Abstract
Craniofacial morphogenesis is accomplished through a complex set of developmental events, most of which are initiated in neural crest cells within the pharyngeal arches. Local patterning cues from the surrounding environment induce gene expression within neural crest cells, leading to formation of a diverse set of skeletal elements. Endothelin-1 (Edn1) is one of the primary signals that establish the identities of neural crest cells within the mandibular portion of the first pharyngeal arch. Signaling through its cognate receptor, the endothelin-A receptor, is critical for patterning the ventral/distal portion of the arch (lower jaw) and also participates with Hox genes in patterning more posterior arches. Edn1/Ednra signaling is highly conserved between mouse and zebrafish, and genetic analyses in these two species have provided complementary insights into the patterning cues responsible for establishing the craniofacial complex as well as the genetic basis of facial birth defect syndromes.
Keywords: endothelin, craniofacial development, neural crest cell, endothelin antagonist, zebrafish, morpholino, knockout mice, transgenic mice, Dlx
INTRODUCTION
Neural crest cells (NCCs) are migratory cells that originate from the neural tube with extensive pluripotency; they populate various regions of the embryo during development and contribute to the formation of multiple organs. These cells and the genetic pathways that regulate them are highly conserved among vertebrates, including those necessary for formation of the lower jaw. In the past 15 years, genetic manipulation of mice and zebrafish have revealed the functional significance of various signaling factors that are crucial for lower jaw development, with most mediating aspects of NCC patterning. In this review, we will discuss one of these pathways, mediated by endothelin-A receptor signaling, through which the identity of NCCs within the developing lower jaw is established.
NEURAL CREST CELL MIGRATION AND DIFFERENTIATION
During early embryogenesis, NCCs arise at the crest of the neural folds along the entire anterior-posterior (A-P) axis of the embryo [Le Douarin and Kalcheim 1999; Le Douarin 1982]. Based on their locations along this axis and their migratory destinations, NCCs can be subdivided into 5 groups: cranial, cardiac, trunk, vagal and sacral [Bronner-Fraser 1995; Le Douarin 1982; Le Douarin et al., 1993; Raible et al., 1992]}. Depending on the class and local patterning signals, NCCs can differentiate into a variety of lineages, including melanocytes, neurons and glia of peripheral ganglia, various connective tissues, as well as smooth muscle of the proximal outflow tract of the heart. In addition, cranial NCCs can form bone and cartilage of the craniofacial skeleton [Couly et al., 1998; Knight and Schilling 2006; Kontges and Lumsden 1996; Noden 1983; Noden 1988; Schilling and Kimmel 1994]. The remainder of this review will focus on cranial NCCs and the patterning signals that regulate their differentiation.
In all vertebrates that have been examined, cranial NCCs initially form at the level of the midbrain and hindbrain along the neural tube. Those NCCs that arise furthest anteriorly form the frontonasal skeleton, whereas NCCs from posterior midbrain and hindbrain move into the pharyngeal arches, paired transient structures on the ventral embryo surface [Couly et al., 1993; Fraser et al., 1990; Le Douarin et al., 1993; Lumsden et al., 1991]. There they receive patterning signals that drive their differentiation into bone, cartilage and connective tissue of the jaw (mandibular, arch 1), middle ear (mandibular, arch 1; hyoid, arch 2) and neck (arches 2–4 in mammals, up to 7 arches in other vertebrates). In addition, cranial NCCs contribute to the cranial ganglia (all arches). While NCCs likely receive patterning signals during migration, much of the signaling necessary for patterning within an arch comes from signals received by NCCs after their arrival at their destinations [Clouthier and Schilling 2004; Couly et al., 1993; Knight and Schilling 2006; Le Douarin and Kalcheim 1999; Le Douarin 1982, Hall, 1982; Trainor 2005]. A number of signaling pathways are involved in NCC patterning within the pharyngeal arches, including bone morphogenetic proteins (Bmps), Wnts, fibroblast growth factors (Fgfs) and retinoic acid (RA) [Chai and Maxson 2006; Helms et al., 2005; Jernvall and Thesleff 2000; Miletich and Sharpe 2004; Yelick and Schilling 2002] (See also the article by Frenz et al.,, 2010 in this issue). However, one signaling pathway that is arguably the key mediator of NCC development and skeletal patterning in the mandibular first arch is that mediated by signaling from the endothelin-A receptor.
ENDOTHELINS AND NEURAL CREST CELL DEVELOPMENT
Among the first factors shown to be involved in pharyngeal arch development were the Hox genes. These genes govern the axial-level specific differentiation of NCCs within pharyngeal arches 2–7, thus establishing segmental identities along the A–P axis within the arches (termed the “Hox code”) [Hunt et al., 1991; Trainor and Krumlauf, 2001]. However, Hox genes are not present in the mandibular arch, and only Hoxa2 is expressed in hindbrain premigratory NCCs that will populate this arch. In addition, in Hoxa2−/− mouse embryos, a subset of first arch-specific structures appear to replace their counterparts in the second arch – so-called mirror-image duplicates [Gendron-Maguire et al., 1993; Rijli et al., 1993]. This has led to the theory that Hox gene expression (primarily from the Hoxa cluster) acts on a ground state (mandibular) patterning program that exists within all arches to specify the distinct morphologies of arches 2–7 [Pasqualetti et al., 2000; Prince and Lumsden 1994; Rijli et al., 1993]. Indeed, targeted deletion of the Hoxa cluster leads to mandibular arch-like structures in the posterior arches in mice, further supporting this hypothesis [Minoux et al., 2009] This is a striking finding, as it was previously thought that this ground pattern existed only in arches one and two. It thus appears that this “mandibular code” was retained by all of the arches during the evolution of the hinged jaw and pharyngeal apparatus, with differential Hox gene expression subsequently providing additional patterning cues to each of the more posterior segments of the head and neck.
So what is the molecular basis of this mandibular program? One critical component is endothelin-1-induced signaling through the endothelin-A receptor. The endothelin family was first identified in 1988 with the cloning of endothelin-1 (Edn1), identified by its potent vasoconstrictive activity [Yanagisawa et al., 1988]. This 21 amino acid ligand is first encoded as a preproendothelin molecule, which is then processed through three proteolytic events [Kido and Sawamura 1998]. Preproendothelin (approximately a 200 amino acid protein) is proteolytically cleaved by a signal peptidase generating pro-endothelin, which is processed by a furin protease (described below) to generate big-endothelin, a 38 amino acid inactive protein. Big-endothelin is then cleaved by one of two endothelin-converting enzyme metalloproteases (Ece-1 and Ece-2) into the 21 amino acid mature active form of endothelin [Emoto and Yanagisawa 1995; Xu et al., 1994]. In mice, there are three ligands (Edn1, 2 and 3), which bind to the two known G-protein coupled receptors - endothelin-A receptor (Ednra) and endothelin-B receptor (Ednrb)) [Yanagisawa, 1994]. Both receptor types bind to all three endothelin isoforms, though each ligand has a specific affinity for a particular receptor [Yanagisawa, 1994]. This family of ligands and receptors is highly conserved across vertebrates [Clouthier and Schilling, 2004], though some differences exist in the number of receptors, including two Ednra receptors in zebrafish [Nair et al., 2007].
While endothelin signaling has dynamic roles in blood pressure regulation in adults [Kedzierski and Yanagisawa, 2001], it also plays important roles in NCC patterning. Edn3/Ednrb signaling is required for normal patterning of melanoblasts and enteric neurons derived from NCCs, with targeted deletion of the mouse Edn3 or Ednrb genes resulting in neonatal lethality around 3 weeks with coat color spotting and intestinal aganglionosis leading to megacolon [Baynash et al., 1994; Hosoda et al., 1994]. Mutations in the Ednrb gene underlie the spontaneous mouse mutants piebald spotting (s) and piebald lethal (sl) [Hosoda et al., 1994] and the spontaneous rat mutant spotting lethal (sl) [Gariepy and Yanagisawa 1996], while mutation of the Edn3 gene is the genetic basis of the spontaneous mouse mutant lethal spotting (ls) [Baynash et al., 1994]. In addition, mutations in the human EDNRB gene are associated with the human genetic disease Hirshsprung syndrome [Puffenberger et al., 1994; Tanaka et al., 1998]. However, none of these mouse or human mutations results in facial defects, illustrating the tight spatial regulation of endothelin signaling during development.
