Abstract
The Schizosaccharomyces pombe dhp1+ gene is an ortholog of the Saccharomyces cerevisiae RAT1 gene, which encodes a nuclear 5′→3′ exoribonuclease, and is essential for cell viability. To clarify the cellular functions of the nuclear 5′→3′ exoribonuclease, we isolated and characterized a temperature-sensitive mutant of dhp1 (dhp1-1 mutant). The dhp1-1 mutant showed nuclear accumulation of poly(A)+ RNA at the restrictive temperature, as was already reported for the rat1 mutant. Interestingly, the dhp1-1 mutant exhibited aberrant chromosome segregation at the restrictive temperature. The dhp1-1 cells frequently contained condensed chromosomes, most of whose sister chromatids failed to separate during mitosis despite normal mitotic spindle elongation. Finally, chromosomes were displaced or unequally segregated. As similar mitotic defects were also observed in Dhp1p-depleted cells, we concluded that dhp1+ is required for proper chromosome segregation as well as for poly(A)+ RNA metabolism in fission yeast. Furthermore, we isolated a multicopy suppressor of the dhp1-1 mutant, referred to as din1+. We found that the gene product of dhp1-1 was unstable at high temperatures, but that reduced levels of Dhp1-1p could be suppressed by overexpressing Din1p at the restrictive temperature. Thus, Din1p may physically interact with Dhp1p and stabilize Dhp1p and/or restore its activity.
INTRODUCTION
In Saccharomyces cerevisiae, two 5′→3′ exoribonucleases have been isolated so far (1–3). One is encoded by the RAT1 gene (4) [also referred as HKE1 (1) and TAP1 (5,6)] and the other is encoded by the XRN1 gene (7) [also referred as DST2 (8), SEP1 (9), KEM1 (10,11) and RAR5 (12)]. They share homologous regions in their amino acid sequences and also similar biochemical properties such as exoribonuclease activity, but have different cellular functions. RAT1 encodes a protein that is predominantly localized in the nucleus (1) and is essential for cell growth (1,4,6). In contrast, XRN1 is not required for cell viability (7–10,12) and the gene product is localized to the cytoplasm (13). Xrn1p functions in turnover of mRNA (14–16) and pre-rRNA (17,18). Interestingly, when Xrn1p was localized in the nucleus by fusion of an NLS, it rescued the temperature-sensitivity of the rat1-1 mutant. In contrast, wild-type Xrn1p did not, indicating that Rat1p and Xrn1p are functionally interchangeable proteins that normally reside and function in the nucleus and cytoplasm, respectively (19). Counterparts of Rat1p and Xrn1p have been identified in Schizosaccharomyces pombe (20,21), mouse (22–24) and human (25,26), indicating that these 5′→3′ exoribonucleases have been conserved throughout eukaryotes.
The RAT1 gene was originally found to be involved in the export of poly(A)+ RNA from the nucleus in S.cerevisiae (4). In the temperature-sensitive rat1-1 mutant poly(A)+ RNA rapidly accumulated in the nucleus after shift-up to the restrictive temperature (4). In addition, the rat1-1 mutant displayed increased levels of a 5′-extended form of 5.8S rRNA, suggesting that the exoribonuclease activity of Rat1p is involved in the 5′-processing step of 5.8S rRNA maturation (4,17). Furthermore, Rat1p has been shown to be involved in the turnover and processing of nuclear RNA such as the degradation of several excised fragments of pre-rRNA spacer regions (27) and 5′-processing of small nucleolar RNAs (snoRNAs) (27–29), which are required for ribosome biosynthesis. However, the relationship between a defect in ribosome biosynthesis and the growth arrest observed for the rat1 mutant remains unclear.
Recent observations have identified a complex of 3′→5′ exoribonucleases, termed the exosome, which consists of at least 10 core subunits (Rrp4p, Rrp40p to Rrp46p, Mtr3p and Csl4p), a nuclear subunit (Rrp6p) and associated factors (Mtr4p, Ski2p, Ski3p and Ski8p) (30,31; reviewed in 32). All of the core subunits display sequence similarity with 3′→5′ exoribonucleases and some possess 3′→5′ exoribonuclease activity (30,31). Conditional defects in subunits of the exosome affect export of poly(A)+ RNA from the nucleus (33) and 3′-processing of 5.8S rRNA (34–39), mRNA (40) and snoRNAs (35,41) at the restrictive temperature. The observations indicate that nuclear 3′→5′ (nuclear exosome) and 5′→3′ (Rat1p) exoribonuclease activities are involved in nucleocytoplasmic RNA trafficking and RNA processing (4,30,41). Besides their involvement in RNA metabolism, a mutation in the S.pombe ortholog of RRP44, termed dis3-54, was shown to result in defective chromosome segregation (42,43), suggesting involvement of nuclear exoribonucleases in chromosome segregation.
