Abstract
In recent years, microRNAs have received greater attention in cancer research. These small, non-coding RNAs could inhibit target gene expression by binding to the 3′ untranslated region of target mRNA, resulting in either mRNA degradation or inhibition of translation. miRNAs play important roles in many normal biological processes; however, studies have also shown that aberrant miRNA expression is correlated with the development and progression of cancers. The miRNAs could have oncogenic or tumor suppressor activities. Moreover, some miRNAs could regulate formation of cancer stem cells and epithelial-mesenchymal transition phenotype of cancer cells which are typically drug resistant. Furthermore, miRNAs could be used as biomarkers for diagnosis and prognosis, and thus miRNAs are becoming emerging targets for cancer therapy. Recent studies have shown that natural agents including curcumin, isoflavone, indole-3-carbinol, 3,3′-diindolylmethane, (−)-epigallocatechin-3-gallate, resveratrol, etc. could alter miRNA expression profiles, leading to the inhibition of cancer cell growth, induction of apoptosis, reversal of epithelial-mesenchymal transition, or enhancement of efficacy of conventional cancer therapeutics. These emerging results clearly suggest that specific targeting of miRNAs by natural agents could open newer avenues for complete eradication of tumors by killing the drug-resistant cells to improve survival outcome in patients diagnosed with malignancies.
Keywords: cancer therapy, chemoprevention, microRNA, natural agents
INTRODUCTION
A group of small non-coding RNAs commonly known as microRNAs (miRNAs) are endogenous single-stranded RNAs of 19–25 nucleotides (~22nt) in length that regulate gene expression by multiple mechanisms. The first miRNA, lin-4, was discovered in 1993 when conducting a genetic analysis in Caenorhabditis elegans (1,2). The lin-4 was found to contain sequences complementary to a repeated sequence element in the 3′ untranslated region (UTR) of lin-14 mRNA, suggesting that miRNA lin-4 could regulate lin-14 mRNA translation via an antisense RNA-RNA interaction (1,2). Several years later, another important miRNA, let-7, was discovered also in Caenorhabditis elegans (3). The let-7 was found to be a 21 nucleotide miRNA that was complementary to elements in the 3′ UTR of the heterochronic genes lin-14, lin-28, lin-41, lin-42 and daf-12 (3). By binding and inhibiting these genes, let-7 could regulate developmental timing in Caenorhabditis elegans. The homologs of the let-7 gene and let-7 miRNA were soon identified in the human, Drosophila, and other animals (4). Since then, more miRNAs have been discovered, and their functions have been investigated as reviewed recently by us and others (5,6). It has been reported that miRNAs are involved in the regulation of many physiological and pathological processes in human, animals, and plants by imperfectly binding to the 3′UTR of target mRNAs leading to translational repression or by target mRNA cleavage because of the imperfect complementarity between miRNA and mRNA.
Emerging evidence suggest that miRNAs play important roles in the development and progression of cancer. Moreover, experimental results have implicated the critical role of miRNAs in the formation of cancer stem cells (CSCs) and the acquisition of epithelial-mesenchymal transition (EMT) of cancer cells which are critically associated with drug resistance and cancer metastasis. Therefore, targeting specific miRNAs could be a novel therapeutic approach for the treatment of cancers, especially by eliminating CSCs or EMT-type cells that are typically drug resistant. Interestingly, it has been reported that miRNAs could be regulated by natural chemopreventive agents (natural agents), leading to the inhibition of cancer cell growth, EMT, drug resistance, and metastasis (7–11). These novel findings suggest that the use of natural agents could open newer avenues for the successful treatment of cancers, especially by combining conventional therapeutics with natural chemopreventive agents that are known to be non-toxic to humans. In this review, we will summarize the current knowledge regarding miRNA biogenesis, miRNAs in cancer, and the novel strategies for targeting miRNAs by natural agents toward better treatment outcome of patients diagnosed with cancers.
BIOGENESIS OF miRNA AND ITS REGULATION ON GENE EXPRESSION
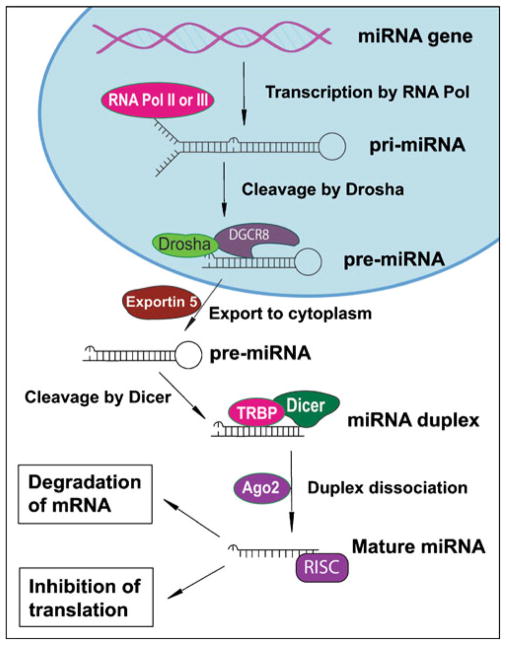
Most miRNA genes are located in intergenic regions more than one kilobase away from annotated genes. Transcription of miRNA genes is mediated by either RNA polymerase II or RNA polymerase III (Fig. 1). The transcription yields primary miRNA (pri-miRNA), which is hundreds to thousands of nucleotides in length (12,13). Most human pri-miRNAs contain a hairpin stem of 33 base-pairs, a terminal loop, and two single-stranded flanking regions upstream and downstream of the hairpin. To become mature miRNA, pri-miRNA undergoes several cleavage and modification steps. The pri-miRNA is first cleaved by the nuclear microprocessor complex formed by the RNase III (Drosha) and the DGCR8 (DiGeorge critical region 8) protein, yielding the precursor miRNA (pre-miRNA) which is about 70 nucleotides in length (14–19). In this process, the double-stranded stem and the unpaired flanking regions are critical for DGCR8 binding and Drosha cleavage. The pre-miRNA is then transported to the cytoplasm by the nuclear export factor Exportin 5, which binds the pre-miRNA (20,21). In the cytoplasm, another RNase III (Dicer) in complex with the double-stranded RNA-binding protein TRBP cleaves the pre-miRNA hairpin and yields an RNA duplex about 22 nucleotides in length (13). Then, under the mediation of Ago2, the RNA duplex is dissociated to single strand: functional guide strand which is complementary to the target mRNA, and the passenger strand, which is degraded (13,22,23). The functional guide strand, mature miRNA, is incorporated into the RNA-induced silencing complex (RISC) and guides RISC to target mRNA (13,22,23). It is commonly accepted that mature miRNAs regulate gene expression by binding to the 3′ untranslated region (3′UTR) of target mRNA, causing degradation of mRNA or inhibition of their translation to functional proteins (Fig. 1) (13,24). However, how the RISC-miRNA complex actually triggers the silencing of mRNA gene expression requires further detailed investigation.
Fig. 1.
Biogenesis of miRNA and its effect on gene expression.
ROLE OF miRNAs IN CANCER
It has been found that most miRNAs are conserved across species (25), indicating that they participate in normal biological processes. Indeed, the expression of miRNAs has been recognized as an integral component of many normal biological behaviors. Experimental studies have shown that miRNAs play important roles in cell proliferation, differentiation, apoptotic cell death, stress resistance, and physiological metabolism (26). However, emerging evidences have also shown that miRNAs are critically involved in the development and progression of cancers (5,6). The aberrant expression of miRNAs has been associated with the stage, progression, and metastasis of cancers (27). Therefore, miRNAs could function as oncogenes or tumor suppressor genes, regulating cellular behavior of cancers. It is known that miRNAs participate in the control of embryonic stem cell differentiation (28–30). More importantly, miRNAs have been found to regulate cancer stem cells and EMT-type cells, which are responsible for drug resistance and cancer metastasis (31–36). The major miRNAs that play important roles in cancers are discussed in the following sections and listed in Table 1 and Fig. 2.
Table 1.
List of miRNAs which have Tumor Suppressor or Oncogenic Activity
| miRNA | Targeted molecules | Altered in tumors | References |
|---|---|---|---|
| Tumor suppressor activity | |||
| let-7 | RAS, PRDM1, HMGA2, c-Myc, E2F, cyclin D2 | Lung, ovarian cancer, breast, and colorectal cancers | (60–64) |
| miR-15/miR-16 | Bcl-2, Wt-1 | Gastric and lung cancers | (65–67) |
| miR-34 | E2F3, Notch1, CDK4, CDK5 | Breast, pancreatic, colon cancers | (31,68,69) |
| miR-17-5p | AIB1, E2F1, p21, BIM | Breast cancer, CLLa | (123–126) |
| miR-29 | MCL-1, TCL-1, DNMT3s | Lung and breast cancers, CLLa, AMLa | (124,127–129) |
| miR-124a | CDK6 | ALLa, medulloblastoma | (130,131) |
| miR-127 | Bcl-6 | Breast cancer, lymphoma | (123) |
| miR-143 | Ras, ERK5 | Gastric and prostate cancers | (132–134) |
| miR-145 | Mucin1, ERG | Breast and gastric cancers | (133,135–138) |
| miR-181 | TCL-1, E2F5, eIF5A | Colorectal cancer | (129) |
| Oncogenic activity | |||
| miR-21 | PTEN, TPM1, Pdcd4, maspin | Colon, breast, pancreatic, lung, prostate, liver, and gastric cancers | (37–48) |
| miR-155 | AT1R, TP53INP1 | Lung and breast cancers, CLLa, AMLa | (37,55–59) |
| miR-17-92 | Tsp1, CTGF, E2F1, AIB1, TGFBR2 | Lung, breast, colon, gastric, pancreatic cancers, lymphomas | (49–54) |
| miR-106a | RB-1 | Colon and gastric cancers | (139,140) |
| miR-373 | LATS2 | Testicular tumor, Gastric cancer | (141–143) |
| miR-197 | ACVR1, TSPAN3, FUS1 | Lung cancer | (144) |
| miR-221 | KIT, p27(Kip1), p57, PTEN | Breast and liver cancers, CLL | (43) |
| miR-222 | KIT, p27(Kip1), p57, PTEN | Breast and liver cancers, CLL | (57) |
| miR-372 | LATS2 | Testicular tumor | (143) |
CLL chronic lymphoblastic leukemia; AML acute myeloid leukemia; ALL acute lymphoblastic leukemia.
