkeley.edu/logo.cgi) was applied for computing the bZIP sequence motif for HCV genotypes 1 to 6 (Fig. 2 and Supporting Information Fig. 1). In general, the bZIP motif displays a pattern of amino acids with hydrophilic and hydrophobic properties, which repeats every seven residues. Residues are typically labelled a to g, apolar amino acids are at positions a and d, and aliphatic or aromatic amino acids at the remaining positions. The leucine at position d is characteristic and highly conserved in most eukaryotic bZIP motifs. Viral proteins are more diverse in evolutionary terms than bacterial or eukaryotic proteins. The NS4B bZIP motif shows a lower frequency of characteristic amino acids but with a high proportion of conserved physico-chemical properties.12
The sequence analysis was used to analyze the amino acid variability within and among different HCV genotypes, as well as to select specific sites within the bZIP motif of HCV genotype 1 for site-directed mutagenesis. The modified sequence was based on the aminoterminal bZIP pattern of HCV genotype 1b, prototype HCVJ.25 We performed multiple, but conservative amino acid mutations at different positions within the bZIP motif to disturb the protein–protein binding functionality while preserving the α-helix structure of this region. Therefore, an alanine transfer was applied for amino acid substitutions at the selected sites.
Homology modeling of the NS4B bZIP dimer structure
Homology modeling was applied to predict a structural model for schematic representation of the homodimer interaction. The model was subsequently used to select amino acid positions for further site-directed mutagenesis experiments. Thereby, we used a published bZIP motif within an amphipathic α-helix of the NS4B aminoterminal part for subsequent homology modelling.12 The bZIP motif was previously predicted using several state-of-the-art in silico methods for transmembrane and secondary structure prediction, as well as consensus procedures. Pairwise sequence-structure alignments of the bZIP sequences of the prototype HCVJ (wild-type) and the mutant sequence with the template structure 1JUN retrieved from Protein Data Bank RCSB PDB (http://www.pdb.org)36 were performed to model the 3D protein structure of the NS4B bZIP wild-type homodimer (Fig. 3), heterodimer (wild-type with mutant bZIP) and the mutant-dimer.36 The template 1JUN is a high-resolution NMR solution structure from the leucine zipper domain of the c-Jun homodimer, a eukaryotic transcription factor. The template was primarily selected because of its homodimer structure. Though the number of identical amino acids between the model and the template sequence is low, the physico-chemical properties are rather conserved. Each alignment was submitted to the WHAT IF (http://swift.cmbi.ru.nl/servers/html/index.html) homology modeling server. The protein structure images were drawn in the Accelrys Discovery Studio ViewerLite (http://www.accelrys.com/).
Calculation of interaction energies
Calculation of interaction energies was aimed in planning subsequent in vitro experiments, not for absolute accuracy of the resulting energies. The calculation of interaction energies for homology modeled 3D-structures of the wild-type dimer, the heterodimer and the mutant-dimer structures was performed by computing minimized energies using the OPLS-AA force field.37 Hereby, solvent effects were approximated by the AGBNP implicit solvent model.38 Afterwards, the interaction energies were calculated between the two chains as well as the interaction energies between all residues mutated in the hetero- and the mutant-dimers (amino acid positions 25, 26, 33, 39, 40, 42, 46, 47, and 49 within NS4B; Fig. 4). The OPLS-AA force field was used for all energy calculations.
Sequencing
Automated sequencing (Applied Biosystems 310 and 3100 DNA Sequencer, Weiterstadt, Germany) was done after labelling of PCR products by performance of one-sided PCR in a GeneAmp™ PE9700 thermocycler (Perkin Elmer, Applied Biosystems, Weiterstadt, Germany) by use of commercial vector primers (pcDNA3T7F, pcDNA3BGHR) according to manufacturer's instructions (Big Dye® Deoxy Terminators, Applied Biosystems, Weiterstadt, Germany).
Site-directed mutagenesis
Site-directed mutagenesis was used to modify residues important for (a) the bZIP hydrophobic core with its leucine-leucine stacking interactions (heptad positions d) and (b) residues at positions important for putative ionic interactions (heptad positions e and g). The new sequence was calculated to (a) disturb the hydrophobic core essential for bZIP functionality and (b) preserve the amphipathic α-helical structure in this region. The nucleotide sequence coding for the modified amino acid bZIP motif was implemented in the NS4B plasmid by repeatedly performed site-directed mutagenesis (QuickChange® Site-Directed Mutagenesis Kit, Stratagene, Heidelberg, Germany). To ensure correct detection of the protein, two different oligonucleotides coding for the FLAG-tag and the MYC-tag were inserted into the native NS4B full-length expression plasmid as well as into the modified NS4B full-length plasmid, performing additional site-directed mutagenesis steps. Tags were inserted at the carboxyterminal end to minimize the risk of potential interference with the aminoterminal bZIP motif.