The dhp1+ gene is an ortholog of RAT1 in S.pombe. Both genes share 40% identity in their deduced amino acid sequences (20). Expression of Dhp1p rescued the temperature-sensitivity of a rat1 mutant (20). In addition, overproduction of Dhp1p alleviated the defects of an xrn1 mutant (slow growth and a reduced level of sporulation) as effectively as did Rat1p (20). These observations indicate that dhp1+ is not only structurally but also functionally homologous to RAT1. The dhp1+ gene disruptant spores germinate but fail to divide, showing that dhp1+ is an essential gene (20). Although these results suggest that the dhp1+ gene product is also an essential nuclear 5′→3′ exoribonuclease, the in vivo function of the Dhp1 protein has remained elusive, as is also the case for Rat1p of S.cerevisiae. In the present communication, we isolated and characterized a temperature-sensitive mutant of dhp1, in order to clarify the cellular functions of the dhp1+ gene in fission yeast. We found that the dhp1+ gene is required for proper chromosome segregation, as well as for poly(A)+ RNA trafficking.
MATERIALS AND METHODS
Strains, media and plasmids
The S.pombe strains used in this study are listed in Table 1. Media and standard genetic procedures for S.pombe were performed as described by Alfa et al. (44). For transcriptional repression of dhp1+ from pREP81-dhp1+, thiamine was added to MM to yield a final concentration of 5 µg/ml. pREP81-dhp1+, pREP41-dhp1+ or pREP41-din1+ was constructed by inserting the DNA fragment coding dhp1+ or din1+ downstream of the weak or medium nmt1 promoter of pREP81 or pREP41 (45).
Table 1. S.pombe strains used.
| Strain |
Genotype |
Source |
| 972 | h– | Our stock |
| MP102 | h+ ade6-M216 leu1 ura4-D18 dhp1-1<<ura4+ | This work |
| JY742 | h+ ade6-M216 leu1 ura4-D18 | Our stock |
| SSP1 | h+/h– ade6-M210/ade6-M216 leu1/leu1 ura4-D18/ura4-D18 dhp1+/dhp1::ura4+ | (20) |
| KP38 | h+ ade6-M216 leu1 his7-lacI-GFP-his7+ lys1-lacO-lys1+ ura4-D18 dhp1-1<<ura4+ | This work |
Sequential deletion analysis of Dhp1p
To generate C-terminal deletion mutants, the Eco47III–EcoRV (ΔC125) or Eco47III–AccIII (ΔC204) fragments of the dhp1+ gene were inserted into the PstI site of pDB248′ (46). For the N-terminal deletion mutant (ΔN91), the Eco47III–EcoT22I fragment lacking the PflMI–NcoI region was inserted into the PstI site of pDB248′ as well. On each plasmid, the mutant gene is transcribed from the intrinsic promoter of dhp1+. Procedures for expressing the deletion mutants in the dhp1 null mutant using SSP1 were previously described (23).
Construction of the dhp1-1 mutant
A 1.8 kb ura4+ gene fragment was inserted into EcoRV site of pH8-2 (20). The resulting plasmid was subjected to restriction digestion by both NdeI and HindIII, and was then used for transformation of the S.pombe strain JY741 (h– ade6-M210 leu1 ura4-D18) to replace the chromosomal dhp1+ gene with the truncated dhp1 mutant gene via homologous recombination. Correct integration was confirmed by Southern blotting. The strain (MP102) was backcrossed once, and used in this study. MP102 was further crossed with MKY7A (h+ leu1 his7-lacI-GFP-his7+ lys1-lacO-lys1+ura4-D18) to generate the KP38 strain used to visualize centromeric DNA–GFP (47).
Determination of growth rate and cell viability
The growth rate in mid-log phase cultures (106–107 cells/ml) at 36°C was monitored in duplicate by removing aliquots at intervals and counting the cell numbers with a haemocytometer. Simultaneously, properly diluted aliquots were plated onto YES plates and incubated for 4 days at 25°C to determine cell viability.