Fig. 2.
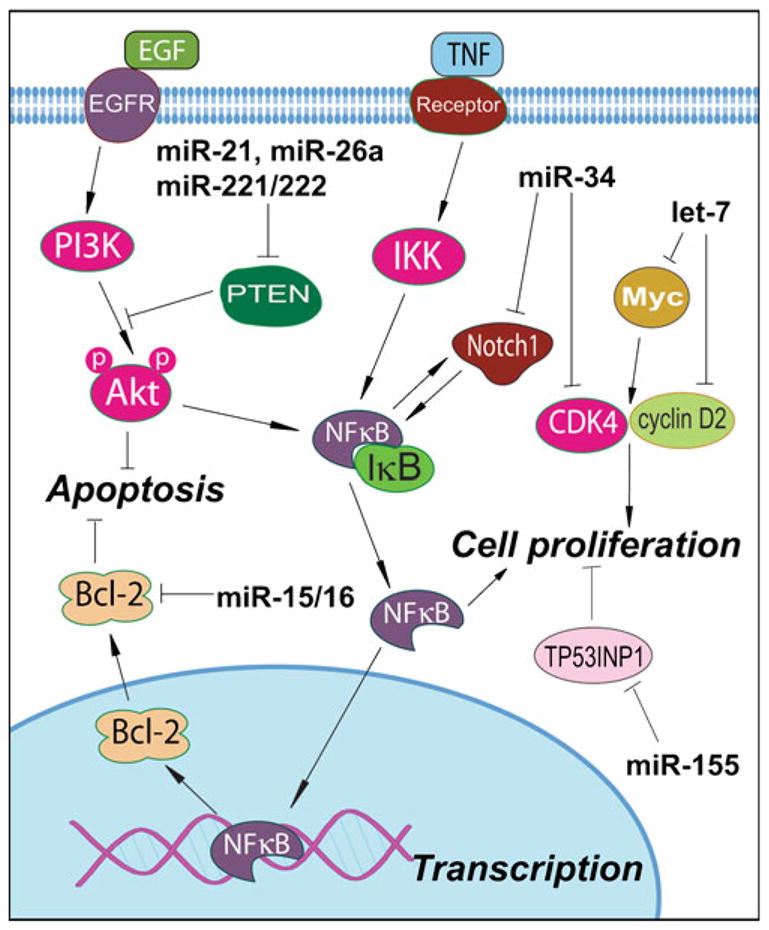
Regulation of cellular signaling by major miRNAs that have tumor suppressor or oncogenic activity.
THE FUNCTIONAL ROLE OF miRNAs AS ONCOGENES
Among many miRNAs, the miR-21 is a well-known miRNA with oncogenic activity (Fig. 2). It has been found that miR-21 is up-regulated in various cancers (37–42). Studies have shown that inhibition of miR-21 caused an increase in apoptosis due to activation of caspases in human glioblastoma cells, indicating the anti-apoptotic role of miR-21 (39). In breast cancer cells, knock-down of miR-21 also caused inhibition of cell growth, down-regulation of Bcl-2, and induction of apoptosis in vitro and in vivo (40). Similar results have been observed in other types of cancer (41–44). These studies suggest that miR-21 is oncogenic; however, no target gene of miR-21 was investigated in these studies. In a study investigating the targets of miR-21, results showed that the 3′UTR of Pdcd4, a novel tumor suppressor, contained the sequence complementary to the sequence of miR-21, suggesting that Pdcd4 is a target of miR-21. Further studies have shown that Pdcd4 was inversely correlated with miR-21 levels in colorectal cancer cell lines and tumor tissues. Moreover, cells transfected with anti-miR-21 showed increased Pdcd4 expression (45), demonstrating that Pdcd4 is a target of miR-21. Recently, several reports confirmed this finding and showed that other molecules, such as bone morphogenetic protein receptor II (BMPRII) and LRRFIP1 (an inhibitor of NF-κB signaling) are also the targets of miR-21 (46–48). Several other targets of miR-21 have been summarized in recent review articles published by us and others (5,6).
Another class of miRNA such as miR-17-92 cluster consists of miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92-1. The miR-17-92 cluster also showed oncogenic activity in various cancers (49–51). Animal study has shown that forced expression of the miR-17-92 cluster and c-myc accelerated tumor development in a mouse B-cell lymphoma model (49). Dews et al. have reported similar results showing that Myc-induced up-regulation of miR-17-92 cluster enhanced tumorigenesis and angiogenesis (52). Recent reports also showed that miR-17-92 was up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors, suggesting that miR-17-92 cluster involved intimately with the sonic hedgehog pathway in cancer development (53,54). These results also suggest that the biological effects of the miR-17-92 cluster are oncogenic (5).
The miR-155 is another oncogenic miRNA (Fig. 2). Studies using miRNA expression profiling have found increased expression of miR-155 in various cancers (37,55–58). Greither et al. have observed a significant correlation between elevated miR-155 expression and low overall survival (p=0.005) in pancreatic cancers (57). Furthermore, patients demonstrating elevated miR-155 expression level in tumor tissue possessed a 6.2-fold increased risk of tumor-related death compared to patients whose tumors showed a lower expression of miR-155 (57). Moreover, aberrant expression of miR-155 has been believed to serve as a biomarker of early pancreatic neoplasia (58). In a study identifying specific targets of miR-155, tumor suppressor protein 53-induced nuclear protein 1 (TP53INP1) has been found to be significantly reduced in pancreatic ductal adenocarcinoma. Transfection with an anti-miR-155 oligonucleotide into pancreatic cancer caused re-expression of TP53INP1 and a significant increase in apoptosis, demonstrating that TP53INP1 is a target of miR-155 (59). In short, the above-mentioned studies clearly suggest the role of some miRNAs as oncogenic (5); however, further research in this area is likely to witness rapid growth as to the role of numerous miRNAs that may be functioning as oncogenic in human malignancies.
THE FUNCTIONAL ROLE OF miRNAs AS TUMOR SUPPRESSORS
It has been found that some miRNAs have tumor-suppressor activity and are down-regulated in cancer (5). One of the most studied miRNA groups with tumor suppressor activity is the let-7 family (Fig. 2). The let-7 family consists of let-7, miR-48, miR-84, and miR-241. Studies on let-7 showed that its expression was down-regulated in various cancers and that let-7 could be used as biological markers for cancer detection and prognosis (60–63). In a study investigating the expression levels of let-7, let-7 has been found to be down-regulated in human lung cancer cell lines and primary human lung cancer tissues (61). Importantly, clinical study showed that the reduced let-7 expression was correlated to shorter post-operative survival in patients diagnosed with lung cancer. The in vitro study showed over-expression of let-7 in the A549 lung cancer cell line, resulting in a 78.6% reduction in the number of colonies (61), suggesting the tumor suppressor effects of let-7. Further research on let-7 showed that human Ras 3′UTRs contain multiple sites complementary to the sequence of let-7 family, suggesting that Ras could be a target of let-7. Moreover, transfection of HepG2 cells with let-7 caused about 70% reduction of Ras, whereas transfection of HeLa cells with anti-sense let-7 molecules resulted in the reduced let-7 level and the increase in RAS level (64), demonstrating that Ras is the target of let-7. Moreover, further studies showed that HMGA2 is another target of let-7 (62).
In addition to let-7, miR-15 and miR-16 have been known to have tumor-suppressor activity (Fig. 2). Recent studies have shown that transient transfection with synthetic miR-16 significantly reduced cell proliferation of 22Rv1, Du145, PPC-1, and PC-3M-luc prostate cancer cells (65). The in vitro studies indicated that miR-16 is likely involved in the suppression of prostate cancer growth by regulating the expression of CDK1 and CDK2, which are associated with cell cycle control and proliferation (65). It has also been reported that the levels of miR-15 and miR-16 were inversely correlated with bcl-2 expression in CLL cells (66). Furthermore, transfection of miR-15 and miR-16 caused a significant reduction in bcl-2 expression and the induction of apoptosis (66), suggesting the effects of miR-15 and miR-16 on apoptotic signaling. In prostate cancer cells, miR-15a and miR-16-1 cluster also targeted CCND1 (encoding cyclin D1) and WNT3A, which promotes several tumorigenic features, such as survival, proliferation and invasion (67). These results suggest the tumor suppressor activity of miR-15 and miR-16.
Another important miRNA, miR-34 has been found to participate in p53 network showing tumor suppressor activity. In an in vitro study, the structure of the miR-34a primary transcript and promoter has been characterized, and the authors have found that miR-34a could be directly transactivated by p53 (68). Other reports also showed similar observation documenting that miR-34 could be induced by p53 signaling, suggesting that miR-34 has tumor suppressor activity, and thus it is an important molecule in the p53 tumor suppressor network (68,69). The increased expression of miR-34a could result in increased apoptosis and also cause changes in the expression of genes related to cell cycle progression, apoptosis, DNA repair, and angiogenesis (68). A recent study showed that miR-34 could inhibit human pancreatic cancer tumor-initiating cells and restore the tumor-suppressing function of the p53 in p53-deficient human pancreatic cancer cells (31), suggesting the anti-cancer effects of miR-34.
