Abstract
Using behavioral and genetic information from the Autism Genetics Resource Exchange (AGRE) data set we developed phenotypes and investigated linkage and association for individuals with and without Autism Spectrum Disorders (ASD) who exhibit expressive language behaviors consistent with a motor speech disorder. Speech and language variables from Autism Diagnostic Interview-Revised (ADI-R) were used to develop a motor speech phenotype associated with non-verbal or unintelligible verbal behaviors (NVMSD:ALL) and a related phenotype restricted to individuals without significant comprehension difficulties (NVMSD:C). Using Affymetrix 5.0 data, the PPL framework was employed to assess the strength of evidence for or against trait-marker linkage and linkage disequilibrium (LD) across the genome. Ingenuity Pathway Analysis (IPA) was then utilized to identify potential genes for further investigation. We identified several linkage peaks based on two related language-speech phenotypes consistent with a potential motor speech disorder: chromosomes 1q24.2, 3q25.31, 4q22.3, 5p12, 5q33.1, 17p12, 17q11.2, and 17q22 for NVMSD:ALL and 4p15.2 and 21q22.2 for NVMSD:C. While no compelling evidence of association was obtained under those peaks, we identified several potential genes of interest using IPA. Conclusion: Several linkage peaks were identified based on two motor speech phenotypes. In the absence of evidence of association under these peaks, we suggest genes for further investigation based on their biological functions. Given that autism spectrum disorders are complex with a wide range of behaviors and a large number of underlying genes, these speech phenotypes may belong to a group of several that should be considered when developing narrow, well-defined, phenotypes in the attempt to reduce genetic heterogeneity.
Electronic supplementary material
The online version of this article (doi:10.1007/s11689-010-9063-2) contains supplementary material, which is available to authorized users.
Keywords: Autism, Linkage analysis, Association analysis, Motor Speech Disorder, Autism genes, Ingenuity Pathway Analysis (IPA), PPL/PPLD
Introduction
Over the past 10 years there has been compelling evidence supporting a genetic basis for autism using a combination of behavioral family studies and genetic linkage and association studies. However, these studies have produced results that are often inconsistent and sometimes contradictory (Newbury et al. 2002). Some linkage studies have identified peaks based on the presence or absence of autism or autism spectrum disorders (ASD), while other studies have concentrated on more specific phenotypic and clinical characterizations such as onset age of first words, family language history, sex of proband, obsessive compulsive and ritualistic behaviors, and social skills [(Alarcón et al. 2002; Auranen et al. 2003; Bradford et al. 2001; Buxbaum et al. 2004; Liu et al. 2008; Shao et al. 2003); See (Abrahams and Geschwind 2010) for a current linkage review].
Of particular interest for several research groups has been the attempt to define and then replicate significant linkage signals using language-based phenotypes in ASD probands with the objective of finding genes that are associated with a specific language-related phenotype. An area on chromosome 7 (q34–36) has been linked to both autism and expressive language impairments. A gene for a contactin associated protein, CNTNAP2, that is down regulated by FOXP2 and is known to influence early brain development in humans, has been associated with both ASD and language (Alarcón et al. 2008; Arking et al. 2008; Vernes et al. 2008). While chromosome 7q continues to be an area of intense interest for both autism and language, other linkage signals have been reported that are also based on language phenotypes in the ASD population. Alarcón et al. (2005) reported linkage on chromosomes 3q and 17q using onset of first words and phrases as the behavioral phenotype while linkage on chromosome 13q21 was reported by Bradford et al. (2001) for ASD probands and family members with a history of language-related problems. Bartlett et al. (2004) identified linkage in the same region for a sample of families with a history of Specific Language Impairment (SLI) without ASD. SLI is a failure to develop language normally without explanatory factors such as low IQ, gross neurological impairment, or inadequate environment. They suggest that although SLI and ASD are distinctly different disorders, both are genetically complex and may share specific susceptibility genes or variants of genes. Spence et al. (2006) stratified expressive language characteristics into word and phrase speech delay in ASD probands and family members in an attempt to better define the language endophenotype and reduce phenotypic heterogeneity. They found evidence for linkage in several already identified locations supporting the idea that more discretely defined characteristics of ASD, specifically language endophenotypes, may improve localization of linkage signals and strengthen existing findings.
Speech and language in ASD
Speech and language impairments constitute a broadly defined area. In their mildest forms they may be characterized by a minor phonological or speech impairment that can affect speech production and possibly reading ability. On the more severe end of the language and speech continuum, a person might be unable to comprehend or process spoken language and/or be non-verbal or unintelligible. This vast scope of speech and language disabilities seen in the ASD population has been documented in detail (Rapin and Dunn 2003; Tager-Flusberg et al. 2005). While some research supports the notion that there may be multiple relations among the language problems seen in SLI and autism, others feel that there is not enough evidence to support a genetic link (Lindgrin et al. 2009) and that ASD and SLI are distinctly different disorders that do not share the same genes.
Previous reports indicate that approximately 50% of all children with autism never acquire functional language by middle childhood (Bailey et al. 1996) while more current estimates place this value as closer to 20% (Lord et al. 2004). Yet little is known about why some individuals, despite years of intervention, never develop language while many others develop enough spoken language to communicate at least minimally. Often the underlying cause is not clear and may be presumed to be a social/interaction issue. But what if language processing problems make incoming verbal information difficult or impossible to understand and severely limit verbal output? Conversely, what if problems with speech output make speech very effortful, resulting in vocalizations that include only vowels sounds or verbalizations that are unintelligible to those around them, as in the case of childhood apraxia of speech (CAS)?
CAS is a motor speech disorder that involves poor motor planning and results in speech output with compromised intelligibility ranging from its most severe form of expressive language production, which is characterized by very limited consonant production, to full phrase production with multiple omissions, substitutions, distortions, and reversals of speech sounds. While good epidemiologic data on the prevalence of CAS is lacking, population estimates derived from referral data suggest that approximately one to two children per 1,000 are affected with CAS (Shriberg et al. 1997). There has been limited study of CAS in terms of its genetic origins, however, in their family and genetic studies of speech sound disorders, Lewis et al. (2004) looked at a small sample of children with a diagnosis of CAS and reported that 59% had at least one parent with some type of speech sound disorder. Moreover, in 86% of the families, at least one nuclear family member reported either a speech sound disorder or a language disorder. In a recent related study of speech sound disorders Lewis et al. (2007) report that 36 of 147 (24%) of parents of children with speech sound disorders also report similar problems as children.
Very little has been reported about individuals with autism whose vocalizations are effortful, unintelligible, or non-existent. One of the few studies (Gernsbacher et al. 2009) was a retrospective study of children’s oral-motor skills that compared toddlers with autism to matched controls. Using videotapes and a detailed questionnaire, they determined that the quality of oral motor skills during the early years was associated with the level of speech intelligibility of the individuals with autism in later years. Minimally verbal older children had poorer oral motor skills as toddlers.
In the current Autism Genetic Resource Exchange (AGRE) dataset, approximately 16% of the individuals who were evaluated with the Autism Diagnostic Interview-Revised (ADI-R) (Lord et al. 1994; Rutter et al. 2003) are non-verbal or minimally verbal at the time of their evaluations. Another 16% of the individuals in the dataset have speech that is unintelligible to most people.
Based on our review of the speech and language characteristics of the subjects in the AGRE database, we suggest that there is a subset of individuals with and without ASD who exhibit an expressive language problem that ranges from being non-verbal to having expressive language that is unintelligible to others and may actually be described as a severe motor speech disorder such as verbal apraxia. As these speech and language behaviors are seen in only a subset of individuals with autism but also seen in individuals who do not meet ASD criteria, we investigated linkage and association for this behavior as part of a broader phenotype.