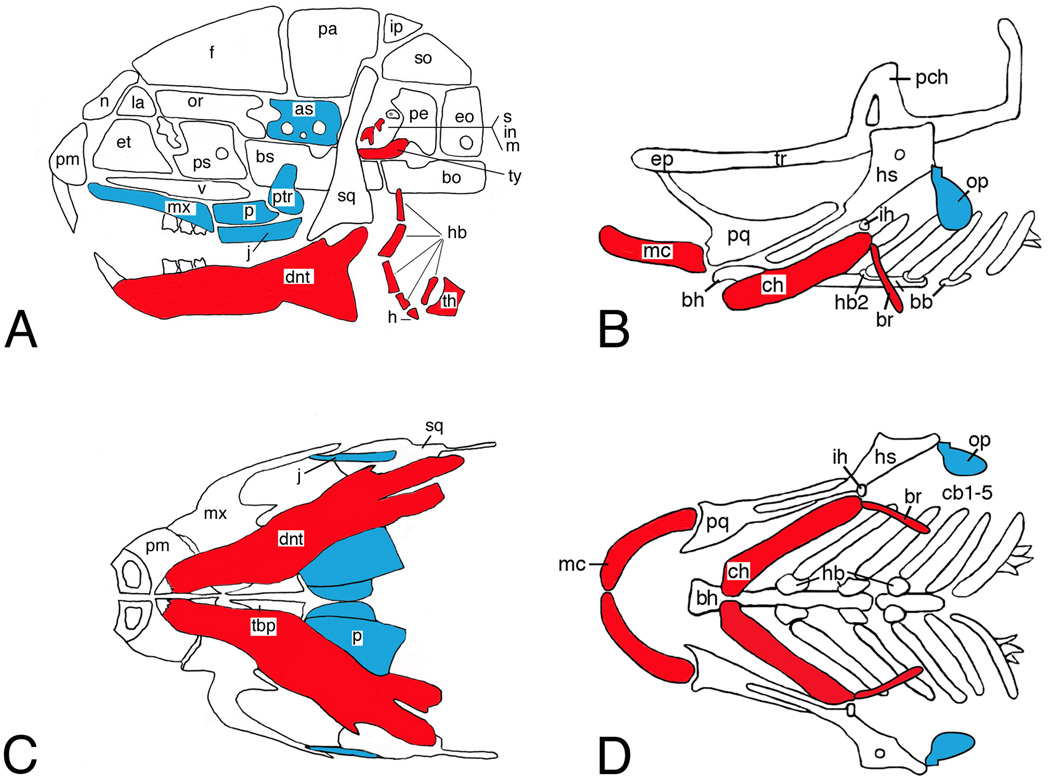
In contrast to Edn3/Ednrb mutants, mice lacking Edn1, Ece1 or Ednra die at birth due to mechanical asphyxia resulting from severe malformation of lower jaw and throat structures [Clouthier et al., 1998; Kurihara et al., 1994; Yanagisawa et al., 1998b]. The observed defects occur in structures derived from the mandibular arch and arches 2–4 (Table I). The most prominent change observed is homeotic transformation of the mandible into a more maxilla-like structure [Ozeki et al., 2004; Ruest et al., 2004]. Multiple other elements derived from the mandibular arch are duplicated including the palatine, pterygoid, jugal and lamina obturans bones and the ala temporalis cartilages (Fig. 1A, C). Other defects include absence of tympanic rings, malleus and incus. In addition, the hyoid bone is moved rostrally and fuses with the pterygoid bones. This fusion constricts the trachea, leading to mechanical asphyxia. Indeed, tracheotomy of Ednra−/− embryos allows survival for at least 24 hours [Clouthier et al., 1998]. Edn1−/−, Ece1−/− and Ednra−/− mutant embryos also have a wide array of cardiac defects, including interruption of the aorta and double outlet right ventricle [Clouthier et al., 1998; Kurihara et al., 1994; Yanagisawa et al., 1998a; Yanagisawa et al., 1998b].
Table 1.
Skeletal defects arising from the first four pharyngeal arches of endothelin-1 or endothelin-A receptor mutant mouse and zebrafish embryos
| Arch | ||||
|---|---|---|---|---|
| Species | 1 | 2 | 3 | 4 |
| Mouse |
Missing / malformed |
Missing / malformed |
Missing / malformed |
Missing / malformed |
| Meckel’s cartilage dentary malleus incus tympanic rings gonial |
lesser hyoid horns body of hyoid |
greater hyoid horns | thyroid cartilage | |
| Duplicated | Duplicated | Duplicated | Duplicated | |
| jugal palatine maxilla pterygoid alisphenoid ala temporalis lamina obturans |
none | none | none | |
|
Missing / malformed |
Missing / malformed |
Missing / malformed |
Missing / malformed |
|
| Zebrafish | Meckel’s cartilage dentary |
opercle branchiostegal rays |
basibranchials* hypobranchials* ceratobranchials* |
basibranchials* hypobranchials* ceratobranchials* |
| Duplicated | Duplicated | Duplicated | Duplicated | |
| none | opercle | none | none | |
Portions of the basibranchials and hypobranchials in zebrafish likely arise from some of the more posterior arches as well.
Figure 1.
Pharyngeal arch morphology in mouse and zebrafish. Lateral (A, B) and ventral (C, D) views of the craniofacial skeletons of embryonic day (E)18.5 (mouse) and 5 days postfertilization larvae (dpf) (fish). Within the mandibular and hyoid arch skeletons, Edn1 signaling is required for distal (ventral) arch cartilage and bone (red) and inhibits formation of more proximal (dorsal) cartilage and bone (blue). as, alisphenoid; bb, basibranchial; bh, basihyal; bo, basioccipital; br, branchiostegal ray; bs, basisphenoid; cb, ceratobranchial; ch, ceratohyal; dnt, dentary; eo, exoccipital; ep, ethmoid plate; et, ethmoid; f, frontal; h, hyoid; hb, hypobranchial; hs, hyosymplectic; ih, interhyal; in, incus; ip, interparietal; j, jugal; la, lacrimal; m, malleus; mc, Meckel’s; mx, maxilla; n, nasal; op, opercle; or, orbital; p, palatine; pa, parietal; pch, prechordal; pe, petrosal; pl, palatine; pm, premaxilla; pq, palatoquadrate; ps, presphenoid; ptr, pterygoid; s, stapes; so, supraoccipital; sq, squamosal; tbp, trabecular basal plate; th, thyroid; tr, trabecula; ty, tympanic ring; v, vomer.
Along with these defects, Ece1−/− embryos also lack epidermal melanocytes and enteric neurons as observed in both Edn3−/− and Ednrb−/− embryos, illustrating that Ece1 is the primary converting enzyme for endothelins during development [Yanagisawa et al., 1998b]. In contrast, Ece2−/− mice are healthy and viable, while a subset of Ece1−/−;Ece2−/− embryos show an earlier cardiac insufficiency resulting in lethality around E12.5 due to defective cardiomyocyte differentiation [Yanagisawa et al., 2000]. A similar phenotype is observed in Ednra−/−;Ednrb−/− mutants, indicating that in the heart Edn1/Ednra and Edn3/Ednrb function redundantly during development, as do Ece1 and Ece2 [Yanagisawa et al., 2000]. There is no known association of Edn1/Ece1/Ednra mutations in human birth defect syndromes, though this is likely due in part to the almost certain neonatal lethality that would occur in the presence of strong loss-of-function mutations in these genes.
ENU-induced mutations in endothelin family members have also been found in zebrafish. The first characterized was a mutation in the endothelin-1 gene (edn1) called sucker or suc/et1 due to the characteristic changes in Meckel’s cartilage that produces a downturned mouth [Miller et al., 2000]. Mutants have defects in both ventral cartilages and bones derived from arches 1 and 2, including severe truncation of Meckel’s cartilage and loss of the ceratohyal (Fig. 1B, D). The joints that separate the dorsal and ventral cartilages are also lost, resulting in cartilage fusions. These fusions also exist between the dentary (mandibular) and maxillary bones derived from the first arch, with the rudimentary dentary bone resembling a maxilla, suggestive of a homeotic transformation similar to that observed in Edn1−/− mouse mutants [Kimmel et al., 2003]. In the posterior branchiostegal ray/opercle dermal bone complex of the second arch, the posterior branchiostegal ray (brp) is almost always absent, though the more medial ray (opm) can be present. This presence is usually associated with an enlargement of the opercle along its dorsal-ventral (DV) axis (analogous to the proximal-distal [PD] axis in mice; see Clouthier and Schilling, 2004) – which is referred to as an opercle-gain phenotype. In some situations, the opercle and branchiostegal ray approach and resemble each other, indicating homeosis has occurred. In contrast to the opercle-gain phenotype, the opercle can also be absent (opercle-loss); in this situation, the medial branchiostegal ray is also usually absent. These differences have been hypothesized to reflect different levels of Edn1 in the arches and are the basis for the morphogen gradient hypothesis that, as discussed below, gives one explanation of how Edn1 regulates patterning along the DV axis of the arches [Kimmel et al., 2003].
ENU mutations in zebrafish have also been identified in three other genes implicated in Edn1 signaling, including furinA (sturgeon, stu), mef2c (hoover, hoo) and plcβ3 (schmerle; she). FurinA, a serine endoprotease encoded by the furinA gene, is a proprotein convertase that processes preproEdn1 into proEdn1 (referred to as Big Edn1, an inactive molecule [Yanagisawa 1994]). Mef2c is a transcription factor that is involved in both bone and cardiac development [Lin et al., 1997; Arnold et al., 2007] and appears to function with Ednra signaling to induce expression of Dlx5 and Dlx6 [Miller et al., 2007; Verzi et al., 2007] (discussed in detail below). Phospholipase-c-beta-3 (Plcβ3; 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta-3) is an enzyme that catalyzes the hydrolysis of phosphatidyl inositol 4,5-bisphosphate into inositol 1,4,5-trisphosphate and diacylglycerol in response to signals including activation of G protein-coupled receptors. As discussed below, Plcβ3 is a likely intracellular mediator of Ednra signaling [Walker et al., 2007]. Suc/edn1 and these three mutants (furinA, mef2c and plcβ3) comprise the anterior arch class of ENU mutants [Piotrowski et al.,, 1996; Schilling et al., 1996]. In furinA mutants, there are minor shape changes in Meckel’s cartilage and the ceratohyal, the most ventral elements of arches 1 and 2, respectively, but joints are lost and both are abnormally fused to dorsal cartilages [Walker et al., 2006]. The branchiostegal rays and opercle also appear abnormally shaped and fused. Similar changes are observed in mef2c mutants, in which joint defects are present and some reduction of posterior ventral cartilages occurs [Miller et al., 2007]. plcβ3 mutants have defects in ventral cartilage length similar in severity to those in edn1 mutant embryos, though the penetrance of these defects is variable [Walker et al., 2007]. In contrast, all three mutants have defects in the joints of the first and second arches. The significance of these differential changes in mutants within this class is discussed below. However, these findings illustrate the conservation of this signaling pathway between mouse and zebrafish in patterning cranial NCCs during facial morphogenesis.