THE ROLE OF miRNAs IN THE REGULATION OF CANCER STEM CELLS AND EPITHELIAL-MESENCHYMAL TRANSITION
Emerging evidence suggests the critical role of miRNAs in the formation of cancer stem cells and the acquisition of EMT phenotype. Specific miRNAs have been found to be the regulators of CSCs and EMT through the regulation of EpCAM (ESA), BMI1, E-cadherin and other molecules (32,70,71), and thus miRNAs appear to play important roles in cancer development, EMT, and metastasis (72,73). We have investigated the expression levels of miR-200 and let-7 in EMT phenotypic pancreatic cancer cells (8). We found that miR-200b, miR-200c, and miR-200a were down-regulated in gemcitabine-resistant cells, which is consistent with the EMT phenotype of gemcitabine-resistant cells. We also found that many members of the let-7 family were down-regulated in gemcitabine-resistant cells (EMT-type cells), and these findings are consistent with the aggressiveness of gemcitabine-resistant pancreatic cancer cells (8). In order to investigate the role of miR-200 family in the reversal of EMT phenotype of gemcitabine-resistant cells, we transfected miR-200a, miR-200b, and miR-200c pre-miRNAs into MiaPaCa-2 cells (gemcitabine resistant) for up to 14 days and found that the re-expression of miR-200 family in the MiaPaCa-2 cells resulted in the up-regulation of epithelial marker E-cadherin and down-regulation of mesenchymal markers including ZEB1 and vimentin both at the mRNA and protein levels (8). After 14 days of transfection, the morphology of miR-200-transfected MiaPaCa-2 cells was partially changed from elongated fibroblastoid to epithelial cobblestone-like appearance, and the cells appeared to grow in close contact with each other (8). These results suggest that the loss of miR-200 family is critical for the acquisition of EMT characteristics and that the re-expression of miR-200 could reverse the EMT phenotype of gemcitabine-resistant cells (8). Moreover, we have also found that re-expression of miR-200b in PDGF-D over-expressing cells (EMT-type cells) led to the reversal of EMT phenotype, which was associated with the down-regulation of ZEB1, ZEB2 and Slug expression, and these results were consistent with increased gene expressions of epithelial markers (35). In addition, PDGF-D over-expressing cells when transfected with miR-200b showed inhibition of cell migration and invasion with concomitant repression of cell adhesion to culture surface and cell detachment (35). In another study, miR-34 showed inhibition of human pancreatic cancer tumor-initiating cells as described earlier (31). These findings suggest that miRNAs play important roles in the formation of CSCs and the maintenance of the CSCs niche, and are also associated with the acquisition of EMT phenotype of human cancers.
RELEVANCE OF miRNAs IN CANCER PATIENTS
In recent years, in vitro and in vivo studies have indicated that miRNAs could be used as diagnostic and prognostic markers. Moreover, recent experiments also showed that miRNAs could be novel targets for cancer therapy as detailed below and also indicated in a recent review article published by our laboratory and others (5,6).
miRNA as Diagnostic and Prognostic Markers
Several studies have focused on profiling miRNA expression in various cancers. The results suggest that different types of cancer could be distinguished by miRNA profiling; hence, miRNA profiling could be diagnostically important. Blenkiron et al. examined miRNA expression in 93 primary human breast tumors using a flow cytometric miRNA expression profiling method (74). They classified the breast tumors as luminal A, luminal B, basal-like, HER2+, and normal-like based on the miRNA expression profiling. Further analysis showed that miRNAs were differentially expressed between these tumor-subtypes and that some of miRNAs were associated with clinicopathological features. These authors have also found that miRNAs could be used to classify basal versus luminal tumor subtypes (74), which is likely to provide additional information for pathological diagnosis. Porkka et al. reported the profiling analysis of different types of prostate tumor and prostate cancer cell lines. They found differential expression of 51 miRNAs between benign tumors and carcinoma tumors. Importantly, they were able to successfully distinguish prostate carcinoma from benign prostatic hyperplasia using clustering analysis of the tumor samples (75). Another study on profiling miRNA expression in prostate cancers also showed five miRNAs (miR-23b, –100, –145, –221 and –222) that were significantly down-regulated in prostate cancer (76). Raponi et al. also identified fifteen miRNAs that were differentially expressed between normal lung and lung squamous cell carcinoma (77). Other investigators also found that the alternations in miRNA expression profiles could be used as markers for cancer diagnosis for various cancers (78–80).
It has been reported that miRNAs could also be used as markers for prediction of cancer prognosis (77,81–83). Approximately 50% of all annotated human miRNAs are located in the areas of genome associated with cancer or fragile sites, suggesting that miRNAs could play critical roles in cancer progression (84). Therefore, the alterations in specific miRNAs could predict the prognosis of cancer. In lung squamous cell carcinoma, several miRNAs including miR-155, let-7, and miR-146a have been found to have prognostic value (77). Among them, miR-146b alone was found to have the strongest prediction accuracy for stratifying prognostic groups. These miRNA signatures were also superior in predicting overall survival (77). In another study on lung adenocarcinomas, miRNA expression profiling and univariate/multivariate analysis were conducted. It was found that high miR-155 and low let-7a expression correlated with poor survival (83). In breast cancer, nine miRNAs, including miR-21, miR-365, miR-181b, let-7f, miR-155, miR-29b, miR-181d, miR-98, and miR-29c, were found to be up-regulated, whereas miR-497, miR-31, miR-355, miR-320, miR-127, and miR-30a-3p were down-regulated (82). Importantly, the most significantly up-regulated miRNA was miR-21, which was correlated with advanced tumor stage, lymph node metastasis, and poor survival of the patients, suggesting that miR-21 could serve as a molecular prognostic marker for breast cancer aggressiveness (82). A similar result was observed in pancreatic cancer. Up-regulation of miR-21 was strongly associated with both a high Ki67 proliferation index marker expression and the presence of liver metastasis (85). In addition, miR-196a-2 has been found to be significantly predictive of survival of patients diagnosed with pancreatic cancer (86). These findings clearly demonstrate the value of miRNA expression as diagnostic and prognostic markers for human cancers.
It is interesting to note that miRNAs are maintained in a protected state in serum and plasma. The miRNAs in plasma could be packaged inside exosomes that are secreted from cells; therefore, they are protected from endogenous RNase activity (87). This property of miRNAs allows the detection of miRNA expression directly from human serum. This increases the value of miRNA expression profiling as biomarkers for cancer detection. Circulating miRNA offers unique opportunities for early diagnosis of cancer and prediction of prognosis. The specificity and simplicity of the reported detection of serum/plasma miRNAs are particularly striking, suggesting that serum-based miRNA detection could emerge as revolutionary sources for cancer diagnosis and prognosis. Lodes et al. recently reported an evaluation of miRNA expression patterns in human serum from the patients diagnosed with prostate, colon, ovarian, breast and lung cancers using a miRNA high density microarray (88). When conducting the microarray data analysis for assessing the presence of miRNAs, all human miRNAs could theoretically be analyzed using either fluorescent or electrochemical signals. Further experimental findings showed that sufficient miRNAs were present in one milliliter of serum for detecting all miRNA expression without amplification techniques, and these authors have also successfully used the miRNA expression patterns, which were found to correctly discriminate between normal and cancer patient samples (88). Zhu et al. measured miRNAs in the serum samples and found that they were able to detect endogenous miR-16, –145, and –155 in all serum samples (89). They also found that women with progesterone receptor-positive tumors had higher miR-155 expression than tumors that were negative for these receptors, suggesting that patients with breast cancers could have differentially expressed miR-155 in the serum dependent on hormone sensitivity of breast cancers (89). In addition, miRNA expression patterns in serum/plasma have also been demonstrated in lymphoma, lung, colorectal, and ovarian cancers (90–92), suggesting the value of serum miRNAs in cancer patients. Circulating miRNAs have also been used as potential biomarkers for drug-induced liver injury (93). Using acetaminophen overdose-induced liver injury in mouse as a model system, Wang et al. have found highly significant differences in the spectrum and levels of miRNAs in both liver tissues and plasma between control and overdosed animals. The miR-122 and miR-192 were enriched in the liver tissue and exhibited dose- and exposure duration-dependent changes in the plasma which correlated with the histopathology of liver degeneration, suggesting the value of miRNAs as biomarkers for drug-induced liver injury (93).
miRNA as a Target for Cancer Therapy
Targeting miRNAs for cancer therapy is an emerging field in human cancer. In recent years, researchers have attempted to alter the expression of miRNAs so that the inhibition of cancer growth and the increase in chemotherapeutic drug sensitivity could be achieved. The strategies for the regulation of miRNAs in cancer could include inactivation of oncogenic miRNAs, activation of tumor suppressor miRNAs, and targeting specific miRNAs to restore drug sensitivity. Several studies have shown that anti-sense oligonucleotides could block the function of miRNAs (94–96). Using synthesized anti-miR-21 oligonucleotides, which is believed to be an oncogenic miRNA, the activity of miR-21 was inhibited (95). Importantly, animal studies have shown that intravenous administration of anti-sense oligonucleotides against miR-16, miR-122, miR-192 and miR-194 resulted in a marked reduction of corresponding miRNA levels in liver, lung, kidney, heart, intestine, fat, skin, bone marrow, muscle, ovaries and adrenals (97). This silencing of endogenous miRNAs was specific, efficient and long-lasting. Furthermore, the activity of miR-122 and its downstream genes was altered upon anti-sense oligonucleotide administration (97). These results suggest that silencing of specific oncogenic miRNAs in vivo could represent a therapeutic strategy for cancer treatment. In addition, using synthetic miRNAs to target mRNAs of oncogenes have also been shown to be efficacious in the inhibition of cancer (98).