Methods
Participants
Subjects in this study were obtained from families who are part of the AGRE database. Unlike many of the other studies that use the AGRE data for linkage analysis and require at least two affected siblings with an ASD diagnosis, we targeted AGRE families who had at least two individuals who were either non-verbal, minimally verbal, or who had speech that was unintelligible to others, regardless of ASD diagnosis.
Responses to specific questions from the ADI-R for all families with available Affymetrix 5.0 data were used (N = 723). Motor-speech phenotypes were then developed to explore linkage and association based on the hypothesis that a subset of individuals on the autism spectrum with little or no expressive language may be part of a distinct phenotype common to autism but also potentially common to other speech and language disorders.
Motor speech phenotypes
Responses by parents and caregivers to the ADI-R were used as variables to develop the phenotypes used for the current analyses. The ADI-R is a semi-structured clinical review for caregivers of children and adults who are suspected of being on the autism spectrum. The ADI-R focuses on three areas of behavior: (a) social interaction; (b) communication; and (c) interests and behaviors that are stereotyped or restricted and repetitive. Variables from the Communication Scale were used to develop the current motor-speech phenotypes and are available in Supplemental Table 1.
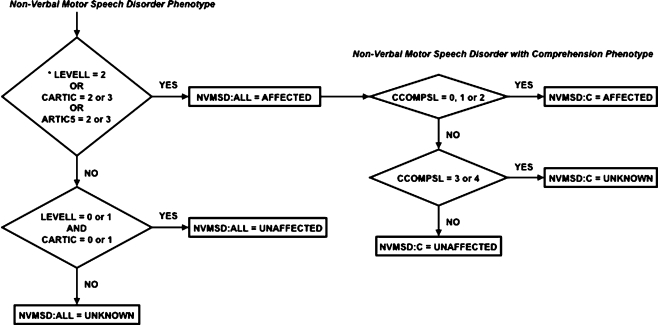
Family members who received the ADI-R (irrespective of their diagnosis of autism, ASD, or not ASD) were included in the evaluation of phenotype status. Any family member who was at least 2 years old and non-verbal or minimally verbal or was at least 4 years old, verbal, but very difficult to understand due to poor sound production, was considered affected for the NVMSD:ALL phenotype (Non-Verbal Motor Speech Disorder:All). The NVMSD:C phenotype (Non-Verbal Motor Speech Disorder:Comprehension) represented a subset of the NVMSD:ALL phenotype and included subjects who were non-verbal or unintelligible but had at least minimal language comprehension (i.e. could at least follow simple directions) as reported in the ADI-R. Figure 1 reflects the decision process used to assign affection status for the two motor speech phenotypes: Non-Verbal, Motor Speech Disorder (NVMSD:ALL) and Non-Verbal, Motor Speech with Comprehension (NVMSD:C).
Fig. 1.
Individuals who lacked functional language (LEVELL = 2) or had poor speech intelligibility (CARTIC or ARTIC5 = 2 or 3) were AFFECTED for the NVMSD:ALL phenotype. If the language level was fair/good (LEVELL = 0 or 1) and current intelligibility was fair/good (CARTIC = 0 or 1) they were labeled UNAFFECTED, otherwise they were labeled UNKNOWN. Note the CARTIC ADI-R score took precedence over the ARTIC5 score (as long as ARTIC5 was not 2 or 3 which was established by the previous test) then its contribution was not considered for this distinction. Only those individuals that were affected according to the NVMSD:ALL phenotype were evaluated on their level of spoken language comprehension (NVMSD:C phenotype). Individuals with some language comprehension (CCOMPSL = 0, 1 or 2) were labeled AFFECTED, those with very little to no comprehension (CCOMPSL = 3 or 4) were labeled UNKNOWN, and the remainder as UNAFFECTED. * Note that individuals had to be at least 2 years old to have a LEVELL score and had to be at least 4 years old to have a CARTIC score but could potentially be older. Also, if an individual scored 2 for the LEVELL variable, they did not receive a score for CARTIC or ARTIC5 since they did not produce enough language to be evaluated.
There were 203 families (1,146 individuals) from the AGRE dataset who had both ADI-R data and Affymetrix 5.0 genotyping data and contained at least two individuals that met our criteria for the NVMSD:ALL phenotype. Among the 427 affected individuals for NVMSD:ALL (79% male), 383 met criteria for the narrow definition of autistic disorder (AD) based on the ADI-R and came from 202 families. The mean age at ADI-R assessment was 98.83 months (s.d. 63.18 months). Of these 203 NVMSD:ALL families, 135 families (778 individuals) contained at least two family members who, irrespective of their final autism diagnosis, met criteria for the NVMSD:C phenotype. Among the 281 affected individuals (80% male), 249 met criteria for the narrow definition of AD based on the ADI-R and came from 133 families. For this phenotype the mean age at ADI-R assessment was 107.02 months (s.d. 52.40 months) (See Supplemental Table 2). While all families were used in the Linkage Disequilibrium (LD) analyses, 35 NVMSD:ALL families were uninformative for linkage (19 in the case of NVMSD:C). This was due, in part, to affected sib-pair families in which the sibs turned out to be MZ twins.
Genotypes
Genotype data were downloaded from the AGRE site for all AGRE families with Affymetrix 5.0 data. Data on 443,106 SNPs were available for download. In preparation for linkage analysis, genotypic data were cleaned for missingness by marker (≤5% missing retained) and by individual (≤15% missing retained) (the average missingness rate was 1.5%, while the highest observed rate of missingness was 11%) and for relationship issues using RelCheck (Broman and Weber 1998) (no families were dropped based on RelCheck identified problems). Data were then screened for Mendel errors and any SNP showing a Mendel error in a particular family was zeroed out for the entire family (the average number of Mendel errors per family was 2,605). However, there were no families excluded due to excessive Mendelian errors.
In preparation for linkage analysis, markers were dropped if the minor allele frequency was <5% or if they showed any signs of departure from Hardy-Weinberg equilibrium (p < 0.05). A subset of these markers was selected at 0.3 cM intervals resulting in a marker map comprising 11,100 SNPs.
For LD analyses, SNPs were dropped if the minor allele frequency was <1%, or if they failed a test of Hardy-Weinberg equilibrium at the p = 10−10 level. This left a total of 263,334 SNPs in the analysis.
Statistical analyses
Analyses were conducted using the software package Kelvin that implements the PPL (posterior probability of linkage) class of models for measuring the strength of genetic evidence (Huang et al. 2006; Vieland 1998; Vieland 2006). Below we report the PPL, the PPLD (posterior probability of trait-marker linkage disequilibrium (LD) and linkage) (Yang et al. 2005) and the PPLD|L (posterior probability of LD given linkage) (Wratten et al. 2009). We report genome-wide PPLD results, and use the PPLD|L to evaluate the evidence for LD under linkage peaks only.
The PPL is parameterized in terms of a general approximating likelihood, and all parameters of the trait model are then integrated out permitting the use of Bayes’ theorem to compute the posterior probability of the hypothesis of interest. Hardy-Weinberg equilibrium has been assumed throughout. The genetic map is based on the Rutgers Combined Linkage-Physical Map, http://compgen.rutgers.edu; (Matise et al. 2007) release 10/09/06. Because Kelvin is at present Elston-Stewart based (Elston and Stewart 1971), the (multipoint) linkage analyses utilized LOD scores computed in Merlin (Lander and Green 1987; Abecasis et al. 2002) as input to PPL calculations (Vieland 1998) using Merlin’s SNP clustering (with r2 ≥ 0.2) to further reduce potential inflation due to residual LD in the marker map. (We experimented with marker effects by varying the density of the map, the particular markers included in the maps, and the r2 threshold for clustering SNPs, and the results were virtually identical in all cases; results not shown.)