TIMING OF EDNRA SIGNALING IN MICE – INSIGHTS INTO FUNCTION
While Ednra signaling is clearly crucial for establishing the identities of NCCs within the jaw and other pharyngeal arches, the timing of its action has, until recently, not been well understood. Ednra expression is first observed in migrating NCCs, suggesting that it could function early in NCC development [Clouthier et al., 1998; Yanagisawa et al., 1998b]. In addition, in mouse chimera experiments, Ednra−/− cells were excluded from the most distal mandibular arch of Ednra−/− <−>+/+ chimeric embryos [Clouthier et al., 2003]. The degree of exclusion was proportional to the percentage of wild type cells present, with an increasing proportion of wild type cells leading to a greater zone of exclusion from the distal tip of the arch. Since NCCs in Ednra mutant mouse embryos can reach the distal arch tip [Abe et al., 2007], one possibility is that loss of Ednra signaling disrupts sorting of NCC after migration. One result in zebrafish that argues against a role in migration is that gross injection of EDN1 into the pharyngeal arches of suc/et-1 mutants following NCC migration is sufficient to rescue cartilage morphogenesis [Miller et al., 2000]. However, it is possible that this signaling can regulate NCC sorting within the arches once NCCs have already migrated.
To address the temporal requirements for Ednra signaling in mouse embryos, the most informative approach has been to use Ednra-specific antagonists during cranial NCC development. In general, this has involved treating wild type embryos with chemical antagonists for a short period of time and then examining molecular or cellular changes in the arches. In the first of two antagonist-based studies in mice, Kurihara and colleagues treated cultured wild type mouse embryos with the Ednra specific antagonist BQ123 between E8.5 and E10.5 [Fukuhara et al., 2004]. When gene expression was assessed in these embryos, Dlx6 and Hand2 expression was downregulated in the mandibular arch only when embryos were cultured with the antagonist between E8.5 and E9.0, suggesting that this is the window during which Ednra signaling is required. Treatment after E9.0 did not alter expression. This confirmed an earlier report in which conditional inactivation of the mouse Ednra gene in the mandibular arch around E10.0 did not disrupt lower jaw development [Ruest et al., 2005].
One drawback to using BQ123 is that this compound has a low oral bioavailability (meaning a low percentage of the unaltered drug reaches the systemic circulation when administered orally), so its use is limited to short term embryo culture. To circumvent this problem, a highly orally bioavailable Ednra selective antagonist (TBC3240; its oral availability is approximately 25% of the administered dose in rats) was used, thus allowing the drug to be administered by gavage to pregnant wild type mice at time points between E8.0 and E10.5 [Ruest and Clouthier 2009]. TBC3240 was administered as either a single dose or two doses separated by 12 hours; as the half-life of the compound is about 7 hours, treating for two time periods would allow longer blockade of Ednra receptors. Embryos were then collected at either E10.5 and subjected to an in situ hybridization panel or collected at E18.5 and bone and cartilage analyzed following staining. In these mice, antagonist treatment resulted in lower jaw defects starting at E8.25, with the maximum affect observed between E8.5 and E9.0, a time period during which NCCs are migrating to and reaching the pharyngeal arches. This included treatment at E9.0/E9.5. However, like the previous study, treatment at E9.5 or later did not cause any defects in bone or cartilage. This was almost certainly not due to the dose of the antagonist being too low, since TBC3214 has a high potency (IC(50) = 40 pM) and the animals were given 100 mg/kg (or 1200 pM, a 30 fold excess). Defects observed in these antagonist-treated embryos were similar to those in Edn1 and Ednra mutant embryos, including duplication of the jugal bone and absence of the malleus, incus and tympanic ring. However, while the mandible was shortened, transformation into a maxilla-like structure was not observed, suggesting that this homeosis event requires loss of Ednra signaling over a longer time period than 24 hours to reprogram the identity of NCCs. Alternatively, Ednra signaling may not have been completely absent during a critical period in these experiments, though as described above, signaling was almost certainly lost for at least part of the treatment. These structural defects were preceded by changes in gene expression, with expression of Dlx3, Dlx5, Dlx6, Hand1 and Hand2 all disrupted between E8.25 and E9.0, though each gene had a very specific temporal pattern of changes. This included reduced Hand1 and Hand2 expression when animals were treated at E8.5 and E9.0 and reduced Dlx5 and Dlx6 when animals were treated at E8.5 and E9.0, E9.0 alone or E9.0 and E9.5.
A conditional inactivation of Ednra was also used to determine the timing of Ednra function, in which the Ednra gene was inactivated at either E8.5 or E9.5 using loxP/Cre technology [Ruest and Clouthier 2009]. To get temporal inactivation of Ednra, the Wnt1-Cre (in which Cre expression occurs in early migrating NCCs) and Hand2-Cre (in which Cre expression is observed in post-migratory NCCs by E9.5) strains were used. In E18.5 Ednrafl/fl;Wnt1-Cre (conditional knockout) embryos, skeletal defects were identical to those found in Edn1 and Ednra mutant embryos, including duplication of the maxilla, jugal, alisphenoid, pterygoid and palatine bones and absence of Meckel’s cartilage, malleus, incus and tympanic rings. This further illustrates the autonomous nature of Ednra signaling in NCCs. Like the structural defects, gene expression changes in E10.5 Ednrafl/fl;Wnt1-Cre embryos were identical to those observed in Edn1 and Ednra mutant embryos, with the expression of both Dlx5, Dlx6, Hand1 and Hand2 reduced in the pharyngeal arches. In contrast, Ednrafl/fl;Hand2-Cre conditional knockout embryos did not exhibit skeletal defects at E18.5. In addition, gene expression at E10.5 was unaffected. This was not likely due to a lack of gene deletion by the Hand2-Cre transgene, since recombination of the Ednra gene in Ednrafl/fl;Hand2-Cre embryos was confirmed by a recombination specific PCR. Taken together with the findings from the antagonist studies, it appears that regulation of gene expression within the mandibular arch shifts from an Ednra-dependent mechanism to a non-Ednra-dependent mechanism around E9.0. As part of this switch, while Ednra signaling through Dlx5/Dlx6 is required for induction of Hand2 expression in the arches [Clouthier et al., 2000; Depew et al., 2002; Ruest et al., 2004], other signaling mechanisms appear to be responsible for maintenance of the Dlx5/Dlx6/Hand2 pathway.
AFTER THE RECEPTOR: INTRACELLULAR SIGNALING
Endothelin receptors can signal through a diverse repertoire of G proteins [Kedzierski and Yanagisawa 2001]. It has been assumed that this diversity could, in part, account for the wide range of responses to endothelin signaling. This is likely true for Ednra signaling during lower jaw development, though the actual nature of the signaling is unclear (Fig. 2). Mouse embryos containing a targeted inactivation of both Gαq and Gα11 die by E11.0 due to cardiac defects leading to heart failure [Offermanns et al., 1998]. However, gene expression analysis in these double knockout embryos prior to death revealed similar changes in early mandibular arch gene expression to those found in Edn1 and Ednra mutant embryos, including downregulation or loss of Hand1, Hand2, Dlx3 and Dlx6 [Ivey et al., 2003], suggesting that Ednra signals through Gαq and Gα11 to pattern NCCs within the mandibular arch. More definitive proof of this came from a conditional inactivation of both Gαq and Gα11 in NCCs using loxP-Cre technology. Neural crest-specific Cre expression was achieved using P0-Cre mice, in which Cre expression in NCCs and their derivatives occurs at least as early as E9.0 [Yamauchi et al., 1999]. In Gαq cko;Gα11cko embryos, defects in mandibular arch-derived structures resembled some of those observed in Edn1 and Ednra mutant embryos, including absence of tympanic rings. The lower jaw was also small and minor duplications of the maxilla and jugal were also reported [Dettlaff-Swierez et al., 2005]. Distal (ventral) mandibular bone development was also more extensive than observed in either Edn1 or Ednra mutants, likely attributable to continued expression of both Dlx5 and Hand2 in the distal arch of Gαq cko;Gα11cko embryos. These findings illustrate that G protein coupling among receptors in the developing face occurs in tight spatial and temporal patterns and that in the distal mandibular arch, Ednra signals through G proteins other than Gαq and Gα11. It is interesting to note that Gαq cko;Gα11cko embryos did not have defects in cardiac NCC derivatives. This is in contrast to Gα12cko;Gα13cko embryos, which had cardiac NCC defects but no facial defects, illustrating a second area of the embryo in which G proteins act in a spatially-restricted fashion.