Fluorescence microscopy
Cells were stained with 4′,6-diamidino-2-phenylindole (DAPI) to visualize DNA in fixed cells and 3,3′-dihexyloxacarbocyanine iodide (DiOC6) to visualize the nuclear envelope. Immunofluorescence microscopy using anti-tubulin antibody YOL1/34 (Sera-lab) and a rhodamine-conjugated goat anti-rat IgG (Immunotech S.A.) as secondary antibody was previously described (48). To visualize centromeric DNA–GFP, the cells (KP38) were fixed in 100% methanol at –70°C overnight, and then subjected to fluorescence microscopy after DAPI staining.
Analysis of localization of poly(A)+ RNA
Localization of poly(A)+ RNA was analyzed by in situ fluorescence hybridization (FISH) as previously described (49).
Isolation of din1+
The S.pombe genomic DNA library was introduced into the dhp1-1 mutant by the lithium acetate method. The transformed cells were incubated at 25°C for 2 days, and then at 35°C for 5 days. Approximately 1.3 × 105 clones were screened and 11 clones that grew at 35°C were obtained. The plasmids were rescued in Escherichia coli and re-introduced into the dhp1-1 cells to confirm their ability to suppress dhp1-1 mutation. Nucleotide sequences of the inserts were determined by using an ABI PRISM 310 genetic analyzer. For the isolation of din1+ cDNA, 5′- or 3′-rapid amplification of cDNA ends (RACE) with the S.pombe cDNA library was performed using the following primers: TGGGATCCAAAGGAGCGCAC for 5′-RACE and TCCTAAATCCGACCCTGATC for 3′-RACE. The nucleotide sequence of the din1+ cDNA was determined as described above.
Western blotting
Rabbit anti-Dhp1p antiserum was obtained by standard procedures. Briefly, fusion proteins of maltose binding protein and full-length Dhp1p were expressed in E.coli. Whole cell lysates were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and the band of the overproduced protein was excised to immunize rabbits.
Fission yeast total soluble protein extracts were prepared using the glass bead method. Protein concentrations were determined by the Protein assay (Bio-Rad) using BSA as a standard. Equal amounts of protein were separated on each of the lanes of an SDS–polyacrylamide gel and transferred to polyvinylidene difluoride membranes, and western blotting was carried out with the Protoblot kit (Promega) with anti-Dhp1p (1:150 dilution) as the primary antibody.
RESULTS
Temperature-sensitivity of a mutant lacking the C-terminal segment of Dhp1p
The dhp1+ gene is essential for cell viability (20). To determine the essential region of Dhp1p necessary for proliferation, truncated forms of the dhp1+ gene were constructed by deleting carboxyl or amino portions of the gene on the LEU2-containing plasmid pDB248′. The plasmids were introduced into the heterozygous dhp1+/dhp1::ura4+ diploid strain SSP1, and the Ura+ and Leu+ haploid progeny, which contained both the chromosomal dhp1 gene inactivated by the ura4+ insertion and the truncated dhp1 mutant gene on the plasmid, were selected for growth on minimal medium plates after sporulation. Dhp1ΔC125p, which had lost the C-terminal 125 amino acids of Dhp1p, rescued the viability of the dhp1::ura4 cells, while neither Dhp1ΔC204p (lacking the C-terminal 204 amino acids) nor Dhp1ΔN91p (lacking the N-terminal 91 amino acids), could do so (Fig. 1). The results indicate that two highly conserved regions, HR1 and HR2, but not the 125 amino acid C-terminal portion, comprise the regions of Dhp1p necessary for cell growth. However, deletion of the C-terminal 125 amino acids of Dhp1p resulted in temperature-sensitive cell growth. When the chromosomal copy of dhp1+ was truncated by the insertion of the ura4+ gene, resulting in the loss of the C-terminal 125 amino acids (Fig. 2A), the mutant cell grew normally below 33°C, but not above 33°C (data not shown). We designated the mutation as dhp1-1. The dhp1-1 mutant ceased to grow and lost viability 4 h after shift-up to 36°C in YES medium (Fig. 2B). In accordance with growth cessation, the mutant Dhp1-1p protein was quickly lost from the cells 3 h after shift-up (Fig. 2C), indicating that Dhp1-1p might be unstable at high temperatures. Introduction of a multicopy plasmid harboring the dhp1-1 mutant gene suppressed the temperature-sensitivity of dhp1-1 (data not shown). These data suggest that the temperature-sensitivity of the dhp1-1 mutant was caused by a quantitative reduction in the amount of Dhp1-1p, possibly due to its instability at the restrictive temperature.