All analyses shown here utilize a simple dichotomous trait model, with parameters α (the standard admixture parameter of Smith (1963) representing the proportion of ‘linked’ pedigrees), p (the disease allele frequency), and the penetrance vector f i (representing the probability that an individual with genotype i develops disease, for i = 1..3). All trait parameters are integrated out of the final statistic; while the gene frequency is integrated over its full range, an ordering constraint is imposed on the penetrances such that f 1 ≥ f 2 ≥ f 3. This model provides a robust approximation for mapping complex traits in terms of the marginal model at each locus, and because the parameters are integrated over, no specific assumptions regarding their values are required. Uniform prior distributions are used for all trait parameters (with adjustment for the ordering constraint). This model implicitly allows for dominant, recessive, and additive models, along with an explicit allowance for heterogeneity. In secondary analyses, we additionally allowed for imprinting or other parent-of-origin effects by allowing the penetrances to depend on the sex of the transmitting parent.
The PPL is on the probability scale. For instance, a PPL of 40% means that there is a 40% probability of a trait gene at the given location based on these data. For biological reasons, the prior probability of linkage at each location is set to 2% (Elston and Lange 1975) so that PPLs >2% indicate (some degree) of evidence in favor of the location as the site of a trait gene, while PPLs <2% represent evidence against the location. The prior probability of LD|L is also set to 2%, so that the prior probability of LD and L is 0.04%.
The PPL and PPLD are measures of statistical evidence, not decision making procedures; therefore there are no “significance levels” associated with them and they are not interpreted in terms of associated error probabilities (Vieland 1998; Royall 1997; Taper and Lele 2004). Similarly, no multiple testing corrections are applied to the PPL or the PPLD, just as one would not “correct” a measure of the temperature made in one location for readings taken at different locations (Vieland 1998). Nevertheless, it may assist readers to have some sense of scale relative to more familiar frequentist test statistics. In a simulation of 10,000 replicates of 200 affected sib pairs per replicate under the null hypothesis (no trait gene at the location being tested) allowing for the observed pattern of missing data, PPLs of 5%, 15%, 25%, 50%, and 80% were associated with Type 1 error probabilities of 0.031, 0.0018, 0.0001, 0.00005, and 0.00001, respectively.
The “null” behavior of the PPLD is moot given the results of the analysis of the experimental data; however, we note that in these same 10,000 replicates no PPLD >5% was observed. Given the sample size, we did not expect to detect LD at unlinked locations in this small set of families assuming low genotypic relative risks (RR’s). However, RR’s under linkage peaks might be expected to be considerably higher in which case power to detect LD under a linkage peak could actually be quite good. But power is entirely a function of the underlying generating model, which remains unknown. For example, fixing the RR at 2.5 and assuming D’ = 0.7 between the trait allele and marker allele, we simulated data under two different models: (a) we assumed locus homogeneity and dominant inheritance; (b) we assumed that only 20% of families carried the associated disease variant and that the mode of inheritance was recessive. In the first case, 96% of replicates showed PPLD ≥20%, 88% showed PPLD ≥50%, and 78% showed PPLD ≥80%. Thus for a model like this, “power” is excellent in this sample and failure to find LD under the linkage peaks is an interesting finding (assuming relatively good marker coverage). In the second case, however, only 2% of replicates showed PPLDs ≥20%, and <1% of replicates showed PPLDs ≥50%. Hence if this latter model is closer to the truth, our failure to detect LD under the linkage peaks may simply reflect the fact that the sample is still quite small.
Identifying susceptibility genes
Following the linkage and LD analyses, we used the Ingenuity Pathway Analysis—IPA software (Ingenuity® Systems, www.ingenuity.com) to identify potential autism susceptibility genes that might fall within our linkage regions. In order to characterize the peaks in our linkage analyses, we used three definitions of peak endpoints. The genome-wide PPL values were ranked (based on calculations done every 1 cM) in ascending order, and the highest 1%, 2.5%, and 5% PPL scores were used to define the narrow, intermediate, and broad regions, respectively. The narrow regions consisted of PPL values greater than 20%, the intermediate regions were greater than 15%, and the broad regions were greater than 5%. The genes within these regions were identified using the UCSC Genome Browser (NCBI Build 36.1, Kent et al. 2002) and were analyzed using the Core Analysis in IPA. The Core Analysis identified the biological functions and/or diseases that were most significant to each linkage analysis. A right-tailed Fisher’s exact test was used to calculate a p-value determining the probability that each biological function and/or disease assigned to that linkage analysis was due to chance alone. We selected genes with functions related to Nervous System Development and Function, Neurological Disorders, Genetic Disorders, and Psychological Disorders from the list of functions with significant p-values as possible candidate genes for our phenotypes.
As a control experiment to assess the uniqueness of our significant findings, we conducted IPA analyses on randomly selected sets of 645 genes, to model the number of genes obtained in our intermediate-peak definition analysis of NVMSD:ALL. We first identified regions of the genome centered about 2% PPL (evidence neither for nor against linkage) under the intermediate NVMSD:ALL scan and randomly selected 645 genes from these regions (Control: Gene Number—C:GN). However, as a total of only 3,549 genes were present in the areas with approximately 2% PPL values, this frequently led to partially overlapping sets of genes. In order to create more independent samples, a second set of control analyses were also conducted by selecting 645 genes at random from the entire genome (Control: Gene Number Genome—C:GNG). We conducted core analyses on 10 C:GN and 10 C:GNG control datasets and compared the results to the gene set defined by our intermediate linkage analysis results of NVMSD:ALL.
Results
Linkage and association
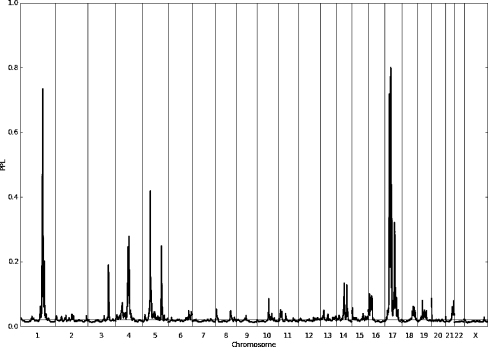
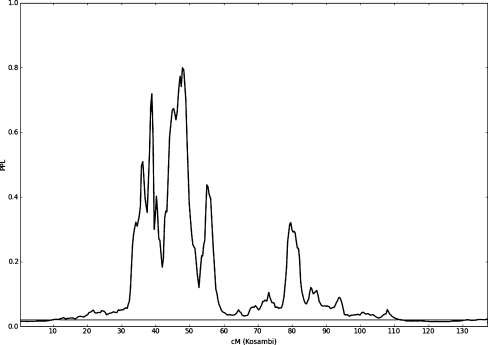
Figure 2 shows genome-wide PPL results for the NVMSD:ALL phenotype. As can be seen, while most of the genome shows evidence against linkage (PPL <2%) or very close to baseline (2%), there are several salient peaks. Table 1 shows all PPL peaks >15% for the NVMSD:ALL phenotype. On chromosome 17 there appear to be multiple peaks (Fig. 3).
Fig. 2.
Genome-wide PPL results for the NVMSD:ALL phenotype. The PPL is on the probability scale. Values >2% represent evidence in favor of linkage, while values <2% represent evidence against linkage.
Table 1.