Figure 2.
Intracellular signaling pathway within cranial neural crest cells mediated by endothelin-1 (Edn1). Preproendothelin-1 coming from cells within the pharyngeal arch environment undergoes two processing events (Furin A/B and endothelin converting enzyme 1/2 (Ece1, 2) before binding to the endothelin-A receptor (Ednra). This initiates signaling through Gαq/Gα11, resulting in phospholipase-β3 activity (Plcβ3) and thus changes in gene expression. Genetic evidence that individual Edn1/Ednra pathway members regulate craniofacial development has come from either mouse (highlighted in orange), zebrafish (highlighted in blue) or both (highlighted in orange/blue).
An interesting aspect of endothelin signaling discussed above is the spatial separation that exists between Ednra and Ednrb function during NCC patterning. While Ednrb is expressed in the mandibular arch (D.E. Clouthier and H. Yanagisawa, unpublished), Ednrb mutant embryos do not have obvious craniofacial defects [Hosoda et al., 1994]. This could imply that Ednrb does not play a role in mandibular arch development or that Ednra and Ednrb function redundantly, with Ednra compensating for Ednrb loss (though the converse is obviously not true). To directly address these possibilities, Kurihara and colleagues recently knocked a mouse Ednrb cDNA into the Ednra locus [Sato et al., 2008a]. Acting in place of Ednra, Ednrb did not rescue the homeotic transformation of the mandible into a maxilla-like structure, suggesting that this change is a result of Ednra-specific signaling. However, Ednrb expression from the Ednra locus did rescue the distal (ventral) most aspect of the mandible, including enhanced alveolar bone around the incisors. This pattern closely resembles that observed in Gαq cko;Gα11cko embryos, suggesting that in this distal region, Ednrb signaling, likely through G proteins other that Gαq and Gα11, can compensate for loss of Ednra signaling. This is not completely surprising, since while Ednrb can couple to Gαq and Gα11 [Cramer et al., 2001; Kedzierski and Yanagisawa 2001; Masaki et al., 1999], Ednrb signaling in enteric nervous system development appears to function by inhibiting PKA (potentially through Gαi), an affect that can be overcome by increasing intercellular levels of cAMP [Barlow et al., 2003]. However, the in vivo function of these receptors is complex, since Ednrb does not compensate for Ednra absence in Ednra mutant embryos [Yanagisawa et al., 2000; Yanagisawa et al., 1998a], suggesting that either Ednrb may not be normally expressed as far distally in the mandibular arch as Ednra or that Ednra expression is required to maintain Ednrb expression in this region. Another aspect likely contributing to this observed specificity is regional expression of G proteins in specific NCC populations (and sub-populations, such as those within the pharyngeal arches). A careful analysis of the spatiotemporal expression patterns of G proteins during arch development is required to address this point.
The major role of Gαq and Gα11 is to activate phospholipase C beta (Plcβ) [Smrcka and Sternweis 1994], which in turns produces inositol triphosphate (IP3) and diacylglycerol from PIP2. This in turn leads to changes in intracellular calcium levels and subsequent biological responses [Cramer et al., 2001]. In mice, targeted inactivation of Plcβ1, 2, 3 and 4 does not cause craniofacial defects [Chakrabarti et al., 2003; Hashimoto et al., 2001; Li et al., 2000; Wang et al., 1998; Xie et al., 1999] , though Plcβ3 inactivation may lead to an early embryonic lethality that would mask later requirements [Wang et al., 1998]. In contrast, in zebrafish the schmerle (she) mutation disrupts Plcβ3 and this causes craniofacial defects reminiscent of disruption of the Edn1 signaling pathway [Walker et al., 2007]. Two independent alleles of she both contain missense mutations in the catalytic domains of Plcβ3 [Walker et al., 2007]. Homozygous mutants display defects in ventral cartilages of the first and second arches and intermediate (joint) defects, but with lower penetrance than observed in edn1 mutants. plcβ3 and edn1 genetically interact, as double heterozygous mutant (plcβ3+/−;edn1+/−) embryos have more severe defects resembling edn1 mutants, consistent with a role for Plcβ3 in Edn1 signaling. Interestingly, unlike edn−/− mutants, expression of hand2 recovers in she mutants by 55 hpf [Miller et al., 2000; Miller et al., 2003a]. One possible explanation for this observation is that Plcβ isoforms may compensate for loss of Plcβ3 during later patterning, which can be addressed in future genetic studies in mouse or zebrafish. Though less likely, it also possible that Ednra in the arches may signal through different G proteins at these later stages, becoming independent of Plcβ3.
THE MOLECULAR BASIS OF DEFECTS IN EDN1/ECE1/EDNRA MUTANTS
Edn1 secreted from surrounding tissues in the pharyngeal arch environment activates Ednra in cranial NCCs. Edn1 is expressed by the pharyngeal arch ectoderm, core paraxial mesoderm and pharyngeal pouch endoderm of the arches [Clouthier et al., 1998; Maemura et al., 1996; Miller et al., 2000; Yanagisawa et al., 1998b] In contrast, Ednra is expressed in NCCs soon after emigration from the neural tube, with expression continuing in the NCC-derived mesenchyme of the arches [Clouthier et al., 1998; Nair et al., 2007; Yanagisawa et al., 1998b]. In zebrafish, a second Ednra gene, termed Ednra2, is also expressed in the arch ectoderm [Nair et al., 2007], the significance of which will be discussed below. In general, Edn1 likely has a short range of action (two to three cells layers from its site of activation) [Yanagisawa 1994]. Thus, arch-derived Edn1 likely binds to the Ednra on neighboring NCCs, leading to the initiation of a genetic cascade responsible for establishing their identities.
Following disruption of Ednra signaling in mouse and zebrafish, the most immediate change is a loss of a Dlx5/Dlx6/Hand2 hierarchical network. Dlx5/Dlx6 expression in the ventral (distal) pharyngeal arches relies on Ednra signaling [Charité et al., 2001; Ozeki et al., 2004; Ruest et al., 2004] and Mef2c activity [Miller et al., 2007; Verzi et al., 2007]. The relationship between Ednra signaling and Mef2c is not clear, though one hypothesis is that Mef2c “interprets” Ednra signaling along a proposed gradient of Edn1 in the pharyngeal arches, thus appropriately regulating Dlx expression in the arches [Miller et al., 2007]. This likely occurs through an enhancer element upstream of Dlx6 [Verzi et al., 2007]. Dlx5 and Dlx6 in turn induce expression of the gene encoding the basic helix-loop-helix transcription factor Hand2, in part through direct binding of Dlx6 to the known arch-specific enhancer of Hand2 [Charité et al., 2000]. However, since expression of Hand2 is not affected in either Dlx5−/− or Dlx6−/− embryos [Jeong et al., 2008], both Dlx5 and Dlx6 likely act interchangeably in inducing Hand2 expression. It should be noted that there is a small Hand2 expression domain in the ventral-most arch of mice and zebrafish that occurs independently of Ednra/Dlx5/Dlx6 [Miller et al., 2003b; Ruest et al., 2004]. Expression of both Dlx3, downstream of Dlx5/Dlx6 [Depew et al., 2002], and Goosecoid (Gsc), downstream of Hand2 [Miller et al., 2003a], are also absent following loss of Ednra signaling. In addition, expression of EphA3 is lost in edn1 mutant zebrafish [Miller et al., 2003a]. A map of these gene expression domains encompassing both mouse and zebrafish is shown in Figure 3. One of the interesting aspects of these domains is that they occur throughout the ventral arch, suggesting that Ednra signaling controls the entire ventral domain. However, the spatial regulation must be tightly controlled, since the expression of other ventrally-restricted genes, including Msx1, is not lost in Ednra mutants [Ruest et al., 2004].
