ϕC31 integrase is a sequence-specific recombinase originating in a bacteriophage of Streptomyces. This system facilitates the genomic integration of plasmid DNA and may have practical application for nonviral gene transfer. In this article by Chavez et al., the authors examine important safety parameters of the ϕC31 system, including kinetics and longevity of transgene expression, both in vitro and in vivo.
Abstract
The ϕC31 integrase system provides genomic integration of plasmid DNA that may be useful in gene therapy. For example, the ϕC31 system has been used in combination with hydrodynamic injection to achieve long-term expression of factor IX in mouse liver. However, a concern is that prolonged expression of ϕC31 integrase within cells could potentially stimulate chromosome rearrangements or an immune response. Western blot and immunofluorescence analyses were performed to investigate the duration of ϕC31 integrase expression in mouse liver. Integrase was expressed within 2 to 3 hr after hydrodynamic injection of a plasmid expressing ϕC31 integrase. Expression peaked between 8 and 16 hr and fell to background levels by 24–48 hr postinjection. Analysis of the amount of integrase plasmid DNA present in the liver over time suggested that the brief period of integrase expression could largely be accounted for by rapid loss of the bulk of the plasmid DNA, as well as by silencing of plasmid expression. PCR analysis of integration indicated that ϕC31 integrase carried out genomic integration of a codelivered attB-containing plasmid by 3 hr after plasmid injection. Integrase was expressed for longer times and at higher levels in transfected cultured cells compared with liver. Inhibitor studies suggested that the enzyme had a short half-life and was degraded by the 26S proteasome. The short duration of integrase expression in liver and rapid integration reaction appear to be features favorable for use in gene therapy.
Introduction
ϕC31 integrase is a sequence-specific recombinase originating in a bacteriophage of Streptomyces. This integrase normally catalyzes the precise unidirectional integration of the phage genome into the bacterial chromosome (Kuhstoss and Rao, 1991; Rausch and Lehmann, 1991). The enzyme recognizes and recombines two ∼30-bp DNA sequences termed attB and attP, without a requirement for additional cofactors (Thorpe and Smith, 1998). ϕC31 integrase has also been shown to direct this reaction efficiently in mammalian cells (Groth et al., 2000). Although mammalian genomes do not contain perfect att sites, they possess related sequences, called pseudo-att sites (Thyagarajan et al., 2001). Integration for gene therapy can be carried out by placing an attB site on a plasmid containing the therapeutic gene, cotransfecting with a plasmid encoding integrase, and taking advantage of integrase-mediated recombination with naturally occurring pseudo-attP sites in the host genome. Because ϕC31 integrase requires a specific, relatively long recognition sequence in order to perform recombination, the number of potential integration sites is orders of magnitude lower than randomly integrating systems such as transposons and retroviruses (Chalberg et al., 2006). Furthermore, the sites appear to be in safe locations (Chalberg et al., 2006). Thus, the risks of insertional mutagenesis are reduced. Furthermore, the ϕC31 integrase system is not limited by vector size (Venken et al., 2006).
The potential of ϕC31 integrase in gene therapy has been demonstrated in numerous studies in animals (reviewed in Calos, 2006). The enzyme is also useful for the creation of transgenic animals (Calos, 2006). To date, there have been no reports of cancer associated with ϕC31 integrase. Transgenic mice producing ϕC31 integrase are viable, disease free, and have normal development and fertility (Belteki et al., 2003; Raymond and Soriano, 2007), and integrase has caused no acceleration of tumors in a MYC model of liver cancer (Woodard et al., 2010b). Nevertheless, at least under the artificial conditions of tissue culture, expression of ϕC31 integrase elevated the frequency of chromosomal rearrangements and large deletions (Chalberg et al., 2006; Ehrhardt et al., 2006; Liu et al., 2006). No evidence of chromosome rearrangements due to ϕC31 expression have been observed in vivo, either in gene therapy studies or in ϕC31 integrase-engineered transgenic animals (Bischof et al., 2007). Even so, prolonged ϕC31 integrase expression would be considered undesirable, because it could increase the probability of chromosomal instability or of an immune response to this prokaryotic protein.
It was unknown how long ϕC31 integrase protein was expressed, either in cultured cells or in the cells of the liver after in vivo plasmid DNA delivery. Integrase longevity in the liver was of particular interest, because of the therapeutic importance of this target tissue for gene therapy and robust preclinical data on therapeutic levels of human factor IX (hFIX) production with ϕC31 integrase in mouse liver (Olivares et al., 2002). To investigate these questions, we determined the time course of ϕC31 integrase protein expression within mouse liver by Western blot analysis, as well as by the relative number of liver cells expressing integrase and hFIX over the same time course by immunofluorescence. In addition, we examined the kinetics of the in vivo integration reaction by PCR and used several methodologies to establish the retention time within mouse liver of injected plasmid DNA. Related studies were carried out in cultured cells. This work provides new data that enhance our understanding of ϕC31 integrase activity and safety.
Materials and Methods
Plasmids
pCSI-HA was constructed by introducing the sequence coding the hemagglutinin (HA) tag, YPYDVPDYA, onto the C-terminal end of the ϕC31 integrase gene by PCR, as described (Keravala et al., 2009). pVFB has been described (Keravala et al., 2009) and carries the human factor IX (hFIX) gene driven by a liver-specific promoter comprising the human α1-antitrypsin promoter with an apolipoprotein E enhancer. pInt-EGFP was made by cloning the ϕC31 integrase gene, carrying an altered stop codon, into the SacI and BamHI sites of pEGFP-N2 (BD Biosciences, San Jose, CA). ϕC31 integrase with an altered stop codon was generated by PCR, using pCSI as template and primers PhiC31-F-aattcccgggtcgacgagctcactagtcgtagggtcgcc and PhiC31-R-cccctcgagggatccgggtgtctcgcaacgccgctacgt, and an annealing temperature of 70°C. The PCR product and pEGFP-N2 were digested with SacI and BamHI, and the integrase gene was ligated into pEGFP-N2 to give pInt-EGFP. pRFP-pericentrin was a gift from the laboratory of S. McConnell (Stanford University, Stanford, CA) and has been described previously (Gillingham and Munro, 2000). Briefly, the plasmid contains a cytomegalovirus (CMV) promoter driving expression of a fusion protein of red fluorescent protein and pericentrin, allowing visualization of the centrosome.