NVMSD:ALL linkage peaks with PPL >15%
| Phenotype NVMSD:ALL (nonverbal motor speech) | PPL | Band name | |
|---|---|---|---|
| Chr | cM | ||
| chr1 | 180 | 0.74 | q24.2 |
| chr3 | 166 | 0.19 | q25.31 |
| chr4 | 109 | 0.28 | q22.3 |
| chr5 | 66 | 0.42 | p12 |
| chr5 | 156 | 0.25 | q33.1 |
| chr17 | 36 | 0.51 | p12 |
| chr17 | 39 | 0.72 | p12 |
| chr17 | 48 | 0.80 | p12 |
| chr17 | 55 | 0.44 | q11.2 |
| chr17 | 80 | 0.32 | q22 |
Fig. 3.
PPL across chromosome 17 for the NVMSD:ALL phenotype.
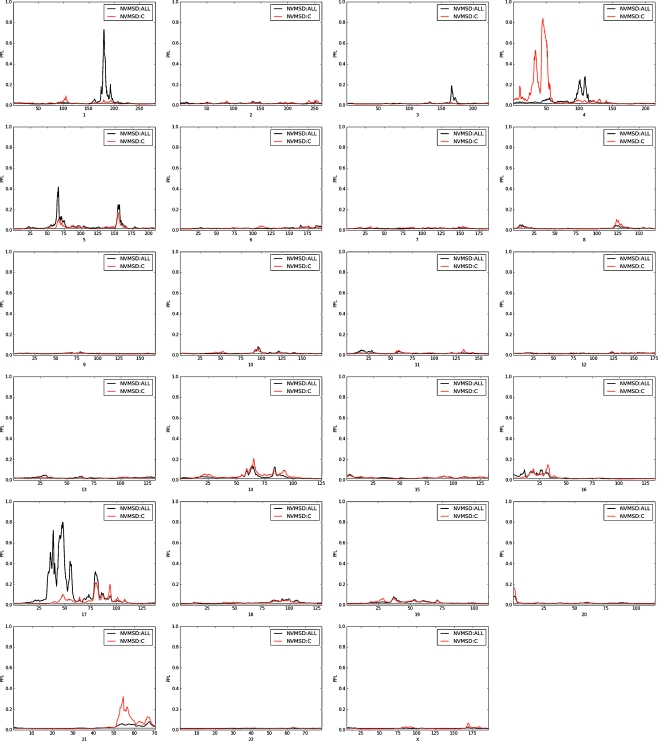
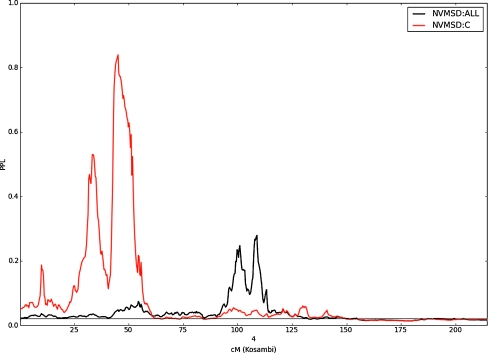
Figure 4 shows results by individual chromosomes for the NVMSD:C phenotype as well as the NVMSD:ALL phenotype. Because the families that are multiplex for NVMSD:C are a subset of those that are multiplex for the NVMSD:ALL phenotype, and because the two phenotypes themselves overlap by design, we expect correlation in the genome scans for the two phenotypes in these families. Moreover, because the NVMSD:C sample is smaller, we would expect to see smaller linkage signals in this group. As Fig. 4 shows, across almost all of the genome, this is exactly what we see: peaks in the same places as in Fig. 2, but lower. One notable exception to this is on 4p15.2, where the large peak in the NVMSD:C analysis (PPL = 84% at 45 cM) is quite far from the NVMSD:ALL peak, as shown in Fig. 5. Also of possible interest are the NVMSD:C peaks on 21q22.2 (PPL = 32% at 55 cM) and 14q24.2 (PPL = 20% at 65 cM).
Fig. 4.
Individual chromosome PPL results for NVMSD:ALL (in black) and NVMSD:C (in red).
Fig. 5.
PPL results across chromosome 4 for NVMSD:ALL (in black) and NVMSD:C (in red).
Notably, an allowance for imprinting did not produce any new peaks or substantially change results at most loci seen under the non-imprinting analyses. In most cases, allowance for imprinting slightly depressed peaks. The exceptions for NVMSD:ALL were on chromosome 17, where the peaks rise to 86% at 48 cM, 64% at 55 cM, and 47% at 80 cM; in all three cases inheritance from the father appeared to be silenced. The exceptions for NVMSD:C were on chromosome 4 (imprinting PPL = 97% at 45 cM), and chromosome 14 (imprinting PPL = 31% at 65 cM); in both of these cases penetrances appeared somewhat higher for paternal alleles but there was no indication of imprinting (full silencing) per se.
The PPLD accumulates evidence against LD as well as in favor of LD. Hence at a SNP that is not in LD with the trait, the larger the sample size the smaller the PPLD will become. For this reason, the smaller NVMSD:C data set yields a noisier GWAS plot around baseline. As discussed above, we were not expecting to see large PPLDs in a sample this size, and indeed, we do not see any. (Supplemental Figure 1)
Of greater interest than the genome-wide PPLDs, however, are the PPLD|L results under the linkage peaks. However, for NVMSD:ALL we did not find any evidence of LD under the linkage peaks. Considering any genomic locations where the PPL was ≥20%, the largest PPLD|L was less than 5%. While the small sample size may make it difficult to detect LD under the peaks, assembling a very large sample of families with this phenotype is difficult. Thus, whether for underlying biological reasons or simply due to practicalities, it does not appear that fine mapping via LD analysis under these linkage peaks is likely to uncover the underlying genes. For NVMSD:C, there were 13 SNPs under the NVMSD:C-specific peak on chromosome 4 (considering all locations where the PPL >20%) which yielded PPLD|L >5%; the maximum PPLD|L was 15%. However, these were distributed across a 20 cM region, complicating the interpretation for this (even smaller) sample. Two of these SNPs fall in genes, and one of these genes (KDNIP4, PPLD|L = 6% at rs1763197, located at physical location 20,692,185 bp or approximately 36.6 cM) is of potential interest for its possible role in regulation of neuronal excitability and interactions between its protein product and presenilin.
IPA core analysis under linkage peaks
Using the output from the core analysis, we selectively identified genes with functions related to Nervous System Development and Function, Neurological Disorders, Genetic Disorders, and Psychological Disorders. For NVMSD:ALL, a total of 25 genes were selected from 261 genes input to IPA for the narrow definition (highest 1% of PPLs), 52 of 645 for the intermediate definition (2.5% highest PPLs), and 62 of 1371 for the broad definition (highest 5% of PPLs). The functions that were most represented overall were neuronal development, myelination, and axonal guidance. Likewise for NVMSD:C, 23 of 111 genes were selected for the narrow definition, 51 of 388 for the intermediate definition, and 147 of 954 for the broad definition. There was an increase of molecules involved in axonal guidance in NVMSD:C. Analyses for both phenotypes reported genes involved in motor function and various psychiatric and neurological disorders including Schizophrenia, Bipolar Disorder, and Alzheimer’s disease (Supplemental Table 3a, b).
IPA core analysis in control regions
The IPA core analysis of the C:GN (control based on gene number) datasets resulted in a different distribution of relevant significant functions than did the analysis of the linkage data. In contrast to the NVMSD:ALL intermediate analysis, each C:GN analysis resulted in several relevant functions with at least 20 contributing genes (Supplemental Table 4b). Likewise, the number of multi-function control candidate genes is different in the C:GN analyses than in our linkage analyses. In our linkage analysis, there was one gene that contributed to 21 functions and the rest of the genes contributed 10 functions or less (Supplemental Table 4a). In the C:GN analyses, there were multiple genes that contributed to more than 15 functions and the rest contributed to 5 or less functions. The most common functions identified in the C:GN analyses were synaptic transmission, neurotransmission, and various psychological disorders (Supplemental Table 4b).