Re-expression of tumor suppressor miRNAs is another important strategy in the field of cancer treatment. We and others have shown that up-regulation of let-7 tumor suppressor miRNA by natural agents or exogenously transfected pre-let-7 could lead to the inhibition of cell growth of lung, liver, and pancreatic cancer cells (8,60), suggesting the value of restoring tumor suppressor miRNAs in cancer treatment. We and others have also found that let-7 and miR-200 are associated with sensitization of cancer cells to chemotherapeutics, and also to radiation therapy (8,99). Up-regulation of let-7 and miR-200 in lung or pancreatic cancer cells restored their radiosensitivity and chemosensitivity (8,99), suggesting that these strategies could become a novel approach for regulating the miRNAs toward enhancing the efficacy of cancer therapy. It is important to note that using “natural agents” that are non-toxic for the regulation of miRNAs, and, thus, increasing therapeutic efficacy could open newer avenues for successful treatment of cancers, and therefore our current understanding of this field is discussed in the following sections.
REGULATION OF miRNA BY NATURAL AGENTS
Since miRNA has been known to play important roles in the progression of cancer, the formation of cancer stem cell, and the acquisition of EMT phenotype, it is reasonable to believe that targeting miRNA could be a novel strategy for cancer treatment. Indeed, more and more experimental studies have shown that knock-down or re-expression of specific miRNA could induce drug sensitivity, inhibit proliferation of cancer cells, and suppress cancer cell invasion and metastasis. Importantly, recent studies have shown that natural agents, including curcumin, isoflavone, I3C, DIM, EGCG, and others, could alter the expression of specific miRNAs, which may lead to increased sensitivity of cancer cells to conventional agents, and thereby inhibition of tumor growth (Table 2 and Fig. 3). These phenomena are consistent with previous study in animals showing that diets could regulate the expression level of miRNAs with up-regulation of miR-705 and miR-1224, and down-regulation of miR-182, miR-183, and miR-199a-3p (100). Most studies regarding the effects of natural agents on the expression of miRNAs were conducted by using miRNA array to uncover the alterations in miRNA expression profile upon natural agent treatment. The specific miRNAs altered by natural agents were subsequently confirmed by RT-PCR. Moreover, the targets of specific miRNAs were mechanistically investigated to reveal the role of specific miRNAs in the regulation of cellular signaling by natural agents. The changes in biological behavior after re-expression or knock-down of specific miRNAs showed the inhibitory effects of natural agents on cancer cells through the regulation of miRNAs. Considering the non-toxic characteristics of natural agents, targeting miRNAs by natural agents combined with conventional chemotherapeutics could be a novel and safer approach for the treatment of cancer. We believe that such strategy could be better in eradication of drug-resistant cells, such as CSCs and EMT-type cells, toward complete cure of patients diagnosed with human malignancies because the elimination of CSCs or EMT-type cells will lead to the elimination of tumor recurrence.
Table 2.
List of miRNAs Altered by Natural Agents
| Natural agents | Up-regulated miRNA | Down-regulated miRNA | References |
|---|---|---|---|
| Curcumin | miR-22, miR-34a, miR-24, miR-181a, miR-21, miR-181b, miR-27a | miR-199a, miR-510, miR-196a, miR-7, miR-15b, miR-195, miR-374, miR-98 | (11) |
| I3C, DIM | miR-200b, miR-200c, let-7b, let-7c, let-7d, let-7e, miR-663, miR-638, miR-122, miR-149 | miR-21, mir-31, miR-130a, miR-146b, miR-377, miR-20b, miR-654, miR-34c | (8) |
| Isoflavone | miR-200b, miR-200c, let-7b, let-7c, let-7d, let-7e, miR-663, miR-146a, miR-374b | miR-34c, miR-376a, miR-196a, miR-320, miR-654, miR-34c, miR-196 | (7,8) |
| EGCG | let-7, miR-16, miR-18b, miR-20a, miR-25, miR-92, miR-93, miR-221, miR-320 | miR-10a, miR-18a, miR-19a, miR-26b, miR-29b, miR-34b, miR-98, miR-129, miR-181d | (117) |
| Resveratrol | miR-146a | (10) | |
| Alcohol, Vit E, folate and retinoids | miR-10a, miR-10b, miR-9, miR-145, miR-30a-3p, miR-152, miR-122a, miR-125b | miR-200a, miR-496, miR-296, miR-30e-5p, miR-362, miR-339, miR-29c, miR-154, miR-10a | (120–122) |
Fig. 3.
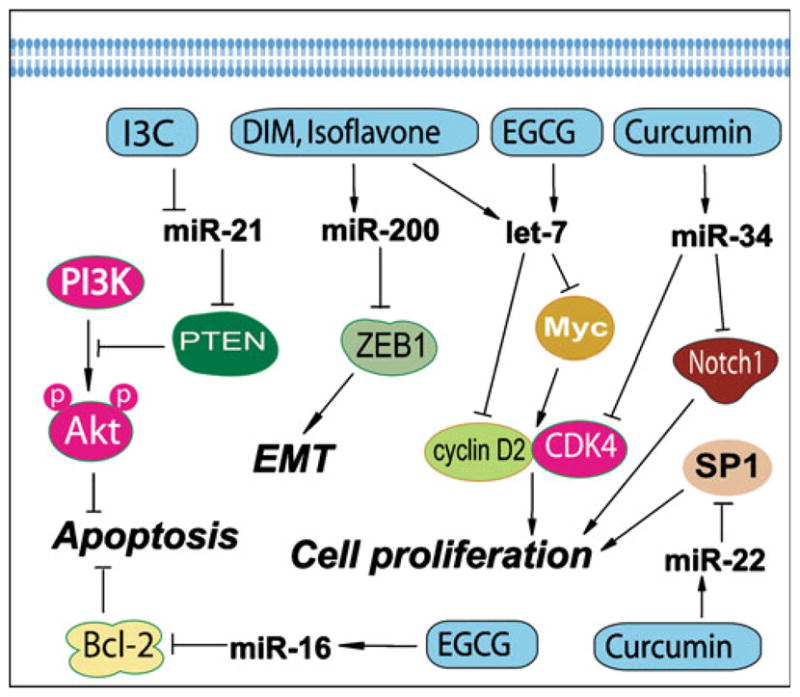
Natural agents alter the expression of miRNAs that are known to regulate cellular signaling and biological behavior.
Indeed, the natural agents and their synthetic analogs have received greater attention in the field of cancer research in recent years. Cancer cells are known to have alterations in multiple cellular signaling pathways. Perhaps this is the reason why specific inhibitors or gene therapies that target only one molecule have typically failed in cancer treatment. Based on the molecular evidence showing that natural agents could target multiple cellular signaling pathways, more and more clinical trials are being conducted to investigate the value of natural agents in human cancer (www.ClinicalTrials.gov). Although these natural compounds are generally consumed fairly regularly, it is important to note that the low water solubility, poor in vivo bioavailability and unacceptable pharmacokinetic profile of these natural agents limit their efficacy as anti-cancer agents for solid tumors (101,102). Therefore, the development of the synthetic analogs of natural agents based on the structure-activity assay and the encapsulation of natural agents with liposome or nanoparticles for enhancing the anti-tumor activity of these natural agents is an exciting area of research. Emerging in vitro and in vivo studies clearly suggest that these analogs and formulations of natural agents could be much more potent for prevention and/or treatment of various cancers (103–105). We believe that cancer chemotherapeutics combined with natural agents that are typically known and chemopreventive agents, which target miRNA and other signaling, could be an important novel strategy for the prevention of tumor progression and/or treatment of cancers.
THE ROLE OF CURCUMIN IN REGULATING miRNAs
Curcumin is a natural compound present in turmeric and has been known to possess both anti-inflammatory and antioxidant effects. Moreover, it has also been studied as a cancer chemopreventive agent in various cancers (106,107). In vitro and in vivo studies have shown its inhibitory effects on cancers through its anti-proliferative and pro-apoptotic activities. It has been known that curcumin regulates the expression of genes that are critically involved in the regulation of cellular signaling pathways, including NF-κB, Akt, MAPK, and other pathways (106,108). These signaling pathways could be regulated by miRNAs. Recently, Sun et al. reported the miRNA expression profiles altered by curcumin in pancreatic cancer cells (11) (Fig. 3 and Table 2). To obtain the miRNA expression profiles, they conducted oligonucleotide microarray analysis in pancreatic cancer cells treated with curcumin. The altered expressions of miRNA on the arrays were validated by real-time PCR. Additionally, potential mRNA targets were analyzed by bioinformatics and further confirmed by flow cytometry experiments. They found that curcumin altered miRNA expression in human pancreatic cancer cells with up-regulation of miR-22 and down-regulation of miR-199a. They also found that up-regulation of miR-22 expression by curcumin or by transfection with pre-miR-22 in BxPC-3 pancreatic cancer cells suppressed the expression of its target genes SP1 transcription factor (SP1) and estrogen receptor 1 (ESR1). Moreover, inhibition of miRNA-22 by anti-sense oligonucleotide enhanced SP1 and ESR1 expression, demonstrating that SP1 and ESR1 are the legitimate target genes of miR-22 (11). These results suggest that curcumin could inhibit the proliferation of pancreatic cancer cells through the regulation of specific miRNAs; however, further in-depth research is needed in order to fully appreciate the beneficial effects of curcumin.