Cell culture and transfection
The expression plasmid containing the cDNA of a full-length HCV genotype 1b NS4B nucleotide sequence was kindly provided by Professor Hak Hotta from the Kobe University, Kobe, Japan.23 Expression of wild-type NS4B proteins with intact bZIP and integrated tags (NS4B-bZIP+/MYC; NS4B-bZIP+/FLAG) and mutant bZIP and integrated tags (NS4B-bZIP-/MYC; NS4B-bZIP-/FLAG) was performed for a single given protein or for combination of different proteins. Approximately 106 HEK 293T cells were transfected with 3μg plasmid DNA suspended in 3μl lipofectamine™ 2000 (Invitrogen, Karlsruhe, Germany) and 150 μl medium (OptiMem®, Invitrogen, Karlsruhe, Germany) for 28 hours. Cells were lysed in 200 μl lysis buffer (1% NP-40, 150mM NaCl, 10mM Tris-HCl pH 7.5, 1mM EDTA, 1mM PMSF, 0.5mM DTT), and insoluble material was removed by centrifugation.
(Co-)immunoprecipitation and immunoblotting
Antibodies (c-Myc 9E19 or OctAProbe F1, both Santa Cruz, Heidelberg, Germany) were allowed to attach for a minimum of 2 hours at +4°C, followed by the addition of protein G-Sepharose (Pharmacia, Karlsruhe, Germany), and completion of the incubation for additional 2 hours at +4°C. Proteins were separated by sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE). After dilution in 30 μl loading buffer (Roti®-Load 1, Carl Roth GmbH, Karlsruhe, Germany) and boiling for 3 min, the samples were separated by SDS-PAGE and electrophoretic transferred to nitrocellulose membranes (Whatman Schleicher & Schell, Dassel, Germany). Non-specific binding sites were blocked by incubation in 5% skim milk in buffer TBST (150mM NaCl; 30mM Tris, pH 7.4; 0.05% Tween20) for 1 hour. Primary antibodies were allowed to bind for a minimum of 2 hours. Detection of immune complexes was performed using horse raddish peroxidase-conjugated antibodies (Bio Rad Laboratories GmbH, München, Germany) and the enhanced chemiluminescence (Pierce, Rockford).
Acknowledgments
The authors thank Hak Hotta from Kobe University, Kobe, Japan, for providing the NS4B expression plasmid. They thank Barbara Schönberger and Guido Plotz for technical advice, Yolanda Martinez for technical assistance, and also appreciates Benno Wölk from the Medizinische Hochschule Hannover (MHH), Hannover, Germany for his helpful discussions.
Glossary
Abbreviations:
- ATF
activating transcription factor
- bZIP
basic leucine zipper
- ER
endoplasmatic reticulum
- HCV
hepatitis C virus
- min
minutes
- NS
nonstructural
- RT-PCR
reverse transcription-polymerase chain reaction
- sec
seconds
- TMD
transmembrane domain.
Supplementary material
References
- 1.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. Hepatitis C—global prevalence (update) Wkly Epidemiol Rec. 2000;75:18–19. [PubMed] [Google Scholar]
- 3.Kato N. Molecular virology of hepatitis C virus. Acta Med Okayama. 2001;55:133–159. doi: 10.18926/AMO/32025. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5:453–463. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- 6.Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 7.Elazar M, Liu P, Rice CM, Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78:11393–11400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lohmann V, Hoffmann S, Herian U, Penin F, Bartenschlager R. Viral and cellular determinants of hepatitis C virus RNA replication in cell culture. J Virol. 2003;77:3007–3019. doi: 10.1128/JVI.77.5.3007-3019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blight KJ, McKeating JA, Rice CM. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol. 2002;76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grobler JA, Markel EJ, Fay JF, Graham DJ, Simcoe AL, Ludmerer SW, Murray EM, Migliaccio G, Flores OA. Identification of a key determinant of hepatitis C virus cell culture adaptation in domain II of NS3 helicase. J Biol Chem. 2003;278:16741–16746. doi: 10.1074/jbc.M212602200. [DOI] [PubMed] [Google Scholar]
- 11.Krieger N, Lohmann V, Bartenschlager R. Enhancement of hepatitis C virus RNA replication by cell culture-adaptive mutations. J Virol. 2001;75:4614–4624. doi: 10.1128/JVI.75.10.4614-4624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Welsch C, Albrecht M, Maydt J, Herrmann E, Welker MW, Sarrazin C, Scheidig A, Lengauer T, Zeuzem S. Structural and functional comparison of the non-structural protein 4B in flaviviridae. J Mol Graph Model. 2007;26:546–557. doi: 10.1016/j.jmgm.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 13.Lundin M, Monne M, Widell A, Von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77:5428–5438. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gouttenoire J, Penin F, Moradpour D. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev Med Virol. 2010;20:117–129. doi: 10.1002/rmv.640. [DOI] [PubMed] [Google Scholar]