To overcome the potential bias introduced by restricting our control analysis to the relatively small percentage of the genome with PPL values tightly centered around 2%, we repeated this analysis using genes selected from the entire genome (C:GNG). A similar overall pattern of significant functions and multi-function candidate genes to the C:GN results was seen in the C:GNG analyses (see Supplemental Table 4c). Like the C:GN results, the most common functions identified in the C:GNG analyses were various psychological disorders and synaptic functionality. It should be noted that genes identified by our linkage analyses were not excluded from the C:GNG datasets (Supplemental Table 4b and c).
Overall, the most common diseases seen in both control analyses were neuropsychiatric disorders, such as Huntington’s Disease and Schizophrenia, and the most common functions were neurotransmission/synaptic transmission and development of neurons and neurites. The presence of these diseases and functions in our control analyses suggest that while they may be related to our phenotypes of interest, these functions are not unique to the core analysis of our linkage study, and may be an artifact of the extensive published research in these areas. Interestingly, only eight specific functions identified by the control analyses overlapped with those identified from the genes from our linkage analysis. Each of these functions (cell death of neuroglia and oligodendrocytes, learning by mice, plasticity of synapse, survival of cortical neurons, development of dentate gyrus, motor neurons, and peripheral nervous system) appeared only once in the control analyses. While there is some commonality in functions, it is important to note that the candidate genes described below are not implicated by functions identified in the control analyses.
Discussion
Linkage and association
We have identified several peaks that represent strong evidence of linkage using two novel and relatively narrow behavioral phenotypes for non-verbal language and motor speech problems; characteristics that are associated with autism but not exclusive to the autism spectrum. While some of the peaks overlap with previously reported linkage locations (Alarcón et al. 2005; Bartlett et al. 2005; Cantor et al. 2005; McCauley et al. 2005; Schellenberg et al. 2006; Yonan et al. 2003), others are novel. In some cases, where results overlap, different behavioral phenotypes have been reported for those peaks. This is not surprising since, by definition, an individual with autism might share behaviors and belong to several phenotypic subgroups within the spectrum as well as share behaviors with individuals who do not meet criteria for ASD. Additionally, there have been multiple studies looking at language and autism using the AGRE sample and the potential for overlapping subjects is impossible to avoid. However, the strength of the peaks and the specificity of the phenotype lend support to the idea that there could be genes of interest under these peaks that warrant further investigation.
We included all individuals from the AGRE data set who had ADI-R diagnostic information and Affymetrix 5.0 genetic data regardless of their final ASD diagnosis. A percentage of those identified as meeting criteria for one or both phenotypes did not meet the ASD cut-off criteria. Yet at some point, they must have demonstrated behaviors compatible with a potential ASD diagnosis or they would not have received the AGRE ASD study battery in the first place. This lends support to the notion that in families with ASD probands, there may be other family members that share behavioral characteristics and genes and fall into some kind of broader autism phenotype.
Evidence of linkage based on PPL values greater than 15% for the NVMSD:ALL phenotype was identified on chromosomes 1q24.2, 3q25.31, 4q22.3, 5p12, 5q33.1, 17p12, 17q11.2, and 17q22. Linkage on 3q25 and 17p supports previous findings in the same area where loci linked to word and phrase speech delays were identified by Alarcón et al. (2005). In that study, the authors identified a region on chromosome 3 (126–170 cM with a peak at 147 cM) that overlaps one of our peaks and was identified with an onset of first words phenotype. In addition, they identified a region on chromosome 17 (13–96 cM) that coincides with one of our peaks and is suggestive of linkage for first words and phrases. Since delayed first words, and/or delayed first phrases might also apply to a number of our probands, because they are nonverbal, one could speculate that there was overlap across our samples. In fact, our finding at chromosome 1q24 (PPL = 74%) was located in the same region as previously reported by our group (Bartlett et al. 2005). In the current study, 88 AGRE families also satisfied the diagnostic criteria for the Bartlett et al. study (i.e., the affected phenotype was based on delayed speech onset in two affected siblings).
For many previous autism linkage studies a more formal diagnostic phenotype was used that ranged from a narrow definition of autistic disorder to an autism spectrum disorder that included Asperger’s disorders and PDD-NOS. Since we already know that a significant number of individuals with autism can be non-verbal or have speech that is significantly unintelligible, it is not surprising that overlapping linkage peaks were observed. McCauley et al. (2005) identified linkage on 17q11 for sib pairs consisting of at least one proband with autistic disorder and one on the spectrum. Using a broader definition of autism, Yonan et al. (2003) identified linkage in the same regions of 17q and 5p as our study. Studying male-only families has been another approach (Cantor et al. 2005; Schellenberg et al. 2006) resulting in linkage peaks on chromosomes 4 and 17. When Schellenberg et al. (2006) stratified their families by male-only, they identified linkage at 4q22 and Cantor et al. (2005) identified the 17q21 region (67 cM) when doing fine mapping of the area. Similarly, our families were enriched for affected male subjects; we had 128 male-only families with the remaining 75 families having at least one female affected for our phenotype, bringing our rate of affected males to approximately 80% in our 203 families. Buxbaum et al. (2004) identified a peak on 1q24 as well and another peak suggestive of linkage on chromosome 4 for an obsessive-compulsive phenotype, thus another example of overlapping linkage based on different phenotypes but behaviors that are part of the ASD profile. In summary, even though samples varied and descriptions of phenotypes differed, the fact that most of our linkage peaks have been previously identified in ASD populations, lends support to the idea that these particular locations are a source for continued investigation.
The NVMSD:C phenotype was created to narrow and better define the motor-speech characteristics that are found in a subset of individuals with ASD as well as other individuals with speech and language impairments. It was based on the premise that probands who have some language comprehension but display minimal or unintelligible expressive language, might belong to a phenotypic group specifically characterized by a motor speech impairment that is seen in apraxia of speech. Using these criteria we identified similar, but weaker, linkage signals to the NVMSD:ALL phenotype, which is not surprising given the smaller sample. Moreover, we hypothesize that our stronger findings with NVMSD:C on chromosomes 4p15.2, 14p15.2, and 21q22.2 might actually be better capturing those individuals who have a more well-defined motor-speech disorder like apraxia.
Notably, we did not find a linkage signal on 7q, a location that has been strongly implicated in linkage and association with both ASD and speech and language impairments (Alarcón et al. 2008; Arking et al. 2008; Vernes et al. 2008; Feuk et al. 2006; Lai et al. 2001). This was true even allowing for imprinting, which has been suggested for FOXP2 on chromosome 7 (Feuk et al. 2006) (Supplemental Figure 2). However, imprinting gene candidates have been reported in regions where we did see evidence for linkage with imprinting. Luedi et al. (2007) report maternal expression of TRIM16 (17p12), TIAF1 (17q11.2), HOXB2 (17q21.32), and HOXB8 (17q21.32). All of these genes match the pattern of imprinting supported by our linkage results on chromosome 17 and so they represent higher priority positional candidates; the homeobox genes are of particular interest due to their role in development patterns in the brain (Matis et al. 2007; Grados et al. 2003; Fanarraga et al. 1997).
Potential genes of interest using IPA
NVMSD:ALL phenotype
When we used IPA to identify potential autism susceptibility genes that might fall within our linkage regions, we identified several genes associated with translation and transcription factors, brain development, nervous system development, and multiple psychiatric disorders. We also took into consideration the overlap of functions between the control and linkage analyses. The candidate genes described below meet our criteria for the IPA core analysis of the linkage regions and have been filtered for overlap with the control analysis functions.