THE ROLE OF ISOFLAVONES IN THE REGULATIION OF miRNAs
Soy isoflavones such as genistein, daidzein, and glycitein are mainly derived from soybean. Isoflavone genistein has been found to inhibit cancer cell growth, invasion, and metastasis in vivo and in vitro (109–111). These effects of isoflavone could be mediated via the regulation of miRNA (Table 2 and Fig. 3). We have investigated the regulatory effects of isoflavone on miRNA in pancreatic cancer cells as discussed below. It has been well known that the aggressiveness of pancreatic cancer is in part due to its intrinsic and extrinsic drug resistance characteristics, which are also associated with the acquisition of EMT. As we have described above, experimental evidence suggests that the processes of EMT are regulated by the expression status of many miRNAs. Therefore, we compared the expression of miRNAs between gemcitabine-sensitive and gemcitabine-resistant pancreatic cancer cells and investigated whether the treatment of cells with isoflavone could affect the expression of miRNAs. We found that the expression of miR-200b, miR-200c, let-7b, let-7c, let-7d, and let-7e was significantly down-regulated in gemcitabine-resistant cells, which showed EMT characteristics such as elongated fibroblastoid morphology, lower expression of epithelial marker E-cadherin, and higher expression of mesenchymal markers such as vimentin and ZEB1 (8). Moreover, we found that re-expression of miR-200 by pre-miR-200 transfection or treatment of gemcitabine-resistant cells with isoflavone resulted in the up-regulation of miR-200 and the down-regulation of ZEB1, slug, and vimentin, which was consistent with morphologic reversal of EMT phenotype leading to epithelial morphology (8). Isoflavone could also induce the expression level of let-7, leading to the inhibition of cancer cell growth (8). These results suggest that isoflavone could function as miRNA regulators, leading to the reversal of EMT phenotype, and that the induction of miR-200 and let-7 by isoflavone could be important for designing novel therapies for cancers.
THE ROLE OF miRNA MODULATION BY I3C AND DIM
Indole-3-carbinol (I3C) and its in vivo dimeric product DIM are produced from naturally occurring glucosinolates found in the family Cruciferae. I3C and DIM have shown inhibitory effects on cancer cell growth through the modulation of genes that are related to the control of cell proliferation, cell cycle, apoptosis, signal transduction, oncogenesis, and transcription regulation (112–114). I3C and DIM could also regulate specific miRNAs which function as tumor suppressors or oncogenes, leading to the inhibition of cancer cell growth (Table 2 and Fig. 3). Melkamu et al. investigated the miRNA expression in lung cancer and examined whether I3C could reverse vinyl carbamate-induced deregulation of miRNA levels in lung tissues of female A/J mice (7). By microarray analysis, they found that miR-21, miR-31, miR-130a, miR-146b, and miR-377 were consistently up-regulated while miR-1 and miR-143 were down-regulated in lung cancer compared to normal lungs (7). Moreover, the up-regulation of miR-21, miR-31, miR-130a, miR-146b, and miR-377 observed in vinyl carbamate-treated animals was abrogated by I3C treatment, suggesting that I3C could inhibit the expression of these oncogenic miRNAs. Further investigation showed that PTEN, PDCD4, and RECK were potential targets of miR-21 (5) and that I3C up-regulated these tumor suppressor genes though inhibition of miR-21 (7). Therefore, targeting miR-21 by I3C could be a novel strategy for cancer treatment.
We have also investigated whether DIM could reverse EMT status by regulation of miRNA in pancreatic cancer (8). Since we found that the expression of miR-200b, miR-200c, let-7b, let-7c, let-7d, and let-7e was significantly down-regulated in gemcitabine-resistant cells with EMT characteristics, we treated the gemcitabine-resistant cells with 25 μM DIM, and we found that the expression of miR-200 and let-7 families were up-regulated in gemcitabine-resistant cells by DIM treatment. Moreover, DIM treatment resulted in the down-regulation of ZEB1, slug, and vimentin, and the morphologic reversal of EMT to epithelial morphology (8). These results clearly showed that I3C or DIM could function as miRNA regulators and that DIM could reverse EMT phenotype, suggesting that DIM could be used for designing novel therapies for pancreatic cancer by specifically targeted inactivation of miRNAs.
THE EFFECT OF EGCG ON miRNAs
EGCG is a major type of green tea polyphenols, and it is known for its antioxidant and anticancer activities in several types of cancer (115,116). EGCG also showed to regulate miRNAs (Table 2 and Fig. 3). Tsang et al. recently reported the influence of EGCG on the expressions of miRNAs in human cancer cells (117). By miRNA microarray analysis, they found that EGCG treatment could modify the expressions of some miRNAs in human hepatocellular carcinoma HepG2 cells. EGCG treatment resulted in the alterations in the expression levels of total 61 miRNAs with up-regulation of 13 miRNAs and down-regulation of 48 miRNAs. Among them, miR-16 was one of the miRNAs up-regulated by EGCG. Importantly, miR-16 has been confirmed to target and inhibit anti-apoptotic protein Bcl-2. Consistent with miRNA data, they found that EGCG treatment down-regulated Bcl-2 and induced apoptosis in HepG2 cells. Further experiment showed that transfection with miR-16 inhibitor suppressed miR-16 expression and attenuated the effect of EGCG on Bcl-2 down-regulation and induction of apoptotic cell death (117). These results demonstrate the role of miR-16 in regulating Bcl-2 and apoptosis and also provide experimental evidence showing that EGCG could inhibit cancer cell growth through the regulation of miRNAs; however, further in-depth studies are needed in multiple human cancer cell lines in order to fully appreciate the targeted effects of EGCG in the regulation of miRNAs.
THE EFFECTS OF RESVERATROL ON miRNAs
Resveratrol (3,5,4′-trihydroxystilbene) is a phytoalexin present in a wide variety of plant species, including grapes, mulberries, and peanuts. Experimental studies have shown that resveratrol inhibits the growth of various cancer cells and induces apoptotic cell death (118,119). Resveratrol could also regulate the expression of miRNA. A study reported by Lukiw et al. has shown that resveratrol analog CAY10512 regulated the expression of miR-146a (10). They found that miR-146a was up-regulated and complement factor H (CFH), an important repressor of the inflammatory response in the brain, was down-regulated in the brain of Alzheimer disease. The sequence of miR-146a was highly complementary to the 3′UTR of CFH, suggesting that CFH is a target of miR-146a. Further experiments showed that transfection of human neural cells with pre-miRNA-146a promoter-luciferase reporter construct in stressed human neural cells showed significant up-regulation of luciferase activity and low level of CFH gene expression, consistent with the observations on the brain of Alzheimer disease. Importantly, treatment of stressed human neural cells with resveratrol analog CAY10512 or an anti-sense oligonucleotide to miRNA-146a could inhibit miR-146a and restore CFH expression levels (10). These data indicate that resveratrol could regulate the expression of specific miRNA and alter the signaling transduction, leading to the alterations in cell physiological behavior, and the results thus far clearly suggest that further research in this area is urgently needed.
EMERGING EVIDENCE ON THE ROLE OF OTHER NATURAL AGENTS IN THE REGULATION OF miRNAs
Other dietary factors have also been found to regulate miRNA expression, resulting in the alterations of target gene expression. To investigate the effects of exposure to environmental factors, such as alcohol, a study has been conducted to examine the expression levels of 509 mature miRNAs in fetal mouse brain with or without prenatal ethanol exposure using miRNA microarray and RT-PCR (120). It has been found that miR-10a, miR-10b, miR-9, miR-145, miR-30a-3p and miR-152 were up-regulated, while miR-200a, miR-496, miR-296, miR-30e-5p, miR-362, miR-339, miR-29c and miR-154 were down-regulated in fetal brain with prenatal ethanol exposure. However, folic acid could down-regulate miR-10a (120), suggesting the effects of dietary factors on miRNA. Dietary vitamin E has been known to regulate gene expression by altering mRNA concentrations. It has been found that vitamin E deficiency could result in reduced hepatic concentrations of miR-122a and miR-125b, which are related to lipid metabolism, cancer, and inflammation (121). Reports also showed that dietary components such as folate and retinoids could exert inhibitory effects on cancer through modulation of miRNA expression (122). All these results demonstrate that miRNA profiling could be useful as biomarkers for cancer prevention, diagnosis, prognosis, and assessing nutritional status in dietary intervention studies (122).
CONCLUDING REMARKS AND PERSPECTIVES
In conclusion, we believe that miRNAs deserve more attention in cancer research (5,6). These small, non-coding RNA molecules could inhibit target gene expression by binding to the 3′UTR of target mRNA, resulting in either mRNA degradation or inhibition of translation. Importantly, aberrant miRNA expression has been correlated with tumor development, cancer progression, the formation of CSCs and the acquisition of EMT phenotype, a process that shares similar molecular characteristics with CSCs. Most importantly, miRNAs have been characterized as the biomarkers for diagnosis and prognosis, and thus miRNAs are becoming attractive targets for cancer therapy. To that end, emerging evidence is beginning to suggest that natural agents could be useful for targeting miRNAs, which in turn could enhance the efficacy of conventional cancer therapies. Therefore, targeting miRNAs by natural agents that are also known as cancer chemopreventive agents could open newer avenues to eradicate tumors toward successful treatment of cancer by eliminating CSCs or EMT-type cells that are increasingly being recognized as the cells causing tumor recurrence after conventional therapies. It is our perspectives that recent reports reviewed above clearly provide tantalizing results showing that natural agents such as curcumin, isoflavone, I3C, DIM, EGCG, resveratrol, etc. could alter miRNA expression profiles, leading to the inhibition of cancer growth, induction of apoptosis, reversal of EMT phenotype, and increasing drug sensitivity. Therefore, regulation of miRNAs by natural agents could be a novel strategy toward designing combination approaches with conventional therapies for the prevention of tumor recurrence and achieving successful treatment outcome of patients diagnosed with cancers.