Mice
Eight- to 10-week-old C57BL/6 mice were ordered from Charles River (Wilmington, MA) or Jackson Laboratory (Bar Harbor, ME). Mice were kept in the Stanford Research Animal Facility and given water and food ad libitum. The Stanford Research Animal Committee approved all animal studies.
Hydrodynamic tail vein injection
A DNA solution was prepared that diluted 20 μg of both pCSI-HA and pVFB in 1.8 ml of Hanks' balanced salt solution (HBSS; Invitrogen, Carlsbad, CA). All DNA was prepared with an EndoFree plasmid maxi kit (Qiagen, Valencia, CA) according to the manufacturer's protocol, with the exception that the DNA pellets were reconstituted in HBSS. To promote tail vein dilation, mice were placed under a heat lamp. Once the vein was dilated, mice were placed in a restrainer (Braintree Scientific, Braintree, MA) and injected with 1.8 ml of the DNA solution over 3–5 sec, using a 3-ml syringe and 27-gauge butterfly needle (BD Biosciences).
Western blotting
Mouse livers were harvested over a time course and kept frozen at −80°C until use. Total liver protein extracts were prepared by adding 1 ml of tissue protein extraction reagent (Thermo Scientific, Rockford, IL) with added protease inhibitor cocktail tablets (Roche, Indianapolis, IN) to 0.1 g of mouse liver tissue and homogenizing, using Pellet Pestles and a Pellet Pestle motor (Daigger, Vernon Hills, IL). Protein sample concentration was determined by the Bradford assay (Bio-Rad, Hercules, CA). Controls, molecular weight markers, and time point samples (150 μg of total protein from liver and 50 μg of total protein from tissue culture cells) were run on 7.5% Tris-HCl Ready Gels (Bio-Rad) and transferred to nitrocellulose Trans-Blot transfer medium (Bio-Rad). Membranes were blocked with 5% nonfat dry milk in 1 × phosphate-buffered saline (PBS). The membranes were cut at the 50-kDa marker. The top half was incubated for 1 hr at room temperature with rat anti-HA high-affinity antibody (cat. no. 11-867-423-001; Roche Applied Science, Indianapolis, IN) diluted 1:1000 in 5% milk solution, and then washed three times for 5 min in 1 × PBS followed by addition of goat anti-rat horseradish peroxidase (HRP)-conjugated secondary antibody (cat. no. 401416; Calbiochem, Darmstadt, Germany) diluted in 1:10,000 milk solution. The remaining half was incubated for 1 hr at room temperature with mouse anti-β-actin (NB600-501; Novus Biologicals, Littleton, CO) diluted 1:1000 in milk solution, and then washed three times for 5 min in 1 × PBS, followed by addition of goat anti-mouse HRP-conjugated secondary antibody (Calbiochem) diluted 1:10,000 in milk solution. The blots were then washed three times in 1 × PBS, and incubated with SuperSignal West Pico substrate (Thermo Scientific, Waltham, MA) and imaged with Kodak BioMax film (Carestream Health, Rochester, NY). Densitometry was performed with ImageJ software (National Institutes of Health, Bethesda, MD).
Polymerase chain reaction
Total DNA was isolated from 25 mg of mouse liver tissue, using a DNeasy blood & tissue kit (Qiagen). For detection of integration at the mpsL1 pseudo-attP site, primers and conditions were used as described (Olivares and Calos, 2003). For integrase gene amplification, 100 ng of total DNA was used as the template to generate a 382-base pair fragment of the ϕC31 integrase gene. The primers used were Int-F (846) (5′-ggcaaccgttatgcgaatcc) and Int-R (505) (5′-ctcgacacgaagaaccttcagc). PCR conditions were as follows: an initial denaturation step at 94°C for 1 min and then 94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec, repeated for 30 cycles, with a final extension of 2 min and a final hold at 4°C.
Southern blot analysis and bacterial colony count
Five micrograms of total liver DNA was digested with NdeI, which cuts the vector once. The digests were electrophoresed through a 0.8% agarose gel and transferred to a nitrocellulose membrane. Integrase DNA was detected after hybridization with a digoxigenin-labeled integrase probe and chemiluminescence (Roche). For the bacterial colony count assay, 0.1–1 μg of total liver DNA was transformed into DH10B Escherichia coli cells, and the cells were spread on ampicillin or kanamycin selection plates and incubated at 37°C overnight. The next day, the numbers of colonies were counted.
Immunofluorescence staining of liver sections
For immunostaining, liver tissues were paraffin embedded and sectioned (Histo-Tec Laboratory, Hayward, CA). The staining procedure was performed as described (Keravala et al., 2009), with the following modifications. For double-stained slides, livers were fixed, after administration of the first secondary antibody, with 4% paraformaldehyde in 1 × PBS for 15 min, washed, reblocked, and incubated with the second primary antibody for 16 hr at 4°C. Next, the second secondary antibody was applied for 1 hr at room temperature. Slides were subsequently mounted with ProLong Gold antifade mounting medium with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen). The antibodies used were rat anti-HA high affinity (cat. no. 11-867-423-001; Roche), goat anti-hFIX (GAFIX-AP; Affinity Biologicals, Ancaster, ON, Canada), Alexa Fluor 488-conjugated goat anti-rabbit (Invitrogen), and Alexa Fluor 594-conjugated goat anti-rat (Invitrogen). All antibody dilutions were 1:500 in serum buffer. Serum buffer consisted of 1 × PBS plus 0.1% Tween 20 and 10% goat serum. Pictures of the stained cells, viewed with an Axioskop 2 plus microscope with a FluoArc power supply, were taken with an AxioCam MRc camera (Zeiss, Thornwood, NY).
Cell culture and protein extraction
HeLa cells (CCL-2; American Type Culture Collection [ATCC], Manassas, VA) were grown in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 9% fetal bovine serum and 1% penicillin–streptomycin (Invitrogen) on 60-mm plates. Plates of cells were transfected with DNA prepared with a Qiagen Maxi kit according to the manufacturer's instructions, using FuGENE 6 (Roche) at a 3:1 reagent (μl)-to-DNA (μg) ratio. For cycloheximide treatment, medium containing cycloheximide at 100 μg/ml (Calbiochem, Beeston, UK) was added to cells 2 days posttransfection. At the time points indicated, plates of cells were harvested for protein as described previously (Woodard et al., 2010a). For cycloheximide plus MG132 treatment, transfected HeLa cells were incubated with cycloheximide (100 μg/ml) and 1, 5, 10, or 15 μM MG132 (Calbiochem), and protein was harvested 10 hr later as described previously.