Our linkage region on 4q22.3 contains NKX6-1, which encodes a transcription factor that binds to AT-rich regions in the promoters of its target genes. NKX6-1 plays an important role in differentiation of motor neurons and the regulation of muscle nerve formation (Lee and Pfaff 2001; Bohl et al. 2008; De Marco Garcia and Jessel 2008). Six gene targets of NKX6-1 (ATOX1, GPX2, HIF1A, HMOX2, IGFBP4, and PHB) fall within the broad linkage regions for the NVMSD:ALL analysis and one gene (ANAPC4) falls within the broad linkage regions for the NVMSD:C analysis.
The linkage peak on 5p (66 cM) contains GHR, which encodes a growth hormone receptor shown to be involved in brain development and neuronal differentiation (Harvey et al. 2001; Harvey et al. 2002; Ransome et al. 2004; Buadet et al. 2007). A second linkage peak on 5q (154–156 cM) contains two candidate genes: DPYSL3, which is involved in neurite outgrowth and guidance and shows a decreased expression in individuals with Down syndrome (Weitzdoerfer et al. 2001) and HTR4, which is a serotonin receptor that has been associated with Schizophrenia, Attention-Deficit Hyperactivity Disorder, and Bipolar Disorder (Hirata et al. 2010; Suzuki et al. 2003; Hayden and Nurnberger 2006; Elia et al. 2009). The regions on 17p and 17q contain NCOR1 and NOS2, which are involved in NOTCH signaling and the NOS pathway, respectively. NOS2, in particular, has been implicated in various neurological disorders such as Alzheimer’s disease, and Amyotrophic Lateral Sclerosis (Colton et al. 2009; Chen et al. 2010). The peak at 17p12 (PPL = 77%) also contains PMP22, which encodes a protein that comprises 2–5% of peripheral nervous system myelin. Most recently, Pinto et al. (2010) identified a rare maternally inherited copy number variation (CNV) that contains PMP22 in an individual with ASD, however this CNV was not experimentally validated.
NVMSD:C phenotype
Our linkage analysis of NVMSD:C resulted in two novel regions, which we further investigated with the core analysis in IPA. Overall, there was a notable increase in genes involved in Down syndrome, which is primarily due to the inclusion of 21q22.2. IPA identified two genes on chromosome 4 involved in axonal guidance: SLIT2 (Hammond et al. 2005) and CRMP1 (Yamashita et al. 2006). SLIT2 is of particular interest as both the SLIT1 and SLIT2 proteins have been identified as selective inhibitors and repellents for dorsally projecting cranial motor axons (Colton et al. 2009). In addition to these axonal guidance genes, the core analysis also identified STIM2 on chromosome 4 (45 cM, PPL = 88%), which regulates calcium entry into neurons (Berna-Erro et al. 2009). Like the NVMSD:ALL analysis, molecules involved in psychiatric disorders such as Schizophrenia, Panic Disorder, and Social Impairment were also identified as genes involved in the NOTCH Signaling and NOS pathways. Both analyses identified molecules that are involved in motor function, however IPA analysis of NVMSD:C did not produce significant findings within regions of our strongest linkage signals.
IPA control analyses
Our control analyses (C:GN and C:GNG) served as a test of the reliability of our IPA analysis which we will use to guide our future investigations of autism genes. As seen in Supplemental Table 4b and c, the numbers of functions and candidate genes obtained for the C:GN and C:GNG analyses were comparable to those obtained in our linkage regions. The functions identified by IPA are not presented in a hierarchical order, which leads to the identification of several similar functions that are, in fact, subsets of one overall function. This is seen commonly in our control analyses (as demonstrated in analysis 9, Supplemental Table 4b) and also occurs in our linkage analyses. While there was some overlap in general function categories, there were only eight specific functions identified in the control analyses that were also identified in the linkage analysis. Despite this overlap in functions, viable candidate genes were identified from the core analysis of our linkage regions after filtering for these common functions. This filtering of the linkage analysis helps to ensure that the functions and genes/molecules identified in the core analysis of the linkage regions are unique to that analysis. Overall, the IPA core analysis used in this study functions primarily as a data reduction tool and was effective in identifying genes in our linkage analysis that require further investigation.
One limitation of this study concerns the variables available to us from the AGRE data set to define a motor speech disorder. To make a clinical diagnosis of such a disorder, a speech language pathologist would use converging information including an extensive language history, a complete oral motor exam, and a comprehensive speech and language assessment that would include specific information about the phonological abilities of each proband. Yet, our strong linkage findings suggest that at this stage of investigation we have defined this disorder well enough to continuing pursuing the genes, gene interactions, and gene variants under the peaks.
Conclusion
We have identified several unique loci, based on two specific motor speech phenotypes, that are present in, but not exclusive to, a subset of individuals within families with autism spectrum disorders. In addition, we have identified several loci that had been previously isolated on the basis of somewhat different diagnostic criteria. Family members who are non-verbal or verbal but have speech that is unintelligible may or may not meet criteria for ASD but may share genes and behaviors that are also seen in other speech and language disorders. Although we found no compelling evidence of association under our linkage peaks, we were able to suggest genes for further investigation based on their biological functions using IPA. It is well recognized that autism spectrum disorders are complex with a wide range of behaviors and, potentially, a large number of underlying genes, so that these particular sets of behaviors might fall into a broader phenotype and further emphasize the need to develop narrow well-defined phenotypes to reduce genetic heterogeneity.
Electronic supplementary materials
Below is the link to the electronic supplementary material.
(DOC 546 kb)
Acknowledgements
We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium* and the participating AGRE families. The Autism Genetic Resource Exchange is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). AGRE GWAS data was provided to AGRE by Dr. Mark Daly and the Autism Consortium. We additionally acknowledge the NIH/NIMH for specific funding for this project awarded to Veronica Vieland (R01 MH76433 and R01 MH086117) and Linda Brzustowicz (R01MH76435 and R01MH070366).
Conflict of Interest Notification Page
There is no copyrighted material in this manuscript and this manuscript has never been published in this form nor is it under review in any other journal. The content has been reviewed and approved by all co-authors.