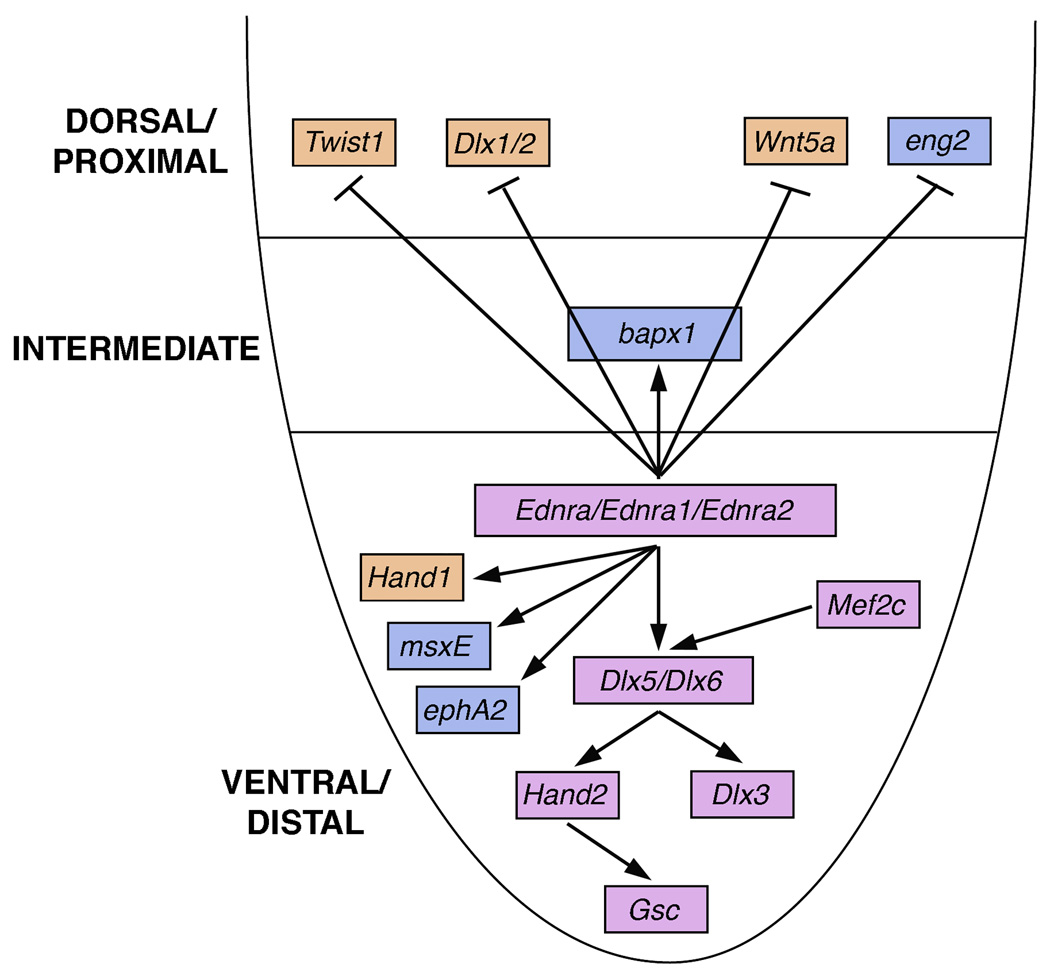
Figure 3.
Schematic of Edn1/Ednra signaling with the mandibular portion of the first pharyngeal arch. Ednra (Ednra1/2 in zebrafish) signaling leads to the induction of gene expression in the ventral (distal) and intermediate domains of the mandibular arch while repressing the expression of dorsal (proximal) genes in the intermediate and ventral arch domains. Many factors induced or repressed by Ednra signaling are conserved between mouse and zebrafish (highlighted in purple), though there are apparent species-specific differences between mouse (highlighted in orange) and zebrafish (highlighted in blue).
This loss of ventral (distal) gene expression is coupled with an expansion of genes normally expressed more dorsally (proximally) in the mandibular and maxillary portions of the first arch. This includes expansion of Dlx1, Dlx2 and Twist1 into the ventral arch, though none enter the small ventral Hand2 domain that occurs independently of Ednra/Dlx5/Dlx6 expression [Ruest et al., 2004]. Wnt5, normally confined to the maxillary portion of the first arch, is also observed in the mandibular region [Ruest et al., 2004]. The basis for these expansions is not clear, though clearly some sort of repressive signal is lost in the absence of Ednra signaling (Fig. 3). In addition, there are minor differences in gene expression patterns between mouse and zebrafish, as zebrafish dlx2 is lost ventrally in edn1 mutants while Dlx2 expression extends distally in Ednra mouse mutants [Miller et al., 2003a; Ruest et al., 2004]. Outside of these limited differences, it appears that overall the Ednra signaling pathway has been highly conserved during evolution, functioning by inducing expression of factors required to establish the identity of ventral (distal) NCCs in the mandibular arch while repressing signals associated with identity of more dorsal (proximal) NCCs.
DLX5 AND DLX6 AS DOWNSTREAM MEDIATORS OF EDNRA SIGNALING
The most prominent mediators of Ednra signaling during NCC patterning with the arches appear to be Dlx5 and Dlx6. Dlx5−/−;Dlx6−/− mutant embryos have similar changes in jaw structure to those observed in Edn1/Ece1/Ednra mutants, including homeosis of the mandible into a maxilla-like structure, duplication of the palatine, pterygoid and lamina obturans and loss of the tympanic ring bones and middle ear structures [Beverdam et al., 2002; Depew et al., 2002]. So what is the role of Dlx5/Dlx6 in arch patterning downstream of Ednra signaling? It has been proposed that Dlx genes within the arches establish DV patterning through a combinatorial code (Dlx code) similar to the role of Hox genes in patterning the segmental identities of arches along the A–P axis [Depew et al., 2002]. The ventral mandibular arch expresses Dlx1–6 while only Dlx1 and Dlx2 are expressed in the maxillary region. This indicates that there may be more redundancy in Dlx function in the ventral arch. This appears true for Dlx5 and Dlx6, since inactivation of either gene alone only results in mild changes in lower jaw structure [Jeong et al., 2008; Robledo et al., 2002]. In addition, Dlx1 and Dlx2 may be able to compensate for loss of Dlx5 or Dlx6, as Dlx1−/−;Dlx6−/−or Dlx2−/−;Dlx6−/− embryos both have more severe changes in lower jaw development and gene expression, some of which (including duplication of the jugal bone and loss of middle ear ossicles and the tympanic ring) resemble those observed in Dlx5−/−;Dlx6−/− mutants [Jeong et al., 2008]. Interestingly, however, Hand2 expression is unchanged in both Dlx1−/−;Dlx6−/− and Dlx2−/−;Dlx6−/− embryos. This further supports the hypothesis that Dlx5 and Dlx6 act redundantly to activate Hand2 and suggests that Hand2 is a crucial mediator of Dlx5/Dlx6 activity during lower jaw development. However, while it has been proposed that the maintenance of Hand2 expression in Dlx1−/−;Dlx6−/− and Dlx2−/−;Dlx6−/− embryos may explain the absence of mandibular homeosis [Jeong et al., 2008], reducing Hand2 expression in the mandibular arch does not lead to mandibular homeosis [Barbosa et al., 2007; Funato et al., 2009]. While persistent, residual Hand2 in these experiments may be sufficient to pattern mandibular NCCs, it is also possible that Hand2 is not solely responsible for establishing NCC identity in the mandibular arch. Since Hand2−/− mouse embryos die around E10.5 from vascular failure [Thomas et al., 1998; Yamagishi et al., 1999], elucidating these functions for Hand2 awaits conditional knockout of the Hand2 gene in NCCs.
A ROLE FOR EDNRA SIGNALING IN ESTABLISHING DV PATTERNING
As illustrated by the analyses of components of Ednra signaling using various loss-of-function approaches (ENU, targeted, antagonist and conditional knockout), the temporal window of essential Ednra activity occurs when cranial NCCs are populating the mandibular arch, consistent with a role in patterning the DV axis of the arch. This occurs in part through an Ednra-mediated morphogenetic domain in the ventral arch in which ventral-specific genes are expressed and more dorsal genes are excluded (reviewed in Clouthier and Schilling, 2004). However, recent findings in zebrafish suggest that the definition of an “endothelin-dependent domain” may be more complex than originally thought.
Zebrafish arches can be divided into three domains (reviewed in Clouthier and Schilling, 2004). The ventral domain gives rise to ventral cartilages, such as Meckel’s cartilage and the ceratohyal, while the dorsal domain gives rise to dorsal elements including the hyosymplectic; in fish the larger dorsal elements are homologous to middle ear bones of mammals. The intermediate domain gives rise to the joint regions between the ventral and dorsal cartilages. The ventral and intermediate domains are marked by region-specific markers (Hand2 and Bapx1, respectively) and it is the identity of these regions that is lost in suc;edn1−/− mutants [Miller et al., 2003a]. As described above, there were four “anterior arch class” mutants isolated from the Tubingen ENU screen [Piotrowski et al., 1996; Schilling 1997; Schilling et al., 1996]. These are now known to be mutations in edn1 [Miller et al., 2000], plcβ3 [Walker et al., 2007], furinA [Walker et al., 2006] and mef2c [Miller et al., 2007]. Based on changes in the branchiostegal ray/opercle dermal bone complex of suc;edn1−/− mutants (the opercle-gain and opercle-loss phenotypes described above), a model has been proposed in which Edn1 acts in a graded manner to pattern the arches (reviewed in Kimmel et al., 2007). In this model, Edn1 concentration is highest in the ventral arch and hence structures in this region (Meckel’s cartilage and ceratohyal) have the highest dependence on Edn1. The more intermediate region of the arch has a lower level of Edn1 and so elements in this region (the joints between dorsal and ventral structures) have a lower requirement for Edn1.
In a simple model, perturbation of a decreasing concentration gradient from ventral to dorsal can initially lead to an expansion of the lower end of the curve - such a model could explain why partial knockdown of edn1 using a morpholino leads to the loss of the ventrally-located branchiostegal ray but an expansion of the dorsal opercle [Kimmel et al., 2003]. This morphogen gradient could also explain why the strong anterior mutants (edn1 and plcβ3) have more pronounced ventral defects while the weaker anterior mutants (furinA and mef2c) have only moderate ventral defects. However, one of the problems with this model is that it does not explain why all four mutants have fully penetrant intermediate domain defects (including joint fusions). This has been suggested to be due to a requirement for Edn1 to properly segregate intermediate and ventral arch domains [Walker et al., 2007], potentially through a positive affect on arch elongation [Walker et al., 2006]. If downregulation of Edn1 occurs, it could lead to slower arch elongation and thus a “high” ventral Edn1 source displaced closer to the intermediate domain (where Edn1 is not normally so high) and thus loss of this domain, while the ventral domain would only be partially affected. When Edn1 is completely lost, both intermediate and ventral domains are lost.