Immunofluorescence staining of cultured cells
NIH/3T3 mouse embryonic fibroblast cells (CRL-1568; ATCC) were seeded onto 6-well plates containing 12-mm-diameter coverslips (VWR, West Chester, PA) coated in poly-l-lysine. Cells were then transfected as described previously. The next evening 25 μM MG132 was added to the indicated cells. Ten hours later cells were fixed by removing the medium and adding 2 ml of ice-cold methanol to each well for 10 min at −20°C. Coverslips were then removed from the wells and placed in a dark humidified chamber. Cells were rehydrated with PBS for 10 min and blocked in PBS-BT (PBS plus 3% bovine serum albumin [BSA], 0.1% Triton X-100, 0.02% sodium azide) for 30 min. The following antibodies were then sequentially incubated on the coverslip, diluted in PBS-BT at the given dilutions: mouse anti-γ-tubulin (GTU-88; Sigma-Aldrich, St. Louis, MO), 1:500; Alexa Fluor 488-conjugated goat anti-mouse (Invitrogen), 1:250; rat anti-HA (3F10; Roche), 1:500; and Alexa Fluor 594-conjugated goat anti-rat (Invitrogen), 1:250. Each antibody was incubated for at least 30 min and then three PBS-BT washes were done. DAPI (0.05 mg/ml; Invitrogen) was added, and cells were mounted onto slides with glycerol-based antifade mounting medium, sealed with nail polish, and allowed to dry. Cells were visualized with a Zeiss Axioskop microscope.
Imaging of live cells
HeLa cells grown in 6-well dishes were transfected with equal amounts of pInt-EGFP and pRFP-pericentrin as described previously. The next morning cells were treated with MG132 or dimethyl sulfoxide (DMSO) alone. Eight hours later, cells were washed three times with DMEM without phenol red (Invitrogen), containing MG132 if indicated. Cells were visualized with a Zeiss Axiovert 200M microscope.
Results
Time course of ϕC31 integrase expression within mouse liver
We wanted to determine the length of time that ϕC31 integrase protein was expressed in vivo within mouse liver, after delivery of plasmid DNA by high-pressure tail vein injection (Liu et al., 1999; Zhang et al., 1999). Two plasmids at 20 μg each were injected, comprising pVFB, an attB-containing hFIX expression plasmid (Keravala et al., 2009), and pCSI-HA (Woodard et al., 2010a), containing the ϕC31 integrase gene with the nine-amino acid hemagglutinin (HA) epitope at the carboxy-terminal end and driven by the human cytomegalovirus (CMV) immediate/early promoter/enhancer. This HA-tagged version of ϕC31 integrase was used for easy detection of the protein and has similar activity to wild-type ϕC31 integrase (Woodard et al., 2010a). Serum levels of hFIX on day 1 were monitored to determine the quality of the injections, with a cutoff of >1000 ng/ml used to indicate adequate transfection. Animals with lower hFIX serum levels were excluded from the study. Mice were then killed over a time course and their livers were extracted for protein and DNA analysis.
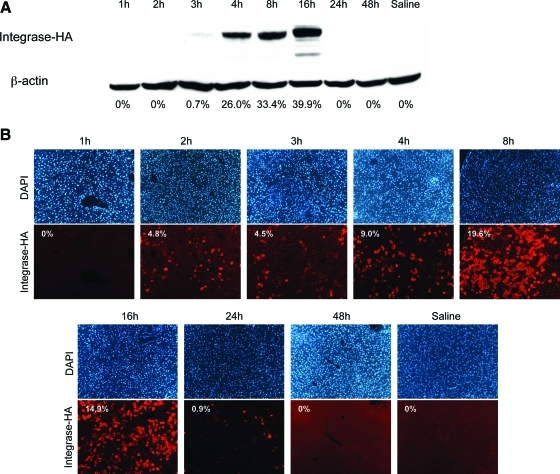
Total protein isolated from mouse livers harvested at various time points was subjected to Western blot analysis (Fig. 1A). These data revealed that ϕC31 integrase was expressed by 3 hr after injection and that protein levels continued to increase up to a peak at 16 hr. By 24 hr postinjection, only trace levels of integrase remained. By 48 hr, integrase was undetectable by Western blot.
FIG. 1.
Time course of ϕC31 integrase protein expression within mouse liver. Twenty micrograms each of pCSI-HA and pVFB were hydrodynamically injected into the tail vein of mice. At various time points over a 6-month time course, the mice were killed, their livers were removed, and total protein and DNA were isolated. (A) Western blot analysis of total protein isolated from mouse liver showed the presence of ϕC31 integrase at 3 hr postinjection. Integrase protein levels increased up to a peak at 16 hr and were undetectable by 24 hr. Below each lane of the blot is the percentage of total ϕC31 integrase protein present, as quantified by densitometry. (B) Immunofluorescence staining for ϕC31 integrase in mouse liver sections obtained over a time course. The top row shows DAPI-stained nuclei, whereas the bottom row shows staining for ϕC31 integrase. Numbers inside the images correspond to the percentage of cells that are expressing ϕC31 integrase protein. Color images available online at www.liebertonline.com/hum.
From the same liver samples that were used for Western blot analysis, sections were prepared for immunofluorescence staining to visualize the number of cells expressing ϕC31 integrase. As seen in Fig. 1B, ϕC31 integrase protein was first detectable within the cells of the liver by 2 hr after injection, with 4.8% of cells positive for integrase. Subsequently, integrase levels rose within the liver, to peak at 19.6 and 14.9% of cells expressing integrase at 8 and 16 hr, respectively, postinjection. A sharp drop-off in the number of ϕC31-positive cells ensued, so that only rare positive cells, 0.9% or less, were detectable after 16 hr. The immunofluorescence analysis was thus in close agreement with the Western blot data.