Figure 1.
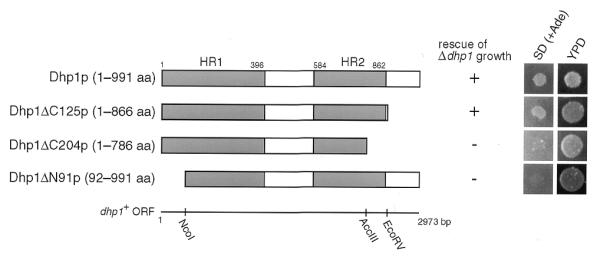
Sequential deletion analysis of Dhp1p. White bars, amino acid sequences of the truncated mutant Dhp1p; shaded bars [HR1 (1–396 amino acids) and HR2 (584–862 amino acids)], conserved regions between Dhp1p, Rat1p and mouse Dhm1p (23). Only relevant restriction sites in the dhp1+ coding sequence are shown. Rescue of the Δdhp1 mutant by a series of truncated Dhp1 proteins was determined using spores from SSP1, which contain the dhp1 alleles expressed from the dhp1 promoter on the pDB248′ vector, depending on whether they can form colonies or not. Aliquots of spore suspensions, which carry plasmids expressing truncated forms of Dhp1p, were spotted on YPD plates and SD plates containing 10 µg/ml adenine [SD (+Ade)] and incubated at 30°C for 5 days. Spores with the dhp1+ gene can form colonies only on YPD plates. Δdhp1 haploid spores (Ura+ Ade–) will form colonies on SD (+Ade) plates only when the dhp1 deletion allele rescues cell growth. The data are presented to show both numbers of colonies that appeared (+) and failed to appear (–) on the plates.
Figure 2.
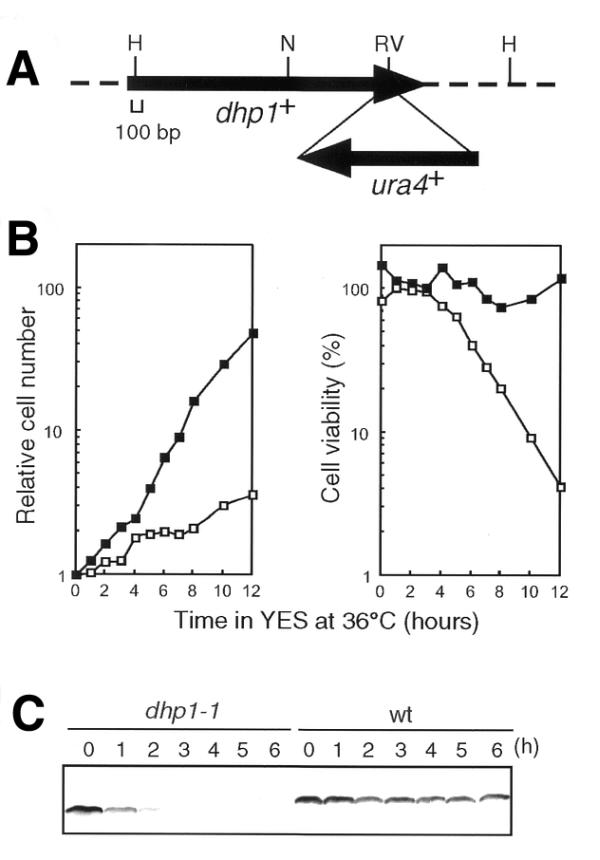
Properties of the dhp1-1 mutation. (A) Construct of the dhp1-1 allele. The ura4+ gene was inserted into the EcoRV site of the dhp1+ gene to generate Dhp1-1p, which lacks 125 amino acids from the C-terminus of Dhp1p. Only relevant restriction sites are shown. H, HindIII; N, NdeI; RV, EcoRV. (B) Growth rate of the dhp1-1 mutant. Wild-type (JY742, closed squares) or dhp1-1 (MP102, open squares) cells grown at 25°C in YES were cultured at 36°C. (C) Instability of Dhp1-1p at the restrictive temperature. Wild-type or the dhp1-1 cells grown at 25°C in YES were cultured at 36°C. Soluble cell lysates prepared from aliquots were separated by 8.3% SDS–PAGE and subjected to western blotting with anti-Dhp1p antiserum.