While gradients may regulate DV patterning within the arches, early experiments showed that injection of human EDN1 into the mandibular arch of suc;edn1−/− zebrafish after cranial NCC migration rescued the Edn1 mutant phenotype [Miller et al., 2000]. While this helped support the argument that loss of Edn1 was the basis for the suc;edn1−/− phenotype, it is hard to understand how such an unlocalized injection of Edn1 can rescue a phenotype that in theory is caused by a disruption of a fine morphogen gradient. Rather, the results suggest more of a permissive role for Edn1 and that ventral and intermediate arch domains have differential requirements. Consistent with this notion, Schilling and colleagues recently presented an alternative possibility, based on work with the two Endra receptors in zebrafish [Nair et al., 2007]. ednra1 is expressed in NCC-derived mesenchyme and the overlying ectoderm while ednra2 is expressed only in the NCC-derived mesenchyme. While knockdown of ednra1 and ednra2 results in defects identical to those observed in suc/edn1−/− embryos, morpholino knockdown of ednra1 alone only disrupts intermediate arch patterning. As ventral arch fate is unaffected, Ednra2 appears sufficient to pattern this domain while additional Ednra signaling is required to pattern the joint or intermediated domain, with this additional signaling coming from an auto-upregulation of Edn1 by Ednra1 within the ectoderm.
It is not clear how these findings relate to the mouse arch, since there is only one known Ednra gene in the mouse [Sakurai et al., 1990]. In addition, a true “intermediate domain” marker that would mark a joint region has not been described. While loss of Bapx1 expression in both zebrafish and chick leads to loss of the jaw joint [Miller et al., 2003a; Wilson and Tucker 2003], targeted deletion of Bapx1 in mice leads to changes in the ossicle/tympanic joint in the middle ear, but not in the joint between the mandible and squamosal bone [Tucker et al., 2004]. This may be due to changes in downstream signaling molecules, as expression of Gdf5 and Gdf6 are unaffected in Bapx1−/− mouse embryos [Tucker et al., 2004]., while Gdf6 expression is lost in zebrafish embryos treated with a Bapx1 morpholino [Miller et al., 2003a]. However, the fact that Bapx1 expression still marks a joint in the mouse craniofacial complex is further evidence of the conserved nature of Ednra signaling in the pharyngeal arches between zebrafish and mice. It would therefore not be surprising to find similar mechanisms (perhaps different levels/sources of Edn1 in the mouse arch leading to activation of different downstream effectors) accomplishing the same domain-specific patterning.
Distinct effects of Ednra signaling on patterning of the ventral and intermediate arch domains illustrate the spatial complexity of this system. Are DV differences in patterning mediated solely by levels of Edn1 expression or differences in competency to respond or both? In zebrafish, injection of high levels of human EDN1 into the arches results in homeotic transformations of dorsal arch structures into ventral arch-like structures, indicating that the dorsal NCC are competent to respond to Edn1 [Kimmel et al., 2007]. This finding has been supported by recent findings in mice in which Edn1 was knocked into the Ednra locus, driving it in NCCs [Sato et al., 2008b]. Ednra expression is normally observed throughout cranial NCCs, whereas Edn1 expression is restricted to the arches posterior to the maxillary prominence [Clouthier et al., 1998; Yanagisawa et al., 1998b]. In the resulting mice, a homeotic transformation of the maxilla into a mandible-like structure occurred. This again suggests that maxillary NCCs are competent to respond to Edn1 and that the exclusion of Edn1 from the maxillary prominence is key to the correct patterning of the maxillary and mandibular prominences. Thus Edn1 appears to be both necessary and sufficient for dictating the DV identities of skeletogenic NCCs, consistent with its role as a key mediator of NCC development and skeletal patterning in the mandibular arch. Future studies to determine both the upstream factors that regulate expression of Edn1 and Ednra, as well as how signaling is interpreted by NCCs in such a subtle and specific fashion, are therefore critical for understanding the developmental basis for craniofacial patterning in vertebrates.
Acknowledgments
Grant information:
Grant Sponsor: NIH Grant Numbers: DE013828 to T. F. S. and DE014181 and DE018899 to D.E.C.
Contributor Information
David E. Clouthier, Department of Craniofacial Biology, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA.
Elvin Garcia, Department of Craniofacial Biology, University of Colorado Denver Anschutz Medical Campus, Aurora, CO 80045, USA.
Thomas F. Schilling, Department of Developmental and Cell Biology, University of California, Irvine, Irvine, CA 92697-2300
REFERENCES
- Abe M, Ruest L-B, Clouthier DE. Fate of cranial neural crest cells during craniofacial development in endothelin-A receptor deficient mice. Int J Dev Biol. 2007:51. doi: 10.1387/ijdb.062237ma. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold MA, Kim Y, Czubryt MP, Phan D, McAnally J, Qi X, Shelton JM, Richardson JA, Bassel-Duby R, Olson EN. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Barbosa AC, Funato N, Chapman S, McKee MD, Richardson JA, Olson EN, Yanagisawa H. Hand transcription factors cooperatively regulate development of the distal midline mesenchyme. Dev Biol. 2007;310:154–168. doi: 10.1016/j.ydbio.2007.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow A, de Graaff E, Pachnis V. Enteric nervous system progenitors are coordinately controlled by the G protein-coupled receptor EDNRB and the receptor tyrosine kinase RET. Neuron. 2003;40:905–916. doi: 10.1016/s0896-6273(03)00730-x. [DOI] [PubMed] [Google Scholar]
- Baynash AG, Hosoda K, Giaid A, Richardson JA, Emoto N, Hammer RE, Yanagisawa M. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 1994;79:1277–1285. doi: 10.1016/0092-8674(94)90018-3. [DOI] [PubMed] [Google Scholar]
- Beverdam A, Merlo GR, Paleari L, Mantero S, Genova F, Barbieri O, Janvier P, Levi G. Jaw transformation with gain of symmetry after Dlx5/Dlx 6 inactivation: mirror of the past. genesis. 2002;34:221–227. doi: 10.1002/gene.10156. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M. Origins and developmental potential of the neural crest. Exp Cell Res. 1995;218:405–417. doi: 10.1006/excr.1995.1173. [DOI] [PubMed] [Google Scholar]
- Chai Y, Maxson JRE. Recent advances in craniofacial morphogenesis. Dev Dyn. 2006;235:2353–2375. doi: 10.1002/dvdy.20833. [DOI] [PubMed] [Google Scholar]
- Chakrabarti S, Liu NJ, Gintzler AR. Reciprocal modulation of phospholipase Cbeta isoforms: adaptation to chronic morphine. Proc Natl Acad Sci USA. 2003 doi: 10.1073/pnas.2335885100. 100(13686–13691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charité J, McFadden DG, Merlo GR, Levi G, Clouthier DE, Yanagisawa M, Richardson JA, Olson EN. Role of Dlx6 in regulation of an endothelin-1-dependent, dHAND branchial arch enhancer. Genes Dev. 2001;15:3039–3049. doi: 10.1101/gad.931701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charité J, McFadden DG, Olson EN. The bHLH transcription factor dHAND controls Sonic hedgehog expression and establishment of the zone of polarizing activity during limb development. Development. 2000;127:2461–2470. doi: 10.1242/dev.127.11.2461. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Schilling TF. Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res (Part C) 2004;72:190–199. doi: 10.1002/bdrc.20007. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Hammer RE, Richardson JA, Yanagisawa M. Cell-autonomous and nonautonomous actions of endothelin-A receptor signaling in craniofacial and cardiovascular development. Dev Biol. 2003;261(2):506–519. doi: 10.1016/s0012-1606(03)00128-3. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Couly GF, Coltey PM, Le Douarin NM. The triple origin of skull in higher vertebrates: a study in chick-quail chimeras. Development. 1993;117:409–429. doi: 10.1242/dev.117.2.409. [DOI] [PubMed] [Google Scholar]
- Couly GF, Grapin-Botton A, Coltey P, Ruhin B, Le Douarin NM. Determination of the identity of the derivatives of the cephalic neural crest: incompatibility between Hox gene expression and lower jaw development. Development. 1998;125:3445–3459. doi: 10.1242/dev.125.17.3445. [DOI] [PubMed] [Google Scholar]