Time course of genomic integration within mouse liver
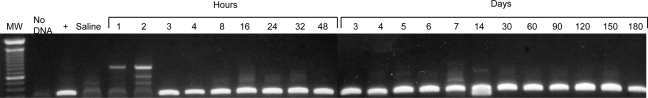
Several studies have shown that in mouse liver, ϕC31 integrase has a preferred integration site within the mouse genome (Olivares et al., 2002; Held et al., 2005; Keravala et al., 2009). This site, termed the mouse pseudo-site L1 (mpsL1) is located on chromosome 2. Integration at this site can be used to monitor ϕC31-mediated integration into the genome by detection of a specific PCR band comprising the junction between the integrated attB plasmid and the chromosomal mpsL1 site. As seen in Fig. 2, this PCR band indicating integration of pVFB, carrying attB and the hFIX gene, was detectable at the mpsL1 site by 3 hr after injection of pVFB and integrase plasmids. This result correlated well with Western blot and immunofluorescence data showing that integrase protein was present within liver cells by 2 to 3 hr after injection (Fig. 1). Taken together, these data indicated that ϕC31 integrase was expressed by 2 hr postinjection and had carried out integration within the mouse genome by 3 hr after injection.
FIG. 2.
Integration of donor plasmid occurs rapidly after plasmid injection. DNA extracted from liver was subjected to PCR using primers that detect the junction between the attB site of the donor pVFB plasmid and mpsL1, a commonly used pseudo-attP site in the mouse genome. Lane 1, molecular weight markers; lane 2, no DNA; lane 3, liver DNA sample known to have integration at mpsL1; lane 4, liver that received saline alone; lanes 5–25, time course of DNA samples from mice receiving pVFB and pCSI-HA, after the specified time in hours (1–48 hr), followed by days (3–180 days).
Time course of retention of integrase plasmid DNA within mouse liver
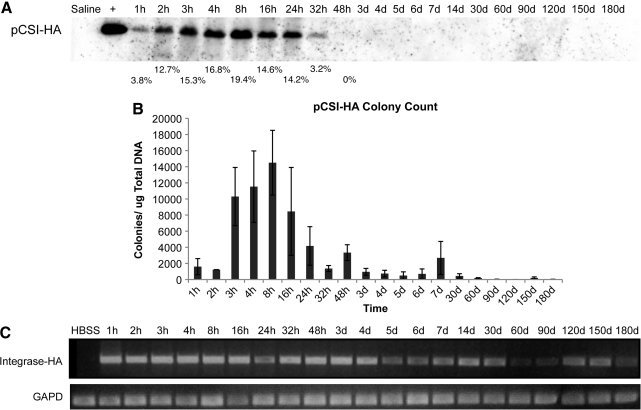
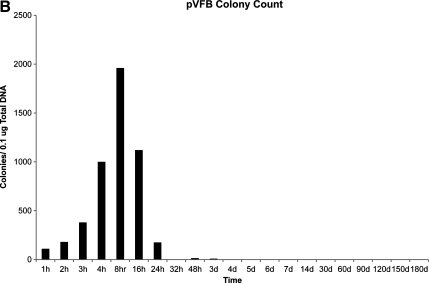
To determine the length of time that plasmid DNA encoding ϕC31 integrase was detectable within the liver of mice injected with pCSI-HA, three separate approaches were employed. First, we conducted Southern blot analysis on total DNA from the mouse liver samples (Fig. 3A). These data revealed that the integrase-encoding plasmid was detectable within the mouse liver at the first time point taken, 1 hr after injection. The amount of plasmid DNA detected within the liver increased over the next four time points, peaking in concentration at 8 hr, and then fell rapidly and was undetectable by Southern blot under these conditions by 48 hr postinjection.
FIG. 3.
Analysis of ϕC31 integrase plasmid DNA retention within mouse liver. ϕC31 integrase expression plasmid pCSI-HA was hydrodynamically injected into the tail vein of mice. At various points over a time course, the mice were killed, their livers were removed, and total DNA was isolated. (A) Southern blot analysis using a probe to the ϕC31 integrase gene showed an increase in signal over the first 8 hr postinjection, followed by a decrease, and a loss of signal by 48 hr. Below each lane of the blot is the percentage of total ϕC31 integrase plasmid present, as quantified by densitometry. (B) Total DNA transformed into bacterial cells showed an increase in colony number over the first 8 hr after injection, followed by a decrease, until few colonies were detected after day 7. (C) PCR detected the presence of the ϕC31 integrase gene over the 180-day time course.
In the second approach, 1 μg of total mouse liver DNA was used to transform E. coli. Because pCSI-HA carries the ampicillin resistance gene, bacterial cells transformed with total liver DNA were spread on ampicillin plates and incubated, and colonies were counted, reflecting the relative amount of unintegrated, circular integrase-encoding plasmid DNA that was present in the samples. The results (Fig. 3B) indicated that pCSI-HA-derived ampicillin-resistant colonies were detected 1 hr after injection, and there was an increase in the number of ampicillin-resistant colonies over the next four time points. Peak numbers of colonies were reached at 8 hr. Colony numbers then fell rapidly to low levels after 24 hr.
Last, we subjected total DNA from the mouse livers to PCR analysis, using primers that would detect the pCSI-HA plasmid, whether extrachromosomal or integrated (Fig. 3C). PCR is an extremely sensitive technique, capable of detecting small amounts of integrase-encoding DNA. Integrase DNA was detectable through the period of 180 days. However, the intensity of the PCR band decreased over time, especially after 4 days, suggesting a reduction in the amount of integrase plasmid present within the livers over time.
Time course of hFIX expression and plasmid retention in mouse liver
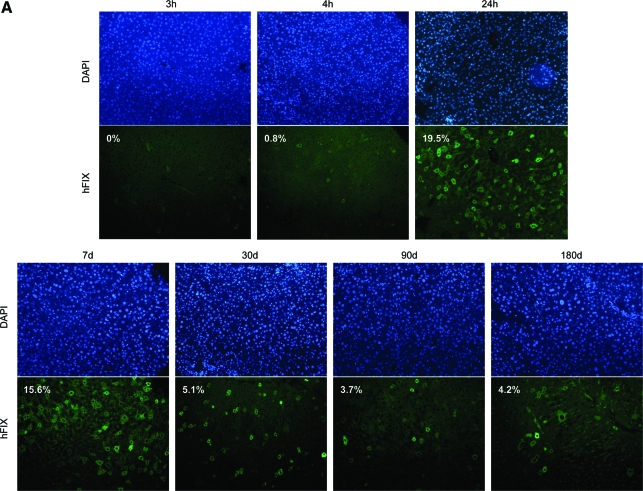
Along with the integrase expression plasmid pCSI-HA, we also introduced pVFB, which expresses hFIX from the liver-specific human α1-antitrypsin promoter (hAAT) and carries an attB site for integration. As seen in Fig. 4A, hFIX protein was first detectable, within 0.8% of the liver cells, by 4 hr after injection. hFIX levels then rose within the liver, to a peak of 19.5% of cells expressing hFIX at 24 hr postinjection. hFIX protein expression dropped to 5.1% of the cells expressing at 30 days postinjection and stabilized at about 4% of the liver cells over the 6 months of the experiment.