Acknowledgments
The authors’ work cited in this review article was funded by grants from the National Cancer Institute, NIH (5R01CA083695, 5R01CA131151, 3R01CA131151-02S109, and 1R01CA132794 awarded to FHS), and a sub-contract award to FHS from the University of Texas MD Anderson Cancer Center through SPORE grant (5P20-CA101936, 3P20CA101936-05 S109) on pancreatic cancer awarded to James Abbruzzese. We also thank Puschelberg and Guido foundations for their generous contribution.
Contributor Information
Yiwei Li, Department of Pathology, Barbara Ann Karmanos Cancer Institute, Wayne State University School of Medicine, Detroit, Michigan, USA.
Dejuan Kong, Department of Pathology, Barbara Ann Karmanos Cancer Institute, Wayne State University School of Medicine, Detroit, Michigan, USA.
Zhiwei Wang, Department of Pathology, Barbara Ann Karmanos Cancer Institute, Wayne State University School of Medicine, Detroit, Michigan, USA.
Fazlul H. Sarkar, Email: fsarkar@med.wayne.edu, Department of Pathology, Barbara Ann Karmanos Cancer Institute, Wayne State University School of Medicine, Detroit, Michigan, USA. Department of Pathology, Karmanos Cancer Institute, Wayne State University School of Medicine, 740 HWCRC, 4100 John R Street, Detroit, Michigan 48201, USA
References
- 1.Lee RC, Feinbaum RL, The AV. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–54. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 2.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–62. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–6. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 4.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–9. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 5.Iorio MV, Croce CM. MicroRNAs in cancer: small molecules with a huge impact. J Clin Oncol. 2009;27:5848–56. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vandenboom Ii TG, Li Y, Philip PA, Sarkar FH. MicroRNA and Cancer: Tiny Molecules with Major Implications. Curr Genomics. 2008;9:97–109. doi: 10.2174/138920208784139555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melkamu T, Zhang X, Tan J, Zeng Y, Kassie F. Alteration of microRNA expression in vinyl-carbamate-induced mouse lung tumors and modulation by the chemopreventive agent indole-3-carbinol. Carcinogenesis. 2009 doi: 10.1093/carcin/bgp208. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, VandenBoom TG, Kong D, Wang Z, Ali S, Philip PA, et al. Up-regulation of miR-200 and let-7 by natural agents leads to the reversal of epithelial-to-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Cancer Res. 2009;69:6704–12. doi: 10.1158/0008-5472.CAN-09-1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pogribny IP, Muskhelishvili L, Tryndyak VP, Beland FA. The tumor-promoting activity of 2-acetylaminofluorene is associated with disruption of the p53 signaling pathway and the balance between apoptosis and cell proliferation. Toxicol Appl Pharmacol. 2009;235:305–11. doi: 10.1016/j.taap.2008.12.021. [DOI] [PubMed] [Google Scholar]
- 10.Lukiw WJ, Zhao Y, Cui JG. An NF-kappaB-sensitive micro RNA-146a-mediated inflammatory circuit in Alzheimer disease and in stressed human brain cells. J Biol Chem. 2008;283:31315–22. doi: 10.1074/jbc.M805371200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun M, Estrov Z, Ji Y, Coombes KR, Harris DH, Kurzrock R. Curcumin (diferuloylmethane) alters the expression profiles of microRNAs in human pancreatic cancer cells. Mol Cancer Ther. 2008;7:464–73. doi: 10.1158/1535-7163.MCT-07-2272. [DOI] [PubMed] [Google Scholar]
- 12.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–60. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 14.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the Microprocessor complex. Nature. 2004;432:231–5. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 15.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–27. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, et al. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–40. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 17.Landthaler M, Yalcin A, Tuschl T. The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Curr Biol. 2004;14:2162–7. doi: 10.1016/j.cub.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 18.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–70. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–9. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 20.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–8. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- 21.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–6. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammond SM, Bernstein E, Beach D, Hannon GJ. An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature. 2000;404:293–6. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 23.Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621–9. doi: 10.1016/j.cell.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 24.Saxena S, Jonsson ZO, Dutta A. Small RNAs with imperfect match to endogenous mRNA repress translation. Implications for off-target activity of small inhibitory RNA in mammalian cells. J Biol Chem. 2003;278:44312–9. doi: 10.1074/jbc.M307089200. [DOI] [PubMed] [Google Scholar]
- 25.Pasquinelli AE, Ruvkun G. Control of developmental timing by micrornas and their targets. Annu Rev Cell Dev Biol. 2002;18:495–513. doi: 10.1146/annurev.cellbio.18.012502.105832. [DOI] [PubMed] [Google Scholar]
- 26.Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–6. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- 27.Nicoloso MS, Spizzo R, Shimizu M, Rossi S, Calin GA. MicroRNAs—the micro steering wheel of tumour metastases. Nat Rev Cancer. 2009;9:293–302. doi: 10.1038/nrc2619. [DOI] [PubMed] [Google Scholar]
- 28.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009 doi: 10.1038/emboj.2009.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dirks PB. MicroRNAs and parallel stem cell lives. Cell. 2009;138:423–4. doi: 10.1016/j.cell.2009.07.025. [DOI] [PubMed] [Google Scholar]
- 30.Wang Y, Russell I, Chen C. MicroRNA and stem cell regulation. Curr Opin Mol Ther. 2009;11:292–8. [PubMed] [Google Scholar]
- 31.Ji Q, Hao X, Zhang M, Tang W, Yang M, Li L, et al. MicroRNA miR-34 inhibits human pancreatic cancer tumor-initiating cells. PLoS One. 2009;4:e6816. doi: 10.1371/journal.pone.0006816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–80. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garzia L, Andolfo I, Cusanelli E, Marino N, Petrosino G, De MD, et al. MicroRNA-199b-5p impairs cancer stem cells through negative regulation of HES1 in medulloblastoma. PLoS One. 2009;4:e4998. doi: 10.1371/journal.pone.0004998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, et al. miR-200 regulates PDGF-D-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–21. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 37.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 38.Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–72. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- 39.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–33. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 40.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 41.Ribas J, Ni X, Haffner M, Wentzel EA, Salmasi AH, Chowdhury WH, et al. miR-21: an androgen receptor-regulated microRNA that promotes hormone-dependent and hormone-independent prostate cancer growth. Cancer Res. 2009;69:7165–9. doi: 10.1158/0008-5472.CAN-09-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Seike M, Goto A, Okano T, Bowman ED, Schetter AJ, Horikawa I, et al. MiR-21 is an EGFR-regulated anti-apoptotic factor in lung cancer in never-smokers. Proc Natl Acad Sci USA. 2009;106:12085–90. doi: 10.1073/pnas.0905234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Park JK, Lee EJ, Esau C, Schmittgen TD. Antisense Inhibition of microRNA-21 or –221 Arrests Cell Cycle, Induces Apoptosis, and Sensitizes the Effects of Gemcitabine in Pancreatic Adenocarcinoma. Pancreas. 2009 doi: 10.1097/MPA.0b013e3181ba82e1. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Huang H, Sun L, Yang M, Pan C, Chen W, et al. MiR-21 indicates poor prognosis in tongue squamous cell carcinomas as an apoptosis inhibitor. Clin Cancer Res. 2009;15:3998–4008. doi: 10.1158/1078-0432.CCR-08-3053. [DOI] [PubMed] [Google Scholar]
- 45.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 46.Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388:539–42. doi: 10.1016/j.bbrc.2009.08.044. [DOI] [PubMed] [Google Scholar]
- 47.Qin W, Zhao B, Shi Y, Yao C, Jin L, Jin Y. BMPRII is a direct target of miR-21. Acta Biochim Biophys Sin (Shanghai) 2009;41:618–23. doi: 10.1093/abbs/gmp049. [DOI] [PubMed] [Google Scholar]
- 48.Li Y, Li W, Yang Y, Lu Y, He C, Hu G, et al. MicroRNA-21 targets LRRFIP1 and contributes to VM-26 resistance in glioblastoma multiforme. Brain Res. 2009;1286:13–8. doi: 10.1016/j.brainres.2009.06.053. [DOI] [PubMed] [Google Scholar]
- 49.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–33. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Diosdado B, van de Wiel MA, Terhaar Sive Droste JS, Mongera S, Postma C, Meijerink WJ, et al. MiR-17-92 cluster is associated with 13q gain and c-myc expression during colorectal adenoma to adenocarcinoma progression. Br J Cancer. 2009;101:707–14. doi: 10.1038/sj.bjc.6605037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Manni I, Artuso S, Careccia S, Rizzo MG, Baserga R, Piaggio G, et al. The microRNA miR-92 increases proliferation of myeloid cells and by targeting p63 modulates the abundance of its isoforms. FASEB J. 2009 doi: 10.1096/fj.09-131847. [DOI] [PubMed] [Google Scholar]