There are no real or potential conflicts of interest that could be seen as having influence on this research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30(1):97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Abrahams BS, Geschwind DH. Genetics of Autism. In: Antonarakis SE, Speicher MR, Motulsky AG, editors. Vogel and Motulsky’s human genetics: problems and approaches. Berlin: Springer-Verlag; 2010. pp. 699–714. [Google Scholar]
- Alarcón M, Cantor RM, Liu J, Gilliam TC, Geschwind DH, Autism Genetic Resource Exchange Consortium Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet. 2002;70(1):60–71. doi: 10.1086/338241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón M, Yonan A, Gilliam TC, Cantor RM, Geschwind DH. Quantitative genome scan and ordered-subsets analysis of autism endophenotypes support language QTLs. Mol Psychiatry. 2005;10(8):747–757. doi: 10.1038/sj.mp.4001666. [DOI] [PubMed] [Google Scholar]
- Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, et al. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, et al. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auranen M, Varilo T, Alen R, Vanhala R, Ayers K, Kempas E, et al. Evidence for allelic association on chromosome 3q25-27 in families with autism spectrum disorders originating from a subisolate of Finland. Mol Psychiatry. 2003;8(10):879–884. doi: 10.1038/sj.mp.4001299. [DOI] [PubMed] [Google Scholar]
- Bailey A, Philips W, Rutter M. Autism: towards an integration of clinical, genetic, neuropsychological, and neurobiological perspectives. J Child Psychol Psychiatry. 1996;37(1):89–126. doi: 10.1111/j.1469-7610.1996.tb01381.x. [DOI] [PubMed] [Google Scholar]
- Bartlett CW, Flax JF, Logue MW, Smith BJ, Vieland VJ, Tallal P, et al. Examination of potential overlap in autism and language loci on chromosome 2, 7, and 13 in two independent samples ascertained for specific language impairment. Hum Hered. 2004;57(1):10–20. doi: 10.1159/000077385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett CW, Goedken R, Vieland VJ. Effects of updating linkage evidence across subsets of data: reanalysis of the autism genetic resource exchange data set. Am J Hum Genet. 2005;76(4):688–695. doi: 10.1086/429345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berna-Erro A, Braun A, Kraft R, Kleinschnitz C, Schuhmann MK, Stegner D, et al. STIM2 regulates capacitive Ca2+ entry in neurons and plays a key role in hypoxic neuronal cell death. Sci Signal. 2009;2(93):ra67. doi: 10.1126/scisignal.2000522. [DOI] [PubMed] [Google Scholar]
- Bohl D, Liu S, Blanchard S, Hocquemiller M, Haase G, Heard JM. Directed evolution of motor neurons from genetically engineered neural precursors. Stem Cells. 2008;26(10):2564–2575. doi: 10.1634/stemcells.2008-0371. [DOI] [PubMed] [Google Scholar]
- Bradford Y, Haines J, Hutcheson H, Gardiner M, Braun T, Sheffield V, et al. Incorporating language phenotypes strengthens evidence of linkage to autism. Am J Med Genet. 2001;105(6):539–547. doi: 10.1002/ajmg.1497. [DOI] [PubMed] [Google Scholar]
- Broman KW, Weber JL. Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet. 1998;63(5):1563–1564. doi: 10.1086/302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buadet ML, Rattray D, Harvey S. Growth hormone and its receptor in projection neurons of the chick visual system: retinofugal and tectobulbar tracts. Neuroscience. 2007;148(1):151–163. doi: 10.1016/j.neuroscience.2007.05.035. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Silverman JM, Keddache M, Smith CJ, Hollander E, Ramoz N, et al. Linkage analysis for autism in a subset families with obsessive-compulsive behaviors: evidence for an autism susceptibility gene on chromosome 1 and further support for susceptibility genes on chromosome 6 and 19. Mol Psychiatry. 2004;9(2):144–150. doi: 10.1038/sj.mp.4001465. [DOI] [PubMed] [Google Scholar]
- Cantor RM, Kono N, Duvall JA, Alvarez Retuerto A, Stone JL, Alarcón M, et al. Replication of autism linkage: fine mapping peak at 17q21. Am J Hum Genet. 2005;76(6):1050–1056. doi: 10.1086/430278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, Northington FJ, Martin LJ. Inducible nitric oxide synthase is present in motor neuron mitochondria and Schwann cells and contributes to disease mechanisms in ALS mice. Brain Struct Funct. 2010;214(2–3):219–234. doi: 10.1007/s00429-009-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colton CA, Wilcock DM, Wink DA, Davis J, Van Nostrand WE, Vitek MP, et al. The effects of NOS2 gene deletion on mice expressing mutated human AbetaPP. J Alzheimers Dis. 2009;15(4):571–587. doi: 10.3233/jad-2008-15405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marco Garcia NV, Jessel TM. Early motor neuron pool identity and muscle nerve trajectory defined by postmitotic restrictions in Nkx6.1 activity. Neuron. 2008;57(2):217–231. doi: 10.1016/j.neuron.2007.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Capasson M, Zaheer Z, Lantieri F, Ambrosini P, Berrettini W, et al. Candidate gene analysis in an on-going genome-wide association study of attention-deficit hyperactivity disorder: suggestive association signals in ADRA1A. Psychiatr Genet. 2009;19(3):134–141. doi: 10.1097/YPG.0b013e32832a5043. [DOI] [PubMed] [Google Scholar]
- Elston RC, Stewart J. A general model for the genetic analysis of pedigree data. Hum Hered. 1971;21(6):523–542. doi: 10.1159/000152448. [DOI] [PubMed] [Google Scholar]
- Elston RC, Lange K. The prior probability of autosomal linkage. Ann Hum Genet. 1975;38(3):341–350. doi: 10.1111/j.1469-1809.1975.tb00619.x. [DOI] [PubMed] [Google Scholar]
- Fanarraga ML, Charite J, Hage WJ, De Graff W, Deschamps J. Hoxb-8 gain-of-function transgenic mice exhibit alterations in the peripheral nervous system. J Neurosci Methods. 1997;71(1):11–18. doi: 10.1016/S0165-0270(96)00122-7. [DOI] [PubMed] [Google Scholar]
- Feuk L, Kalervo A, Lipsanen-Nyman M, Skaug J, Nakabayashi K, Finucane B, et al. Absence of a paternally inherited FOXP2 gene in developmental verbal dyspraxia. Am J Hum Genet. 2006;79(5):965–972. doi: 10.1086/508902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gernsbacher MA, Sauer EA, Geye MH, Schewiegert EK, Hill-Goldsmith H. Infant and toddler oral- and manual-motor skills predict later fluency in autism. J Child Psychol Psychiatry. 2009;4(1):43–50. doi: 10.1111/j.1469-7610.2007.01820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grados MA, Walkup J, Walford S. Genetics of obsessive-compulsive disorders: new findings and challenges. Brain Dev. 2003;25(Suppl 1):S55–S61. doi: 10.1016/S0387-7604(03)90010-6. [DOI] [PubMed] [Google Scholar]
- Hammond R, Vivancos V, Naeem A, Chilton J, Mambetisaeva E, Andrews W, et al. Slit-mediated repulsion is a key regulator of motor axon pathfinding in the hindbrain. Development. 2005;132(20):4483–4495. doi: 10.1242/dev.02038. [DOI] [PubMed] [Google Scholar]
- Harvey S, Johnson CD, Sanders EJ. Growth hormone in neural tissues of the chick embryo. J Endocrinol. 2001;169(3):487–498. doi: 10.1677/joe.0.1690487. [DOI] [PubMed] [Google Scholar]
- Harvey S, Lavelin I, Pines M. Growth hormone (GH) action in the brain: neural expression of a GH-response gene. J Mol Neurosci. 2002;18(1–2):89–95. doi: 10.1385/JMN:18:1-2:89. [DOI] [PubMed] [Google Scholar]
- Hayden EP, Nurnberger JI., Jr Molecular genetics of bipolar disorder. Genes Brain Behav. 2006;5(1):85–95. doi: 10.1111/j.1601-183X.2005.00138.x. [DOI] [PubMed] [Google Scholar]
- Hirata Y, Souza RP, Lieberman JA, Meltzer HY, Kennedy JL. Lack of association between HTR4 gene polymorphisms and schizophrenia in case-control and family-based samples. Psychiatry Res. 2010;175(1–2):176–178. doi: 10.1016/j.psychres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Huang Y, Segre A, O’Connell J, Wang H, Vieland V. KELVIN: a 2nd generation distributed multiprocessor linkage and linkage disequilibrium analysis program. Presented at the annual meeting of The American Society of Human Genetics. New Orleans, Louisiana, 2006.