FIG. 4.
Analysis of hFIX expression in mouse liver. (A) Immunofluorescence staining for hFIX in mouse liver sections obtained over a time course. Twenty micrograms each of pCSI-HA and pVFB were hydrodynamically injected into the tail vein of mice. At various time points over a 6-month time course, the mice were killed and their livers were removed, sectioned, and stained with anti-hFIX antibody. The top row shows DAPI-stained nuclei, whereas the bottom row shows staining for hFIX. Numbers inside the images correspond to the percentage of cells that were expressing hFIX protein. (B) Total DNA transformed into bacterial cells showed an increase in colony numbers over the first 8 hr after injection, followed by a decrease, until few colonies were detected after day 3. Color images available online at www.liebertonline.com/hum.
To analyze persistence of pVFB DNA in the liver, 0.1 μg of total mouse liver DNA was used to transform E. coli. The kanamycin resistance gene is carried by pVFB. Therefore, bacterial cells transformed with total liver DNA were spread on kanamycin-containing plates and incubated, and colonies were counted. The colony numbers reflected the relative amount of unintegrated, intact hFIX plasmid that remained in the samples. Kanamycin-resistant colonies derived from pVFB were detected 1 hr postinjection (Fig. 4B). The number of kanamycin-resistant colonies increased over the next four time points. At 8 hr postinjection, peak numbers of colonies were reached. After 24 hr, colony numbers fell rapidly to low levels.
Time course of ϕC31 integrase expression and plasmid retention in cultured cells
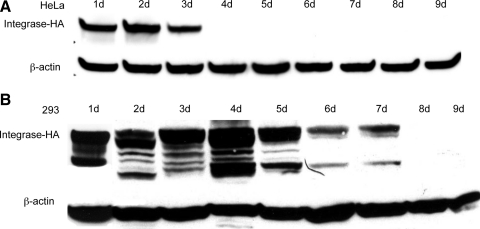
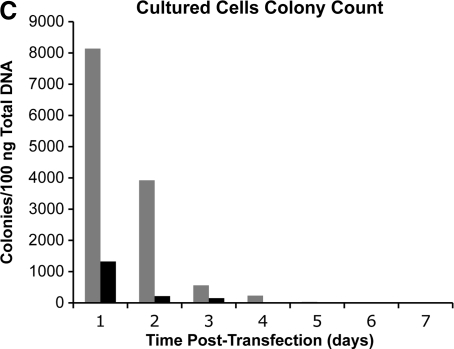
We were interested in whether the longevity of integrase expression in cultured cells was similar to what was observed in liver. Two human cell lines, HeLa and 293, were transfected with the pCSI-HA plasmid. Cells were harvested over a time course, total cell protein extracts were prepared, and ϕC31 integrase protein was detected by Western blot analysis using an anti-HA antibody. The results are shown in Fig. 5A and B. ϕC31 integrase was expressed at high levels for 3 days in HeLa cells and for 1 week in 293 cells. Furthermore, the levels of integrase protein present, as judged by the intensity of the signal on the Western blot, normalized to the amount of total protein, were approximately 3-fold higher in the cultured cells, compared with the integrase levels in liver (Fig. 1A).
FIG. 5.
Duration of integrase expression and plasmid retention in two human cell lines. One microgram of pCSI-HA was transfected into two tissue culture cell lines. At various time points over a 9-day time course, the cells were harvested and total protein and DNA were isolated. (A) Western blot analyses of total protein isolated from HeLa cells showed the presence of ϕC31 integrase protein at 1 day posttransfection. Integrase protein levels were stably expressed over a 3-day period, until they dropped below detection levels on day 4. (B) Western blot analyses of total protein isolated from 293 cells showed the presence of ϕC31 integrase protein at 1 day posttransfection. Integrase protein levels reached a peak 4 days posttransfection and then declined over a 3-day period, until they dropped below detection levels on day 8. β-Actin was used as a loading control. (C) Total DNA transformed into bacterial cells from HeLa (solid columns) and 293 cells (shaded columns) showed the maximal colony number 1 day after transfection, followed by a rapid decline, until few colonies were detected after day 6.
To analyze how much integrase plasmid DNA remained in the cells during the time course, total DNA was isolated from cells at each time point. The resulting DNA (0.1 μg) was transformed into E. coli, and bacteria were grown on ampicillin plates. Ampicillin-resistant colonies were counted after 24 hr. Colony numbers reflect the amount of free plasmid DNA present in the cells. The results (Fig. 5C) indicated that integrase plasmid DNA became less abundant in parallel with the decline in integrase expression in HeLa cells, consistent with progressive plasmid loss during cell division being a primary cause of loss of integrase expression. In 293 cells, ϕC31 integrase protein levels remained high, even as plasmid pCSI-HA levels fell, consistent with the strong and prolonged expression of transfected DNA characteristic of 293 cells.