- 15.Lundin M, Lindstrom H, Gronwall C, Persson MA. Dual topology of the processed hepatitis C virus protein NS4B is influenced by the NS5A protein. J Gen Virol. 2006;87:3263–3272. doi: 10.1099/vir.0.82211-0. [DOI] [PubMed] [Google Scholar]
- 16.Cho NJ, Dvory-Sobol H, Lee C, Cho SJ, Bryson P, Masek M, Elazar M, Frank CW, Glenn JS. Identification of a class of HCV inhibitors directed against the nonstructural protein NS4B. Sci Transl Med. 2010;2:15ra6. doi: 10.1126/scitranslmed.3000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouttenoire J, Castet V, Montserret R, Arora N, Raussens V, Ruysschaert JM, Diesis E, Blum HE, Penin F, Moradpour D. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J Virol. 2009;83:6257–6268. doi: 10.1128/JVI.02663-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hugle T, Fehrmann F, Bieck E, Kohara M, Krausslich HG, Rice CM, Blum HE, Moradpour D. The hepatitis C virus nonstructural protein 4B is an integral endoplasmic reticulum membrane protein. Virology. 2001;284:70–81. doi: 10.1006/viro.2001.0873. [DOI] [PubMed] [Google Scholar]
- 19.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76:5974–5984. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77:5487–5492. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao L, Aizaki H, He JW, Lai MM. Interactions between viral nonstructural proteins and host protein hVAP-33 mediate the formation of hepatitis C virus RNA replication complex on lipid raft. J Virol. 2004;78:3480–3488. doi: 10.1128/JVI.78.7.3480-3488.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu GY, Lee KJ, Gao L, Lai MM. Palmitoylation and polymerization of hepatitis C virus NS4B protein. J Virol. 2006;80:6013–6023. doi: 10.1128/JVI.00053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong WY, Nagano-Fujii M, Hidajat R, Deng L, Takigawa Y, Hotta H. Physical interaction between hepatitis C virus NS4B protein and CREB-RP/ATF6beta. Biochem Biophys Res Commun. 2002;299:366–372. doi: 10.1016/s0006-291x(02)02638-4. [DOI] [PubMed] [Google Scholar]
- 24.Strauss HM, Keller S. Pharmacological interference with protein–protein interactions mediated by coiled-coil motifs. Handb Exp Pharmacol. 2008;186:461–482. doi: 10.1007/978-3-540-72843-6_19. [DOI] [PubMed] [Google Scholar]
- 25.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Jones DM, Patel AH, Targett-Adams P, McLauchlan J. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J Virol. 2009;83:2163–2177. doi: 10.1128/JVI.01885-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Einav S, Elazar M, Danieli T, Glenn JS. A nucleotide binding motif in hepatitis C virus (HCV) NS4B mediates HCV RNA replication. J Virol. 2004;78:11288–11295. doi: 10.1128/JVI.78.20.11288-11295.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paredes AM, Blight KJ. A genetic interaction between hepatitis C virus NS4B and NS3 is important for RNA replication. J Virol. 2008;82:10671–10683. doi: 10.1128/JVI.00875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Namba K, Naka K, Dansako H, Nozaki A, Ikeda M, Shiratori Y, Shimotohno K, Kato N. Establishment of hepatitis C virus replicon cell lines possessing interferon-resistant phenotype. Biochem Biophys Res Commun. 2004;323:299–309. doi: 10.1016/j.bbrc.2004.08.091. [DOI] [PubMed] [Google Scholar]
- 31.Wu CH, Apweiler R, Bairoch A, Natale DA, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ, Mazumder R, O'Donovan C, Redaschi N, Suzek B. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Combet C, Garnier N, Charavay C, Grando D, Crisan D, Lopez J, Dehne-Garcia A, Geourjon C, Bettler E, Hulo C, Le Mercier P, Bartenschlager R, Diepolder H, Moradpour D, Pawlotsky JM, Rice CM, Trepo C, Penin F, Deleage G. euHCVdb: the European hepatitis C virus database. Nucleic Acids Res. 2007;35:D363–D366. doi: 10.1093/nar/gkl970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Galtier N, Gouy M, Gautier C. SEAVIEW and PHYLO_Wtwo graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- 36.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 38.Gallicchio E, Levy RM. AGBNP: an analytic implicit solvent model suitable for molecular dynamics simulations and high-resolution modeling. J Comput Chem. 2004;25:479–499. doi: 10.1002/jcc.10400. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