- Cramer H, Schmenger K, Heinrich K, Horstmeyer A, Boning H, Breit A, Piiper A, Lundstrom K, Muller-Esterl W, Schroeder C. Coupling of endothelin receptors to the ERK/MAP kinase pathways: Roles of palmitoylation and G(alpha)q. Eur J Biochem. 2001;268:5449–5459. doi: 10.1046/j.0014-2956.2001.02486.x. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Dettlaff-Swierez DA, Wettschureck N, Moers A, Huber K, Offermanns S. Characteristic defects in neural crest cell-specific Galpahq/Galph11- and Galpha12/Galpha13-deficient mice. Dev Biol. 2005;282:174–182. doi: 10.1016/j.ydbio.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Emoto N, Yanagisawa M. Endothelin converting enzyme-2: a membrane-bound, phosphoramidon-sensitive metalloprotease with acidic pH optimum. J Biol Chem. 1995;70:15262–15268. doi: 10.1074/jbc.270.25.15262. [DOI] [PubMed] [Google Scholar]
- Fraser SE, Keynes RJ, Lumsden AGS. Segmentation in the chick embryo hindbrain is defined by cell lineage restriction. Nature. 1990;344:431–435. doi: 10.1038/344431a0. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Kurihara Y, Arima Y, Yamada N, Kurihara H. Temporal requirement of signaling cascade involving endothelin-1/endothelin receptor type A in branchial arch development. Mech Dev. 2004;121:1223–1233. doi: 10.1016/j.mod.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Funato N, Chapman SL, McKee MD, Funato H, Morris JA, Shelton JM, Richardson JA, Yanagisawa H. Hand2 controls osteoblast differentiation in the branchial arch by inhibiting DNA binding of Runx2. Development. 2009;136:615–625. doi: 10.1242/dev.029355. [DOI] [PubMed] [Google Scholar]
- Gariepy CE, Yanagisawa M. Null mutation of endothelin receptor type B gene in spotting lethal rats causes aganglionic megacolon and white coat color. Proc Natl Acad Sci U S A. 1996;93:867–872. doi: 10.1073/pnas.93.2.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron-Maguire M, Mallo M, Zhang M, Gridley T. Hoxa-2 mutant mice exhibit homeotic transformation of skeletal elements derived from cranial neural crest. Cell. 1993;75:1317–1331. doi: 10.1016/0092-8674(93)90619-2. [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Miyata M, Watanabe M, Kano M. Roles of phospholipase Cbeta4 in synapse elimination and plasticity in developing and mature cerebellum. Mol Neurobiol. 2001;23:69–82. doi: 10.1385/MN:23:1:69. [DOI] [PubMed] [Google Scholar]
- Helms JA, Cordero D, Tapadia MD. New insights into craniofacial morphogenesis. Development. 2005;132:851–861. doi: 10.1242/dev.01705. [DOI] [PubMed] [Google Scholar]
- Hosoda K, Hammer RE, Richardson JA, Baynash AG, Cheung JC, Giaid A, Yanagisawa M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell. 1994;79:1267–1276. doi: 10.1016/0092-8674(94)90017-5. [DOI] [PubMed] [Google Scholar]
- Hunt P, Wilkinson DG, Krumlauf R. Patterning the vertebrate head: murine Hox 2 genes mark distinct subpopulations of premigratory and migrating cranial neural crest. Development. 1991;112:43–50. doi: 10.1242/dev.112.1.43. [DOI] [PubMed] [Google Scholar]
- Ivey K, Tyson B, Ukidwe P, McFadden DG, Levi G, Olson EN, Srivastava D, Wilkie TM. Gαq and Gα11 proteins mediate endothelin-1 signaling in neural crest-derived pharyngeal arch mesenchyme. Dev Biol. 2003;255:230–237. doi: 10.1016/s0012-1606(02)00097-0. [DOI] [PubMed] [Google Scholar]
- Jeong J, Li X, McEvilly RJ, Rosenfeld MG, Lufkin T, Rubenstein JLR. Dlx genes pattern mammalian jaw primordium by regulating both lower jaw-specific and upper jaw-specific genetic programs. Development. 2008;135:2905–2916. doi: 10.1242/dev.019778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- Kedzierski RM, Yanagisawa M. Endothelin system: the double-edged sword in health and disease. Annu Rev Pharmacol Toxicol. 2001;41:851–876. doi: 10.1146/annurev.pharmtox.41.1.851. [DOI] [PubMed] [Google Scholar]
- Kido T, Sawamura T. The processing pathway of endothelin-1 production. Cardiovasc Pharmacol. 1998;31 Suppl 1:S13–S15. doi: 10.1097/00005344-199800001-00006. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130(7):1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Walker MB, Miller CT. Morphing the hyomandibular skeleton in development and evolution. J Exp Zool (Mol Dev Evol) 2007;308B:609–624. doi: 10.1002/jez.b.21155. [DOI] [PubMed] [Google Scholar]
- Knight RD, Schilling TF. Cranial neural crest and development of the head skeleton. Adv Exp Med Biol. 2006;589:120–133. doi: 10.1007/978-0-387-46954-6_7. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao W-H, Kamada N, Jishage K, Ouchi Y, Azuma S, Toyoda Y, Ishikawa T, Kumada M, Yazaki Y. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Le Douarin N, Kalcheim C. The Neural Crest. Cambridge: University of Cambridge Press; 1999. [Google Scholar]
- Le Douarin NM. The Neural Crest. Cambridge: Cambridge Univ. Press; 1982. [Google Scholar]
- Le Douarin NM, Ziller C, Couly GF. Patterning of neural crest derivatives in the avian embryo: in vivo and in vitro studies. Dev Biol. 1993;159:24–49. doi: 10.1006/dbio.1993.1219. [DOI] [PubMed] [Google Scholar]
- Li Z, Jiang H, Xie W, Zhang Z, Smrcka AV, Wu D. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287:1046–1049. doi: 10.1126/science.287.5455.1046. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwartz L, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcriptional factor MEF2c. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden A, Sprawson N, Graham A. Segmental origin and migration of neural crest cells in the hindbrain region of the chick embryo. Development. 1991;113:1281–1291. doi: 10.1242/dev.113.4.1281. [DOI] [PubMed] [Google Scholar]
- Maemura K, Kurihara H, Kurihara Y, Oda H, Ishikawa T, Copeland NG, Gilbert DJ, Jenkins NA, Yazaki Y. Sequence analysis, chromosomal location, and developmental expression of the mouse preproendothelin-1 gene. Genomics. 1996;31:177–184. doi: 10.1006/geno.1996.0029. [DOI] [PubMed] [Google Scholar]
- Masaki T, Ninomiya H, Sakamoto A, Okamoto Y. Structural basis of the function of endothelin receptor. Mol Cell Biochem. 1999;190:153–156. [PubMed] [Google Scholar]
- Miletich I, Sharpe PT. Neural crest contribution to mammalian tooth formation. Birth Defects Res C Embryo Today. 2004;72(2):200–212. doi: 10.1002/bdrc.20012. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K-H, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3838. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miller CT, Swartz ME, Khuu PA, Walker MB, Eberhart JK, Kimmel CB. mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish. Dev Biol. 2007;308:144–157. doi: 10.1016/j.ydbio.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DY, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003a;130(7):1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Miller CT, Yelon D, Stainier DYR, Kimmel CB. Two endothelin 1 effectors, hand2 and bapx1, pattern ventral pharyngeal cartilage and the jaw joint. Development. 2003b;130(7):1353–1365. doi: 10.1242/dev.00339. [DOI] [PubMed] [Google Scholar]
- Minoux M, Antonarakis GS, Kmita M, Duboule D, Rijli FM. Rostral and caudal pharyngeal arches share a common neural crest ground pattern. Development. 2009;136:637–645. doi: 10.1242/dev.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Li W, Cornell R, Schilling TF. Requirements for endothelin type-A receptors and endothelin-1 signaling in the facial ectoderm for the patterning of skeletogenic neural crest cells in zebrafish. Development. 2007;134:335–345. doi: 10.1242/dev.02704. [DOI] [PubMed] [Google Scholar]
- Noden DM. The role of the neural crest in patterning of avian cranial skeletal, connective, and muscle tissues. Dev Biol. 1983;96:144–165. doi: 10.1016/0012-1606(83)90318-4. [DOI] [PubMed] [Google Scholar]
- Noden DM. Interactions and fates of avian craniofacial mesenchyme. Development. 1988;103:121–140. doi: 10.1242/dev.103.Supplement.121. [DOI] [PubMed] [Google Scholar]
- Offermanns S, Zhao LP, Gohla A, Sarosi I, Simon MI, Wilkie TM. Embryonic cardiomyocyte hypoplasia and craniofacial defects in G alpha q/G alpha 11-mutant mice. EMBO J. 1998;17:4304–4312. doi: 10.1093/emboj/17.15.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozeki H, Kurihara Y, Tonami K, Watatani S, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech Dev. 2004;121(4):387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Pasqualetti M, Ori M, Nardi I, Rijli FM. Ectopic Hoxa2 induction after neural crest migration results in homeosis of jaw elements in Xenopus. Development. 2000;127:5367–5378. doi: 10.1242/dev.127.24.5367. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Schilling TF, Brand M, Jiang Y-J, Heisenberg C-P, Beuchle D, Grandel H, van Eeden FJM, Furutani-Seiki M, Granato M, Haffter P, Hammerschmidt M, Kane DA, Kelsh RN, Mulllins MC, Odenthal J, Warga RM, Nusslein-Volhard C. Jaw and branchial arch mutants in zebrafish II. anterior arches and cartilage differentiation. Development. 1996;123:345–356. doi: 10.1242/dev.123.1.345. [DOI] [PubMed] [Google Scholar]