Inhibitor studies to elucidate ϕC31 integrase half-life and degradation location
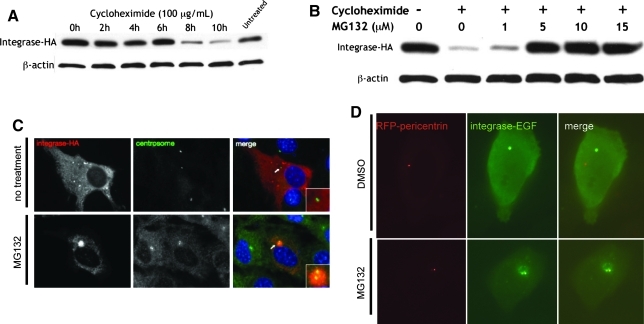
To elucidate the half-life of ϕC31 integrase protein in mammalian cells and to investigate its site of degradation, we used the ribosomal inhibitor cycloheximide (Colombo et al., 1965) and the proteasomal inhibitor MG132 (Palombella et al., 1994). Transfected HeLa cells were treated with cycloheximide and then lysed every 2 hr to obtain protein. The amount of integrase protein present in the same amount of total protein lysate as determined by the Bradford method was examined by immunoblotting, using an antibody to the HA tag. The pool of integrase protein was found to remain constant until 6 hr after ribosomal inhibition was initiated, followed by a decrease to barely detectable levels at 8 hr postinhibition (Fig. 6A). Therefore, the half-life of ϕC31 integrase was estimated to be between 6 and 8 hr (Fig. 6A). This relatively short half-life led us to investigate whether ϕC31 integrase might be actively degraded by the 26S proteasome.
FIG. 6.
ϕC31 integrase protein is degraded within 8 hr by the 26S proteasome. (A) HeLa cells transfected with pCSI-HA were treated with cycloheximide (100 μg/ml) to inhibit protein production and harvested every 2 hr at the indicated times (0–10 hr after adding cycloheximide). Protein levels in the cells were greatly diminished by 8 hr. (B) HeLa cells transfected with pCSI-HA were treated with either no cycloheximide or with cycloheximide at 100 μg/ml (– or+, respectively) and the MG132 concentration as indicated (0, 1, 5, 10, or 15 μg/ml) to inhibit the 26S proteasome. When cells were treated with both inhibitors, the pool of integrase was stabilized. In both (A) and (B), the immunoblot was cut in half at 50 kDa after blocking and incubated with either anti-HA or anti-β-actin primary antibodies. (C) NIH/3T3 cells transfected with pCSI-HA were either untreated (top) or treated with 10 μM MG132 for 10 hr (bottom), methanol-fixed, and stained on coverslips with both anti-HA (integrase-HA, red) and anti-γ-tubulin (centrosome, green) primary antibodies. (D) HeLa cells were cotransfected with pRFP-pericentrin (red) and pPhi-EGFP (green), treated with either 10 μM MG132 (bottom) or the same volume of the solvent DMSO (top) for 8 hr, and live imaged. Only when the 26S proteasome was inhibited could ϕC31 integrase be seen at the centrosome, a site of concentration for the proteasomal machinery. Thus, ϕC31 integrase appeared to be quickly degraded by the 26S proteasome at the centrosome in cultured mammalian cells.
After 10 hr of cycloheximide treatment, most of the integrase protein was gone. By contrast, in HeLa cells receiving both cycloheximide and 5–15 μM levels of the proteasomal inhibitor MG132 for 10 hr, an amount equal to the starting amount of integrase protein was still present in the cell (Fig. 6B). This result suggested that the 26S proteasome was responsible for the degradation of ϕC31 integrase.
A well-established site of proteasome localization is the centrosome (Wigley et al., 1999). The centrosome acts as an organizing structure and common meeting point for the active proteasome, chaperone proteins, and misfolded or ubiquitinated protein substrates (Fabunmi et al., 2000). To further establish proteasomal degradation of ϕC31 integrase, we imaged untreated and proteasome-inhibited cells in an attempt to colocalize the enzyme and the centrosome. NIH/3T3 cells were used because they are flatter than HeLa cells, allowing for better visualization. Staining for integrase-HA in untreated cells revealed what appeared to be inclusion bodies throughout the cytoplasm (Fig. 6C). This result implied that proteasomal degradation of integrase might occur through nonspecific, low levels of ubiquitination of the misfolded aggregates, as has been the case with other proteins (Dantuma and Lindsten, 2010). Indeed, integrase-HA and the centrosomal marker γ-tubulin colocalized in the proteasome-inhibited cells (Fig. 6C). To reduce potential artifacts and confirm these results, we also used a live-cell imaging approach. Constructs expressing two fusion proteins, integrase–EGFP and RFP–pericentrin, were transfected into HeLa cells. The latter protein marks the centrosome in live cells (Gillingham and Munro, 2000). Once again, ϕC31 integrase was localized to the centrosome in proteasome-inhibited cells (Fig. 6D).
Discussion
In this study, we determined the kinetics of ϕC31 integrase protein expression within mouse liver. Western blot and immunofluorescence analyses indicated that the majority of ϕC31 integrase protein expression was confined to a 22-hr period after hydrodynamic injection (hours 2–24) (Fig. 1). The 2-hr lag time between plasmid injection and the accumulation of sufficient ϕC31 integrase protein for detection is in agreement with the observations of Rossmanith and colleagues (2002), where β-galactosidase activity was detected in mouse liver 2 hr after hydrodynamic injection. Over the next 6 hr (hours 2–8), we observed a sharp rise in the ϕC31 integrase protein levels within the liver, as seen by Western blot and immunofluorescence staining (Fig. 1A and B). Peak expression levels were reached at 8–16 hr, when 19.6 and 14.9% of the hepatocytes were positive for ϕC31 integrase protein. This elevation in ϕC31 integrase protein levels correlated with increased intracellular pCSI-HA levels as seen by Southern blot and colony count assay (Fig. 3A and B). It therefore appeared that after hydrodynamic injection, plasmid DNA accumulated within the liver over an 8-hr period.
Western blot and immunofluorescence data showed that the ϕC31 integrase protein levels were fairly constant from hours 8–16 (Fig. 1A and B). However, there was a large drop in expression between 16 and 24 hr after injection. Southern blot analysis and colony count data showed that after 8 hr, pCSI-HA levels began to drop somewhat, but were fairly stable up to 24 hr (Fig. 3A and B). Therefore, the drop in expression between 16 and 24 hr was not due predominantly to plasmid loss, but was more likely due to silencing of the CMV promoter. One hour after ϕC31 integrase protein was first detected in the liver and within 3 hr of injection, pVFB integration was observed by PCR (Fig. 2). The increased concentration of plasmid pCSI-HA within the liver over the first 8 hr after injection (Fig. 5) could be due to the accumulation of free plasmid circulating in the blood within the liver. High pressure and volume during the injection process results in transport of plasmid through the fenestrae of the sinusoidal endothelial cells, where the plasmid DNA enters the space of Disse and is probably taken up by the hepatocytes directly (Fig. 3A and B). The ϕC31 protein expression profile followed a time course similar to that of the plasmid, but showed a delay in peak expression that correlated well with the proposed 6- to 8-hr half-life of the enzyme. After hour 16, there was a rapid loss of pCSI-HA plasmid from the liver. This was most likely due to lysosomal degradation of the plasmid as shown by Lecocq and colleagues (2003). Because of the sensitive nature of PCR, we were able to detect the presence of the integrase gene for the full 6 months of the experiment (Fig. 3C), probably reflecting trace amounts of randomly integrated plasmid DNA.