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Lander ES, Green P. Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA. 1987;84(8):2363–2367. doi: 10.1073/pnas.84.8.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Pfaff SL. Transcriptional networks regulating neuronal identity in the developing spinal cord. Nat Neurosci. 2001;4(Suppl):1183–1191. doi: 10.1038/nn750. [DOI] [PubMed] [Google Scholar]
- Lewis BA, Freebairn LA, Hansen AJ, Taylor HG, Iyengar S, Shriberg D. Family pedigrees of children with suspected childhood apraxia of speech. J Commun Disord. 2004;37(2):157–175. doi: 10.1016/j.jcomdis.2003.08.003. [DOI] [PubMed] [Google Scholar]
- Lewis BA, Freebairn LA, Hansen AJ, Miscimarra L, Iyengar S, Taylor HG. Speech and language skills of parents of children with speech sound disorders. Am J Speech Lagn Pathol. 2007;16(2):108–118. doi: 10.1044/1058-0360(2007/015). [DOI] [PubMed] [Google Scholar]
- Lindgrin KA, Folstein SE, Tomblin JB, Tager-Flusberg H. Language and reading abilities of children with autism spectrum disorders and specific language impairment and their first-degree relatives. Autism Res. 2009;2(1):22–38. doi: 10.1002/aur.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XQ, Paterson AD, Szatmari P, Autism Genome Project Consortium Genome-wide linkage analyses of quantitative and categorical autism subphenotypes. Biol Psychiatry. 2008;64(7):561–570. doi: 10.1016/j.biopsych.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism diagnostic interview-revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24(5):659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Lord C, Risi S, Pickles A. Trajectory of language development in autism spectrum disorders. In: Rice M, Warren SF, editors. Developmental language disorders: from phenotypes to etiologies. Mahwah: Erlbaum; 2004. [Google Scholar]
- Luedi PP, Dietrich FS, Weidman JR, Bosko JM, Jirtle RL, Hartemink AJ. Computational and experimental identification of novel human imprinted genes. Genome Res. 2007;17(12):1723–1730. doi: 10.1101/gr.6584707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matis C, Oury F, Remacle S, Lampe X, Gofflot F, Picard JJ, et al. Identification of Lmo1 as part of a Hox-dependent regulatory network for hindbrain pattering. Dev Dyn. 2007;236(9):2675–2684. doi: 10.1002/dvdy.21266. [DOI] [PubMed] [Google Scholar]
- Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17(12):1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley JL, Li C, Jiang L, Olson LM, Crockett G, Gainer K, et al. Genome-wide and ordered-subset linkage analyses provide support for autism loci on 17q and 19p with evidence of phenotypic and interlocus genetic correlates. BMC Med Genet. 2005;6:1. doi: 10.1186/1471-2350-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newbury DF, Bonora E, Lamb JA, Fisher SE, Lai CS, Baird G, et al. FOXP2 is not a major susceptibility gene for autism or specific language impairment. Am J Hum Genet. 2002;70(5):1318–1327. doi: 10.1086/339931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransome MI, Goldshmit Y, Bartlett PF, Waters MJ, Turnley AM. Comparative analysis of CNS populations in knockout mice with altered growth hormone responsiveness. Eur J Neurosci. 2004;19(8):2069–2079. doi: 10.1111/j.0953-816X.2004.03308.x. [DOI] [PubMed] [Google Scholar]
- Rapin I, Dunn M. Update on the language disorders of individuals on the autistic spectrum. Brain Dev. 2003;25(3):166–172. doi: 10.1016/S0387-7604(02)00191-2. [DOI] [PubMed] [Google Scholar]
- Royall R. Statistical evidence: a likelihood paradigm. London; 1997.
- Rutter M, Le Couteur A, Lord C. The autism diagnostic interview, revised (ADI-R) Los Angeles: Western Psychological Services; 2003. [Google Scholar]
- Schellenberg GD, Dawson G, Sung YJ, Estes A, Munson J, Rosenthal E, et al. Evidence for multiple loci from a genome scan of autism kindreds. Mol Psychiatry. 2006;11(11):1049–1060. doi: 10.1038/sj.mp.4001874. [DOI] [PubMed] [Google Scholar]
- Shao Y, Cuccaro ML, Hauser ER, Raiford KL, Menold MM, Wolpert CM, et al. Fine mapping of autistic disorder to chromosome 15q11-q13 by use of phenotypic subtypes. Am J Hum Genet. 2003;72(3):539–548. doi: 10.1086/367846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriberg LD, Aram DM, Kwiatkowski J. Developmental apraxia of speech: I. Descriptive and theoretical perspectives. J Speech Lang Hear Res. 1997;40(2):273–285. doi: 10.1044/jslhr.4002.273. [DOI] [PubMed] [Google Scholar]
- Smith CA. Testing for heterogeneity of recombination fraction values in human genetics. Ann Hum Genet. 1963;27:175–182. doi: 10.1111/j.1469-1809.1963.tb00210.x. [DOI] [PubMed] [Google Scholar]
- Spence SJ, Cantor RM, Chung L, Kim S, Geschwind DW, Alarcón M. Stratification based on language-related endophenotypes in Autism: attempt to replicate reported linkage. Am J Med Genet Par B Neuropsychiatr Genet. 2006;141B(6):591–598. doi: 10.1002/ajmg.b.30329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Iwata N, Kitamura Y, Kitajima T, Yamanouchi Y, Ikeda M, et al. Association of a haplotype in the serotonin 5-HT4 receptor gene (HTR4) with Japanese schizophrenia. Am J Med Genet B Neuropsychiatr Gen. 2003;121B(1):7–13. doi: 10.1002/ajmg.b.20060. [DOI] [PubMed] [Google Scholar]
- Tager-Flusberg H, Paul R, Lord C. Language and communication in autism. In: Volkmar FR, Paul R, Klin A, Cohen DJ, editors. Handbook of autism and pervasive developmental disorders. New York: Wiley; 2005. pp. 335–364. [Google Scholar]
- Taper M, Lele S. The nature of scientific evidence. Chicago: Univ Chicago Press; 2004. [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, et al. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359(22):2337–2345. doi: 10.1056/NEJMoa0802828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieland VJ. Bayesian linkage analysis, or: how I learned to stop worrying and love the posterior probability of linkage. Am J Hum Genet. 1998;63(4):947–954. doi: 10.1086/302076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieland VJ. Thermometers: something for statistical geneticists to think about. Hum Hered. 2006;6(13):144–156. doi: 10.1159/000093775. [DOI] [PubMed] [Google Scholar]
- Weitzdoerfer R, Fountoulakis M, Lubec G. Aberrant expression of diphydropyrimidinase related proteins-2, -3 and -4 in fetal Down syndrome brain. J Neural Transm Suppl. 2001;61:95–107. doi: 10.1007/978-3-7091-6262-0_8. [DOI] [PubMed] [Google Scholar]
- Wratten NS, Memoli H, Huang Y, Dulencin AM, Matteson PG, Cornacchia MA, et al. Identification of a Schizophrenia-associated functional noncoding variant in NOS1AP. Am J Psychiatry. 2009;166(4):434–441. doi: 10.1176/appi.ajp.2008.08081266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita N, Uchida Y, Ohshima T, Hirai S, Nakamura F, Taniguchi M, et al. Collapsin response mediator protein 1 mediates reelin signaling in cortical neuronal migration. J Neurosci. 2006;26(51):13357–13362. doi: 10.1523/JNEUROSCI.4276-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Huang J, Logue MW, Vieland VJ. The posterior probability of linkage allowing for linkage disequilibrium and a new estimate of disequilibrium between a trait and a marker. Hum Hered. 2005;59(4):210–219. doi: 10.1159/000086699. [DOI] [PubMed] [Google Scholar]
- Yonan AL, Alarcón M, Cheng R, Magnusson PK, Spence SJ, Palmer AA, et al. A genomewide screen of 345 families for autism-susceptibility loci. Am J Hum Genet. 2003;73(4):886–897. doi: 10.1086/378778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOC 546 kb)