- Prince V, Lumsden A. Hoxa-2 expression in normal and transposed rhombomeres: independent regulation in the neural tube and neural crest. Development. 1994;120:911–923. doi: 10.1242/dev.120.4.911. [DOI] [PubMed] [Google Scholar]
- Puffenberger EG, Hosoda K, Washington SS, Nakao K, deWit D, Yanagisawa M, Chakravarti A. A missense mutation of the endothelin-B receptor gene in multigenic Hirschsprung's disease. Cell. 1994 doi: 10.1016/0092-8674(94)90016-7. [DOI] [PubMed] [Google Scholar]
- Raible DW, Wood A, Hodson W, Henion PD, Weston JA, Eisen JS. Segregation and early dispersal of neural crest cell in the embryonic zebrafish. Dev Dyn. 1992;195:29–42. doi: 10.1002/aja.1001950104. [DOI] [PubMed] [Google Scholar]
- Rijli FM, Mark M, Lakkaraju S, Dierich A, Dolle P, Chambon P. A homeotic transformation is generated in the rostral branchial region of the head by disruption of Hoxa-2, which acts as a selector gene. Cell. 1993;75:1333–1349. doi: 10.1016/0092-8674(93)90620-6. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, Lufkin T. The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 2002;16:1089–1101. doi: 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L-B, Kedzierski R, Yanagisawa M, Clouthier DE. Deletion of the endothelin-A receptor gene within the developing mandible. Cell Tissue Res. 2005;319:447–454. doi: 10.1007/s00441-004-0988-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Clouthier DE. Elucidating timing and function of endothelin-A receptor signaling during craniofacial development using neural crest cell-specific gene deletion and receptor antagonism. Dev Biol. 2009;328:94–108. doi: 10.1016/j.ydbio.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest LB, Xiang X, Lim KC, Levi G, Clouthier DE. Endothelin-A receptor-dependent and - independent signaling pathways in establishing mandibular identity. Development. 2004;131(18):4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai T, Yanagisawa M, Takuwa Y, Miyazaki H, Kimura S, Goto K, Masaki T. Cloning of a cDNA encoding a non-isopeptide-selective subtype of the endothelin receptor. Nature. 1990;348:732–735. doi: 10.1038/348732a0. [DOI] [PubMed] [Google Scholar]
- Sato T, Kawamura Y, Asai R, Amano T, Uchijima Y, Dettlaff-Swiercz DA, Offermanns S, Kurihara Y, Kurihara H. Recombinase-mediated cassette exchange reveals the selective use of Gq/G11-dependent and -independent endothelin 1-endothelin type A receptor signaling in pharyngeal arch development. Development. 2008a;135:755–765. doi: 10.1242/dev.012708. [DOI] [PubMed] [Google Scholar]
- Sato T, Kurihara Y, Asai R, Kawamura Y, Tonami K, Uchijima Y, Heude E, Ekker M, Levi G, Kurihara H. An endothelin-1 switch specifies maxillomandibular identity. Proc Natl Acad Sci U S A. 2008b;105:18806–18811. doi: 10.1073/pnas.0807345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF. Genetic analysis of craniofacial development in the vertebrate embryo. Bioessays. 1997;19(6):459–468. doi: 10.1002/bies.950190605. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Kimmel CB. Segment and cell type lineage restrictions during pharyngeal arch development in the zebrafish embryo. Development. 1994;120:483–494. doi: 10.1242/dev.120.3.483. [DOI] [PubMed] [Google Scholar]
- Schilling TF, Piotrowski T, Grandel H, Brand M, Heisenberg CP, Jiang YJ, Beuchle D, Hammerschmidt M, Kane DA, Mullins MC, van Eeden FJ, Kelsh RN, Furutani-Seiki M, Granato M, Haffter P, Odenthal J, Warga RM, Trowe T, Nusslein-Volhard C. Jaw and branchial arch mutants in zebrafish I: branchial arches. Development. 1996;123:329–344. doi: 10.1242/dev.123.1.329. [DOI] [PubMed] [Google Scholar]
- Smrcka AV, Sternweis PC. Regulation of purified subtypes of phosphatidylinositol-specific phospholipase C beta by G protein alpha and beta gamma subunits. J Biol Chem. 1994;268 9667-8674. [PubMed] [Google Scholar]
- Tanaka H, Moroi K, Iwai J, Takahashi H, Ohnuma N, Hori S, Takimoto M, Nishiyama M, Masaki T, Yanagisawa M, Sekiya S, Kimura S. Novel mutations of the endothelin B receptor gene in patients with Hirshsprung's disease and their characterization. J Biol Chem. 1998;273:11378–11383. doi: 10.1074/jbc.273.18.11378. [DOI] [PubMed] [Google Scholar]
- Thomas T, Kurihara H, Yamagishi H, Kurihara Y, Yazaki Y, Olson EN, Srivastava D. A signaling cascade involving endothelin-1, dHAND and Msx1 regulates development of neural-crest-derived branchial arch mesenchyme. Development. 1998;125:3005–3014. doi: 10.1242/dev.125.16.3005. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Specification of neural crest cell formation and migration in mouse embryos. Semin Cell Dev Biol. 2005 doi: 10.1016/j.semcdb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Krumlauf R. Hox genes, neural crest cells and branchial arch patterning. Curr Opin Cell Biol. 2001;13(6):698–705. doi: 10.1016/s0955-0674(00)00273-8. [DOI] [PubMed] [Google Scholar]
- Tucker AS, Watson RP, Lettice LA, Yamada G, Hill RE. Bapx1 regulates patterning in the middle ear: altered regulatory role in the transition from the proximal jaw during vertebrate evolution. Development. 2004;131:1235–1245. doi: 10.1242/dev.01017. [DOI] [PubMed] [Google Scholar]
- Verzi MP, Agarwar P, Brown C, McCulley DJ, Schwarz JJ, Black BL. The transcription factor MEF2C is required for craniofacial development. Dev Cell. 2007;12:645–652. doi: 10.1016/j.devcel.2007.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Swartz ME, Eberhart JK, Kimmel CB. phospholipase C, beta 3 is required for Endothelin1 regulation of pharyngeal arch patterning in zebrafish. Dev Biol. 2007;304:194–207. doi: 10.1016/j.ydbio.2006.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker MB, Miller CT, Talbot JC, Stock DW, Kimmel CB. Zebrafish furin mutants reveal intricacies in regulating Endothelin1 signaling in craniofacial patterning. Dev Biol. 2006;295:194–205. doi: 10.1016/j.ydbio.2006.03.028. [DOI] [PubMed] [Google Scholar]
- Wang S, Gebre-Medhin S, Betsholtz C, Stalberg P, Zhou Y, Larsson C, Weber G, Feinstein R, Oberg K, Gobl A, Skogseid B. Targeted disruption of the mouse phospholipase C beta3 gene results in early embryonic lethality. FEBS Let. 1998;441:261–265. doi: 10.1016/s0014-5793(98)01518-x. [DOI] [PubMed] [Google Scholar]
- Wilson J, Tucker AS. Fgf and Bmp signals repress the expression of Bapx1 in the mandibular mesenchyme and control the position of the developing jaw joint. Dev Biol. 2003;266:138–150. doi: 10.1016/j.ydbio.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Xie W, Samoriski GM, McLaughlin JP, Romoser VA, Smrcka A, Hinkle PM, Bidlack JM, Gross RA, Jiang H, Wu D. Genetic alteration of phospholipase C beta3 expression modulates behavioral and cellular responses to mu opioids. Proc Natl Acad Sci USA. 1999;96:10385–10390. doi: 10.1073/pnas.96.18.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Emoto N, Giaid A, Slaughter C, Kaw S, deWit D, Yanagisawa M. ECE-1: a membrane-bound metalloprotease that catalyzes the proteolytic activation of big endothelin-1. Cell. 1994;78:473–485. doi: 10.1016/0092-8674(94)90425-1. [DOI] [PubMed] [Google Scholar]
- Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science. 1999;283:1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Abe K, Mantani A, Hitoshi Y, Suzuki M, Osuzu F, Kuratani SKY. A novel transgenic technique that allows specific marking of the neural crest cell lineage in mice. Dev Biol. 1999;212:191–203. doi: 10.1006/dbio.1999.9323. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Emoto N, Williams SC, Takeda S, Clouthier DE, Yanagisawa M. Disruption of ECE-1 and ECE-2 reveals a role for endothelin-converting enzyme-2 in murine cardiac development. J Clin Invest. 2000;105:1373–1382. doi: 10.1172/JCI7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Hammer RE, Richardson JA, Williams SC, Clouthier DE, Yanagisawa M. Role of Endothelin-1/Endothelin-A receptor-mediated signaling pathway in the aortic arch patterning in mice. J Clin Invest. 1998a;102(1):22–33. doi: 10.1172/JCI2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, de Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998b;125(5):825–836. doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M. The endothelin system: a new target for therapeutic intervention. Circulation. 1994;89:1320–1322. doi: 10.1161/01.cir.89.3.1320. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Kurihara H, Kimura S, Tomobe Y, Kobayashi M, Mitsui Y, Yazaki Y, Goto K, Masaki T. A novel potent vasoconstrictor peptide produced by vascular endothelial cells. Nature. 1988;332:411–415. doi: 10.1038/332411a0. [DOI] [PubMed] [Google Scholar]
- Yelick PC, Schilling TF. Molecular dissection of craniofacial development using zebrafish. Crit Rev Oral Biol Med. 2002;13(4):308–322. doi: 10.1177/154411130201300402. [DOI] [PubMed] [Google Scholar]