In addition to determining the time course of ϕC31 integrase expression, we also determined the expression of hFIX over time. The hFIX gene was driven by the liver-specific human α1-antitrypsin promoter (hAAT), not the CMV promoter used to express ϕC31 integrase. As can be seen in Fig. 4, hFIX was first detectable in the liver at 4 hr postinjection, with 0.8% of the cells expressing. This was 2 hr after ϕC31 integrase expression began. We believe the difference in the temporal expression patterns of hFIX and ϕC31 integrase to be due to the different promoters driving each gene. hFIX expression dropped from approximately 20% to 4% of cells and remained at this level long-term, implying an integration frequency of about 20%, which is sufficient for long-term therapeutic production of hFIX expression in vivo (Olivares et al., 2002). It is likely that most of this hFIX expression was derived from integrated pVBF plasmid, because little or no free hFIX plasmid remained at these times (Fig. 4B). It is notable that the longevity of pVBF plasmid DNA was likely shortened by its coexistence with an integrase plasmid, as we have shown previously (Olivares et al., 2002). This effect is probably due to removal of some hFIX free plasmid by integration, combined with more rapid degradation of the rest through binding by integrase and subsequent rapid degradation (Fig. 6).
We also demonstrated that ϕC31 integrase was expressed in tissue culture cells for up to 1 week after transfection, had a half-life of 6–8 hr, and was degraded by the 26S proteasome (Figs. 5 and 6). One might hypothesize that the longer term expression of ϕC31 integrase within cultured cells could lead to undesired events such as chromosomal rearrangements, and work in tissue culture cells has revealed such events (Chalberg et al., 2006; Ehrhardt et al., 2006; Liu et al., 2006). The larger quantity and longer duration of integrase expression after in vitro transfection that was uncovered here may be responsible for a higher frequency of aberrant events in cultured cells, compared with in vivo. The artificial conditions of cell culture, including elevated oxygen concentration, may also contribute (Atkuri et al., 2007).
Cytosolic proteins are most often degraded by the 26S proteasome, with half-lives ranging from only a few minutes (p53) to several days (cardiac myosin) (Khoo et al., 2009; Dantuma and Lindsten, 2010). However, many proteins, such as those that are extracellular, membrane-bound, or only monoubiquitinated, are degraded by the lysosomal pathway (Raiborg et al., 2003). Because ϕC31 integrase is a protein that evolved for expression in Streptomyces species, but has been artificially expressed in mammalian cells for the purposes of gene therapy, it was not known whether it might possess signals or other cues that would target it to a particular protein degradation pathway. Here, we demonstrated that ϕC31 integrase was expressed quickly in great quantity from the constitutive CMV promoter in the mammalian cells, resulting in inclusion bodies of misfolded protein throughout the cytoplasm (Fig. 6C). This misfolded fraction, comprising half of the total pool of cytoplasmic ϕC31 integrase in transfected HeLa cells, could also be detected by Western blot in the insoluble protein fraction (Woodard et al., 2010a). Rather than the inclusion bodies being endocytosed and lysosomally degraded, stabilization of the amount of integrase and colocalization with the centrosome under conditions of MG132 treatment point toward proteasomal degradation of the ϕC31 integrase along a time course generally recognized as short (half-life less than 10 hr) (Berger et al., 1988). This short half-life may be desirable for gene therapy, in that the length of exposure of the mammalian genome to the integrase is limited, reducing the need for a system of integrase regulation.
Acknowledgments
C.L.C. was supported by a Dean's Postdoctoral Fellowship from Stanford University School of Medicine, and L.E.W. was supported by grant CA09302 from the NCI. This work was supported by NIH grant HL68012 to M.P.C. M.P.C. is an inventor on Stanford-owned patents covering ϕC31 integrase.
References
- Atkuri K. Herzenberg L. Niemi A. Cowan T. Herzenberg L. Importance of culturing primary lymphocytes at physiological oxygen levels. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4547–4552. doi: 10.1073/pnas.0611732104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belteki G. Gertsenstin M. Ow D.W. Nagy A. Site-specific cassette exchange and germline transmission with mouse ES cells expressing the ϕC31 integrase. Nat. Biotechnol. 2003;21:321–324. doi: 10.1038/nbt787. [DOI] [PubMed] [Google Scholar]
- Berger J. Eisenhauer D. Taylor A. Intracellular protein degradation in cultured bovine lens epithelial cells. In Vitro Cell Dev. Biol. 1988;24:990–994. doi: 10.1007/BF02620871. [DOI] [PubMed] [Google Scholar]
- Bischof J. Maeda R.K. Hediger M. Karch F. Basler K. An optimized transgenesis system for Drosophila using germ-line-specific ϕC31 integrases. Proc. Natl. Acad. Sci. U.S.A. 2007;104:3312–3317. doi: 10.1073/pnas.0611511104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M.P. The ϕC31 integrase system for gene therapy. Curr. Gene Ther. 2006;6:633–645. doi: 10.2174/156652306779010642. [DOI] [PubMed] [Google Scholar]
- Chalberg T.C. Portlock J.L. Olivares E.C. Thyagarajan B. Kirby P. Hillman R.T. Hoelters J. Calos M.P. Integration specificity of phage ϕC31 integrase in the human genome. J. Mol. Biol. 2006;357:28–48. doi: 10.1016/j.jmb.2005.11.098. [DOI] [PubMed] [Google Scholar]
- Colombo B. Felicetti L. Baglioni C. Inhibition of protein synthesis by cycloheximide in rabbit reticulocytes. Biochem. Biophys. Res. Commun. 1965;18:389–395. doi: 10.1016/0006-291x(65)90719-9. [DOI] [PubMed] [Google Scholar]