- 52.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–5. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Northcott PA, Fernandez L, Hagan JP, Ellison DW, Grajkowska W, Gillespie Y, et al. The miR-17/92 polycistron is up-regulated in sonic hedgehog-driven medulloblastomas and induced by N-myc in sonic hedgehog-treated cerebellar neural precursors. Cancer Res. 2009;69:3249–55. doi: 10.1158/0008-5472.CAN-08-4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Uziel T, Karginov FV, Xie S, Parker JS, Wang YD, Gajjar A, et al. The miR-17 92 cluster collaborates with the Sonic Hedgehog pathway in medulloblastoma. Proc Natl Acad Sci USA. 2009;106:2812–7. doi: 10.1073/pnas.0809579106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–51. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 56.Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–61. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- 57.Greither T, Grochola LF, Udelnow A, Lautenschlager C, Wurl P, Taubert H. Elevated expression of microRNAs 155, 203, 210 and 222 in pancreatic tumors is associated with poorer survival. Int J Cancer. 2009 doi: 10.1002/ijc.24687. [DOI] [PubMed] [Google Scholar]
- 58.Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009;8:340–6. doi: 10.4161/cbt.8.4.7338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gironella M, Seux M, Xie MJ, Cano C, Tomasini R, Gommeaux J, et al. Tumor protein 53-induced nuclear protein 1 expression is repressed by miR-155, and its restoration inhibits pancreatic tumor development. Proc Natl Acad Sci USA. 2007;104:16170–5. doi: 10.1073/pnas.0703942104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–22. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 61.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 62.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–30. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bussing I, Slack FJ, Grosshans H. let-7 microRNAs in development, stem cells and cancer. Trends Mol Med. 2008;14:400–9. doi: 10.1016/j.molmed.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 64.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–47. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 65.Takeshita F, Patrawala L, Osaki M, Takahashi RU, Yamamoto Y, Kosaka N, et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via down-regulation of multiple cell-cycle genes. Mol Ther. 2009 doi: 10.1038/mt.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–9. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc Natl Acad Sci USA. 2008;105:5166–71. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–52. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth. Cancer Res. 2007;67:8433–8. doi: 10.1158/0008-5472.CAN-07-1585. [DOI] [PubMed] [Google Scholar]
- 70.Gregory PA, Bracken CP, Bert AG, Goodall GJ. MicroRNAs as regulators of epithelial-mesenchymal transition. Cell Cycle. 2008;7:3112–8. doi: 10.4161/cc.7.20.6851. [DOI] [PubMed] [Google Scholar]
- 71.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spahn M, Kneitz S, Scholz CJ, Nico S, Rudiger T, Strobel P, et al. Expression of microRNA-221 is progressively reduced in aggressive prostate cancer and metastasis and predicts clinical recurrence. Int J Cancer. 2009 doi: 10.1002/ijc.24715. [DOI] [PubMed] [Google Scholar]
- 73.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, et al. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T. MicroRNA expression profiling in prostate cancer. Cancer Res. 2007;67:6130–5. doi: 10.1158/0008-5472.CAN-07-0533. [DOI] [PubMed] [Google Scholar]
- 76.Tong AW, Fulgham P, Jay C, Chen P, Khalil I, Liu S, et al. MicroRNA profile analysis of human prostate cancers. Cancer Gene Ther. 2009;16:206–16. doi: 10.1038/cgt.2008.77. [DOI] [PubMed] [Google Scholar]
- 77.Raponi M, Dossey L, Jatkoe T, Wu X, Chen G, Fan H, et al. MicroRNA classifiers for predicting prognosis of squamous cell lung cancer. Cancer Res. 2009;69:5776–83. doi: 10.1158/0008-5472.CAN-09-0587. [DOI] [PubMed] [Google Scholar]
- 78.Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007;120:1046–54. doi: 10.1002/ijc.22394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yang H, Kong W, He L, Zhao JJ, O’Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–33. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 80.Mattie MD, Benz CC, Bowers J, Sensinger K, Wong L, Scott GK, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. doi: 10.1186/1476-4598-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang QZ, Xu W, Habib N, Xu R. Potential uses of microRNA in lung cancer diagnosis, prognosis, and therapy. Curr Cancer Drug Targets. 2009;9:572–94. doi: 10.2174/156800909788486731. [DOI] [PubMed] [Google Scholar]
- 82.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 84.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. J Clin Oncol. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 86.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–8. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 87.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA. 2008;105:10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One. 2009;4:e6229. doi: 10.1371/journal.pone.0006229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhu W, Qin W, Atasoy U, Sauter ER. Circulating microRNAs in breast cancer and healthy subjects. BMC Res Notes. 2009;2:89. doi: 10.1186/1756-0500-2-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Resnick KE, Alder H, Hagan JP, Richardson DL, Croce CM, Cohn DE. The detection of differentially expressed microRNAs from the serum of ovarian cancer patients using a novel real-time PCR platform. Gynecol Oncol. 2009;112:55–9. doi: 10.1016/j.ygyno.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 91.Chen X, Ba Y, Ma L, Cai X, Yin Y, Wang K, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 92.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–5. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 93.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, et al. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc Natl Acad Sci USA. 2009;106:4402–7. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hutvagner G, Simard MJ, Mello CC, Zamore PD. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004;2:E98. doi: 10.1371/journal.pbio.0020098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Meister G, Landthaler M, Dorsett Y, Tuschl T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. RNA. 2004;10:544–50. doi: 10.1261/rna.5235104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Orom UA, Kauppinen S, Lund AH. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–41. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- 97.Krutzfeldt J, Rajewsky N, Braich R, Rajeev KG, Tuschl T, Manoharan M, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–9. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 98.Tsuda N, Ishiyama S, Li Y, Ioannides CG, Abbruzzese JL, Chang DZ. Synthetic microRNA designed to target glioma-associated antigen 1 transcription factor inhibits division and induces late apoptosis in pancreatic tumor cells. Clin Cancer Res. 2006;12:6557–64. doi: 10.1158/1078-0432.CCR-06-0588. [DOI] [PubMed] [Google Scholar]
- 99.Weidhaas JB, Babar I, Nallur SM, Trang P, Roush S, Boehm M, et al. MicroRNAs as potential agents to alter resistance to cytotoxic anticancer therapy. Cancer Res. 2007;67:11111–6. doi: 10.1158/0008-5472.CAN-07-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dolganiuc A, Petrasek J, Kodys K, Catalano D, Mandrekar P, Velayudham A, et al. MicroRNA Expression Profile in Lieber-DeCarli Diet-Induced Alcoholic and Methionine Choline Deficient Diet-Induced Nonalcoholic Steatohepatitis Models in Mice. Alcohol Clin Exp Res. 2009 doi: 10.1111/j.1530-0277.2009.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sharma RA, Euden SA, Platton SL, Cooke DN, Shafayat A, Hewitt HR, et al. Phase I clinical trial of oral curcumin: biomarkers of systemic activity and compliance. Clin Cancer Res. 2004;10:6847–54. doi: 10.1158/1078-0432.CCR-04-0744. [DOI] [PubMed] [Google Scholar]
- 102.Garcea G, Berry DP, Jones DJ, Singh R, Dennison AR, Farmer PB, et al. Consumption of the putative chemopreventive agent curcumin by cancer patients: assessment of curcumin levels in the colorectum and their pharmacodynamic consequences. Cancer Epidemiol Biomarkers Prev. 2005;14:120–5. [PubMed] [Google Scholar]
- 103.Padhye S, Banerjee S, Chavan D, Pandye S, Swamy KV, Ali S, et al. Fluorocurcumins as Cyclooxygenase-2 Inhibitor: Molecular Docking, Pharmacokinetics and Tissue Distribution in Mice. Pharm Res. 2009 doi: 10.1007/s11095-009-9955-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Takahashi M, Uechi S, Takara K, Asikin Y, Wada K. Evaluation of an oral carrier system in rats: bioavailability and antioxidant properties of liposome-encapsulated curcumin. J Agric Food Chem. 2009;57:9141–6. doi: 10.1021/jf9013923. [DOI] [PubMed] [Google Scholar]