- Dantuma N. Lindsten K. Stressing the ubiquitin/proteasome system. Cardiovasc. Res. 2010;85:263–271. doi: 10.1093/cvr/cvp255. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A. Engler J. Xu H. Cherry A. Kay M. Molecular analysis of chromosomal rearrangements in mammalian cells after ϕC31-mediated integration. Hum. Gene Ther. 2006;17:1077–1094. doi: 10.1089/hum.2006.17.1077. [DOI] [PubMed] [Google Scholar]
- Fabunmi R. Wigley W. Thomas P. Demartino G. Activity and regulation of the centrosome-associated proteasome. J. Biol. Chem. 2000;275:409–413. doi: 10.1074/jbc.275.1.409. [DOI] [PubMed] [Google Scholar]
- Gillingham A. Munro S. The PACT domain, a conserved centrosomal targeting motif in the coiled-coil proteins AKAP450 and pericentrin. EMBO Rep. 2000;1:524–529. doi: 10.1093/embo-reports/kvd105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A.C. Olivares E.C. Thyagarajan B. Calos M.P. A phage integrase directs efficient site-specific integration in human cells. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5995–6000. doi: 10.1073/pnas.090527097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held P.K. Olivares E.C. Aguilar C.P. Finegold M. Calos M.P. Grompe M. In vivo correction of murine hereditary tyrosinemia type I by ϕC31 integrase-mediated gene delivery. Mol. Ther. 2005;11:399–408. doi: 10.1016/j.ymthe.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Keravala A. Lee S. Thyagarajan B. Olivares E.C. Gabrovsky V. Woodard L.E. Calos M.P. Mutational derivatives of ϕC31 integrase with enhanced efficiency and specificity. Mol. Ther. 2009;17:112–120. doi: 10.1038/mt.2008.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoo K. Mayer S. Fersht A. Effects of stability on the biological function of p53. J. Biol. Chem. 2009;284:30974–30980. doi: 10.1074/jbc.M109.033183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhstoss S. Rao R.N. Analysis of the integration function of the streptomycete bacteriophage ϕC31. J. Mol. Biol. 1991;222:897–908. doi: 10.1016/0022-2836(91)90584-s. [DOI] [PubMed] [Google Scholar]
- Lecocq M. Andrianaivo F. Warnier M. Wattiaux-De Coninck S. Wattiauz R. Jadot M. Uptake by mouse liver and intracellular fate of plasmid DNA after a rapid tail vein injection of a small or a large volume. J. Gene Med. 2003;5:142–156. doi: 10.1002/jgm.328. [DOI] [PubMed] [Google Scholar]
- Liu F. Song Y.K. Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Liu J. Jeppesen I. Nielsen K. Jensen T.G. ϕC31 integrase induces chromosomal aberrations in primary human fibroblasts. Gene Ther. 2006;13:1188–1190. doi: 10.1038/sj.gt.3302789. [DOI] [PubMed] [Google Scholar]
- Olivares E.C. Hollis R.P. Chalberg T.W. Meuse L. Kay M.A. Calos M.P. Site-specific genomic integration produces therapeutic factor IX levels in mice. Nat. Biotechnol. 2002;20:1124–1128. doi: 10.1038/nbt753. [DOI] [PubMed] [Google Scholar]
- Olivares E.C. Calos M.P. Phage ϕC31 integrase-mediated site-specific integration for gene therapy. Gene Ther. Regul. 2003;2:103–120. [Google Scholar]
- Palombella V. Rando O. Goldberg A. Maniatis T. The ubiquitin–proteasome pathway is required for processing the NF-κB1 precursor protein and the activation of NF-κB. Cell. 1994;78:8773–8785. doi: 10.1016/s0092-8674(94)90482-0. [DOI] [PubMed] [Google Scholar]
- Raiborg C. Rusten T. Stenmark H. Protein sorting into multivesicular endosomes. Curr. Opin. Cell Biol. 2003;4:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- Rausch H. Lehmann M. Structural analysis of the actinophage ϕC31 attachment site. Nucleic Acids Res. 1991;19:5187–5189. doi: 10.1093/nar/19.19.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond C.S. Soriano P. High-efficiency FLP and ϕC31 site-specific recombination in mammalian cells. PLoS One. 2007;2:e162. doi: 10.1371/journal.pone.0000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossmanith W. Chabicovsky M. Herkner K. Schulte-Hermann R. Cellular gene dose and kinetics of gene expression in mouse livers transfected by high-volume tail-vein injection of naked DNA. DNA Cell Biol. 2002;21:837–845. doi: 10.1089/104454902320908496. [DOI] [PubMed] [Google Scholar]
- Thorpe H.M. Smith M.C.M. In vitro site-specific integration of bacteriophage DNA catalyzed by a recombinase of the resolvase/invertase family. Proc. Natl. Acad. Sci. U.S.A. 1998;95:5505–5510. doi: 10.1073/pnas.95.10.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thyagarajan B. Olivares E.C. Hollis R.P. Ginsburg D.S. Calos M.P. Site-specific genomic integration in mammalian cells mediated by phage ϕC31 integrase. Mol. Cell. Biol. 2001;21:3926–3934. doi: 10.1128/MCB.21.12.3926-3934.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venken K.J.T. He Y. Hoskins R.A. Bellen H.J. P[acman]: A BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–1751. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- Wigley W. Fabunmi R. Lee M. Marino C. Muallem S. Demartino G. Thomas P. Dynamic association of proteasomal machinery wtih the centrosome. J. Cell Biol. 1999;145:481–490. doi: 10.1083/jcb.145.3.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard L. Hillman R. Keravala A. Lee S. Calos M. Effect of nuclear localization and hydrodynamic delivery-induced cell divisin on ϕC31 integrase activity. Gene Ther. 2010a;17:217–226. doi: 10.1038/gt.2009.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodard L. Keravala A. Jung W. Wapinski O. Yang Q. Felsher D. Calos M. Impact of hydrodynamic injection and ϕC31 integrase on tumor latency in amouse model of MYC-induced hepatocellular carcinoma. PLoS One. 2010b;5:e11367. doi: 10.1371/journal.pone.0011367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G. Budker V. Wolff J.A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum. Gene Ther. 1999;10:1735–1737. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]