- 105.Anand P, Nair HB, Sung B, Kunnumakkara AB, Yadav VR, Tekmal RR, et al. Design of curcumin-loaded PLGA nano-particles formulation with enhanced cellular uptake, and increased bioactivity in vitro and superior bioavailability in vivo. Biochem Pharmacol. 2010;79:330–8. doi: 10.1016/j.bcp.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 106.Mukhopadhyay A, Bueso-Ramos C, Chatterjee D, Pantazis P, Aggarwal BB. Curcumin downregulates cell survival mechanisms in human prostate cancer cell lines. Oncogene. 2001;20:7597–609. doi: 10.1038/sj.onc.1204997. [DOI] [PubMed] [Google Scholar]
- 107.Shao ZM, Shen ZZ, Liu CH, Sartippour MR, Go VL, Heber D, et al. Curcumin exerts multiple suppressive effects on human breast carcinoma cells. Int J Cancer. 2002;98:234–40. doi: 10.1002/ijc.10183. [DOI] [PubMed] [Google Scholar]
- 108.Sarkar FH, Li Y. Cell signaling pathways altered by natural chemopreventive agents. Mutat Res. 2004;555:53–64. doi: 10.1016/j.mrfmmm.2004.04.015. [DOI] [PubMed] [Google Scholar]
- 109.Barnes S. The chemopreventive properties of soy isoflavonoids in animal models of breast cancer. Breast Cancer Res Treat. 1997;46:169–79. doi: 10.1023/a:1005956326155. [DOI] [PubMed] [Google Scholar]
- 110.Li Y, Sarkar FH. Down-regulation of invasion and angiogenesis-related genes identified by cDNA microarray analysis of PC3 prostate cancer cells treated with genistein. Cancer Lett. 2002;186:157–64. doi: 10.1016/s0304-3835(02)00349-x. [DOI] [PubMed] [Google Scholar]
- 111.Dixon RA, Ferreira D. Genistein. Phytochemistry. 2002;60:205–11. doi: 10.1016/s0031-9422(02)00116-4. [DOI] [PubMed] [Google Scholar]
- 112.Chinni SR, Li Y, Upadhyay S, Koppolu PK, Sarkar FH. Indole-3-carbinol (I3C) induced cell growth inhibition, G1 cell cycle arrest and apoptosis in prostate cancer cells. Oncogene. 2001;20:2927–36. doi: 10.1038/sj.onc.1204365. [DOI] [PubMed] [Google Scholar]
- 113.Garikapaty VP, Ashok BT, Tadi K, Mittelman A, Tiwari RK. 3, 3′-Diindolylmethane downregulates pro-survival pathway in hormone independent prostate cancer. Biochem Biophys Res Commun. 2006;340:718–25. doi: 10.1016/j.bbrc.2005.12.059. [DOI] [PubMed] [Google Scholar]
- 114.Li Y, Li X, Sarkar FH. Gene expression profiles of I3C- and DIM-treated PC3 human prostate cancer cells determined by cDNA microarray analysis. J Nutr. 2003;133:1011–9. doi: 10.1093/jn/133.4.1011. [DOI] [PubMed] [Google Scholar]
- 115.Mukhtar H, Ahmad N. Green tea in chemoprevention of cancer. Toxicol Sci. 1999;52:111–7. doi: 10.1093/toxsci/52.2.111. [DOI] [PubMed] [Google Scholar]
- 116.Katiyar SK, Afaq F, Perez A, Mukhtar H. Green tea polyphenol (−)-epigallocatechin-3-gallate treatment of human skin inhibits ultraviolet radiation-induced oxidative stress. Carcinogenesis. 2001;22:287–94. doi: 10.1093/carcin/22.2.287. [DOI] [PubMed] [Google Scholar]
- 117.Tsang WP, Kwok TT. Epigallocatechin gallate up-regulation of miR-16 and induction of apoptosis in human cancer cells. J Nutr Biochem. 2009 doi: 10.1016/j.jnutbio.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 118.Kundu JK, Surh YJ. Cancer chemopreventive and therapeutic potential of resveratrol: mechanistic perspectives. Cancer Lett. 2008;269:243–61. doi: 10.1016/j.canlet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 119.Whyte L, Huang YY, Torres K, Mehta RG. Molecular mechanisms of resveratrol action in lung cancer cells using dual protein and microarray analyses. Cancer Res. 2007;67:12007–17. doi: 10.1158/0008-5472.CAN-07-2464. [DOI] [PubMed] [Google Scholar]
- 120.Wang LL, Zhang Z, Li Q, Yang R, Pei X, Xu Y, et al. Ethanol exposure induces differential microRNA and target gene expression and teratogenic effects which can be suppressed by folic acid supplementation. Hum Reprod. 2009;24:562–79. doi: 10.1093/humrep/den439. [DOI] [PubMed] [Google Scholar]
- 121.Gaedicke S, Zhang X, Schmelzer C, Lou Y, Doering F, Frank J, et al. Vitamin E dependent microRNA regulation in rat liver. FEBS Lett. 2008;582:3542–6. doi: 10.1016/j.febslet.2008.09.032. [DOI] [PubMed] [Google Scholar]
- 122.Davis CD, Ross SA. Evidence for dietary regulation of microRNA expression in cancer cells. Nutr Rev. 2008;66:477–82. doi: 10.1111/j.1753-4887.2008.00080.x. [DOI] [PubMed] [Google Scholar]
- 123.Robertus JL, Harms G, Blokzijl T, Booman M, de Jong D, van Imhoff G, et al. Specific expression of miR-17-5p and miR-127 in testicular and central nervous system diffuse large B-cell lymphoma. Mod Pathol. 2009;22:547–55. doi: 10.1038/modpathol.2009.10. [DOI] [PubMed] [Google Scholar]
- 124.Mraz M, Malinova K, Kotaskova J, Pavlova S, Tichy B, Malcikova J, et al. miR-34a, miR-29c and miR-17-5p are downregulated in CLL patients with TP53 abnormalities. Leukemia. 2009;23:1159–63. doi: 10.1038/leu.2008.377. [DOI] [PubMed] [Google Scholar]
- 125.Fontana L, Fiori ME, Albini S, Cifaldi L, Giovinazzi S, Forloni M, et al. Antagomir-17-5p abolishes the growth of therapy-resistant neuroblastoma through p21 and BIM. PLoS One. 2008;3:e2236. doi: 10.1371/journal.pone.0002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hossain A, Kuo MT, Saunders GF. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol Cell Biol. 2006;26:8191–201. doi: 10.1128/MCB.00242-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Park SY, Lee JH, Ha M, Nam JW, Kim VN. miR-29 miRNAs activate p53 by targeting p85 alpha and CDC42. Nat Struct Mol Biol. 2009;16:23–9. doi: 10.1038/nsmb.1533. [DOI] [PubMed] [Google Scholar]
- 128.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–40. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pekarsky Y, Santanam U, Cimmino A, Palamarchuk A, Efanov A, Maximov V, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–3. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 130.Agirre X, Vilas-Zornoza A, Jimenez-Velasco A, Martin-Subero JI, Cordeu L, Garate L, et al. Epigenetic silencing of the tumor suppressor microRNA Hsa-miR-124a regulates CDK6 expression and confers a poor prognosis in acute lymphoblastic leukemia. Cancer Res. 2009;69:4443–53. doi: 10.1158/0008-5472.CAN-08-4025. [DOI] [PubMed] [Google Scholar]
- 131.Pierson J, Hostager B, Fan R, Vibhakar R. Regulation of cyclin dependent kinase 6 by microRNA 124 in medulloblastoma. J Neurooncol. 2008;90:1–7. doi: 10.1007/s11060-008-9624-3. [DOI] [PubMed] [Google Scholar]
- 132.Clape C, Fritz V, Henriquet C, Apparailly F, Fernandez PL, Iborra F, et al. miR-143 interferes with ERK5 signaling, and abrogates prostate cancer progression in mice. PLoS One. 2009;4:e7542. doi: 10.1371/journal.pone.0007542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Takagi T, Iio A, Nakagawa Y, Naoe T, Tanigawa N, Akao Y. Decreased expression of microRNA-143 and -145 in human gastric cancers. Oncology. 2009;77:12–21. doi: 10.1159/000218166. [DOI] [PubMed] [Google Scholar]
- 134.Chen X, Guo X, Zhang H, Xiang Y, Chen J, Yin Y, et al. Role of miR-143 targeting KRAS in colorectal tumorigenesis. Oncogene. 2009;28:1385–92. doi: 10.1038/onc.2008.474. [DOI] [PubMed] [Google Scholar]
- 135.Sachdeva M, Mo YY. MicroRNA-145 Suppresses Cell Invasion and Metastasis by Directly Targeting Mucin 1. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-09-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Spizzo R, Nicoloso MS, Lupini L, Lu Y, Fogarty J, Rossi S, et al. miR-145 participates with TP53 in a death-promoting regulatory loop and targets estrogen receptor-alpha in human breast cancer cells. Cell Death Differ. 2009 doi: 10.1038/cdd.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Cho WC, Chow AS, Au JS. Restoration of tumour suppressor hsa-miR-145 inhibits cancer cell growth in lung adenocarcinoma patients with epidermal growth factor receptor mutation. Eur J Cancer. 2009;45:2197–206. doi: 10.1016/j.ejca.2009.04.039. [DOI] [PubMed] [Google Scholar]
- 138.Wang S, Bian C, Yang Z, Bo Y, Li J, Zeng L, et al. miR-145 inhibits breast cancer cell growth through RTKN. Int J Oncol. 2009;34:1461–6. [PubMed] [Google Scholar]
- 139.Xiao B, Guo J, Miao Y, Jiang Z, Huan R, Zhang Y, et al. Detection of miR-106a in gastric carcinoma and its clinical significance. Clin Chim Acta. 2009;400:97–102. doi: 10.1016/j.cca.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 140.Diaz R, Silva J, Garcia JM, Lorenzo Y, Garcia V, Pena C, et al. Deregulated expression of miR-106a predicts survival in human colon cancer patients. Genes Chromosomes Cancer. 2008;47:794–802. doi: 10.1002/gcc.20580. [DOI] [PubMed] [Google Scholar]
- 141.Lee KH, Goan YG, Hsiao M, Lee CH, Jian SH, Lin JT, et al. MicroRNA-373 (miR-373) post-transcriptionally regulates large tumor suppressor, homolog 2 (LATS2) and stimulates proliferation in human esophageal cancer. Exp Cell Res. 2009;315:2529–38. doi: 10.1016/j.yexcr.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 142.Huang Q, Gumireddy K, Schrier M, le Sage C, Nagel R, Nair S, et al. The microRNAs miR-373 and miR-520c promote tumour invasion and metastasis. Nat Cell Biol. 2008;10:202–10. doi: 10.1038/ncb1681. [DOI] [PubMed] [Google Scholar]
- 143.Voorhoeve PM, le Sage C, Schrier M, Gillis AJ, Stoop H, Nagel R, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Adv Exp Med Biol. 2007;604:17–46. doi: 10.1007/978-0-387-69116-9_2. [DOI] [PubMed] [Google Scholar]
- 144.Du L, Schageman JJ, Subauste MC, Saber B, Hammond SM, Prudkin L, et al. miR-93, miR-98, and miR-197 regulate expression of tumor suppressor gene FUS1. Mol Cancer Res. 2009;7:1234–43. doi: 10.1158/1541-7786.MCR-08-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]