Nuclear accumulation of poly(A)+ RNA in the dhp1-1 mutant
The rat1-1 mutant was reported to accumulate poly(A)+ RNA in the nucleus (4). To test whether the dhp1+ gene is also involved in poly(A)+ RNA metabolism in fission yeast, we performed FISH to analyze the localization of poly(A)+ RNA in the dhp1-1 mutant. The dhp1-1 cells grown at 26°C were transferred to 37°C for 2 h, and then subjected to in situ hybridization with biotin-labeled oligo(dT)50 and FITC–avidin. As shown in Figure 3, nuclear accumulation of poly(A)+ RNA was observed in the dhp1-1 cells at the restrictive temperature. In contrast, poly(A)+ RNA was distributed throughout the wild-type cells under the same conditions. Thus, the dhp1+ gene product is involved in poly(A)+ RNA metabolism such as mRNA export from the nucleus or processing of poly(A)+ RNA in the nucleus, as was also shown for Rat1p in budding yeast.
Figure 3.
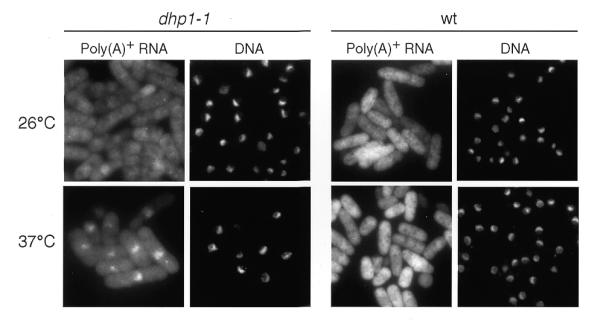
Analysis of poly(A)+ RNA localization in the wild-type (wt) and dhp1-1 cells. Wild-type (972) or dhp1-1 (MP102) cells grown at 26°C were cultured at 37°C for 2 h. The cells were fixed and analyzed by in situ hybridization with a biotin-labeled oligo (dT)50 probe. Hybridized signals were detected by FITC-conjugated avidin. Left panels show the poly(A)+ RNA distribution, right panels (DNA) show the cells stained with DAPI.
Requirement of the dhp1+ gene product for proper chromosome segregation
To characterize the temperature-sensitive dhp1-1 mutant, the gross nuclear structure was analyzed in the dhp1-1 cells at the restrictive temperature (Fig. 4). When the mutant was grown at 25°C, the majority of the cells exhibited interphase-like cell morphology, as observed for wild-type cells incubated at 25 or 36°C (data not shown). After a 3 h incubation at 36°C, however, cells exhibiting a variety of mitotic defects emerged in the dhp1-1 mutant (Fig. 4B). This corresponded to the period during which cell viability also began to decrease (Fig. 2B). Some cells contained condensed chromosomes, which often formed the three blocks of nuclear material that are characteristic of the haploid karyotype of S.pombe (Fig. 4A). In other cells, the three blocks of condensed chromosomes appeared to be unequally pulled apart by the mitotic spindle (Fig. 4C) when separation of sister chromatids appeared to be hindered as reported for dis mutants (43). To investigate whether dhp1-1 was deficient in sister chromatid separation, the Cen1–GFP system was used to visualize the centromere DNA of the chromosomes in the dhp1-1 mutant (Fig. 4D). At the restrictive temperature, a single Cen1–GFP dot was frequently detected in only one of the three blocks of condensed chromosomes as well as in the unequally separated chromosomes (Fig. 4D, ‘Nondisjunction’). Another fraction of the cells exhibits two Cen1–GFP signals in each nuclei, which seem to have executed normal separation of chromosomes (Fig. 4B and D, ‘Normal’). On the contrary, the cells lacking disjunction of Cen1–GFP signals were not seen in the wild-type cells. These data suggest that disjunction of sister chromatids was impaired in the dhp1-1 mutant. After prolonged incubation, the portion of cells with condensed chromosome decreased, while the fraction of the cells with a displaced nucleus increased (Fig. 4B). Anuclear cells were also observed. DiOC6 staining of the cells revealed that the nuclear envelope seemed to be normal in the dhp1-1 cells at the restrictive temperature (data not shown). These results indicate that the dhp1-1 mutant is defective in the normal execution of chromosome segregation. Taken together, it is suggested that the dhp1-1 cells undergo a transient arrest in the cell number increase soon after shift-up (Fig. 2B, left), followed by cell division without chromosome segregation (Fig. 4A–D) and finally lose viability (Fig. 2B, right) in the period 3–4 h after shift-up.
Figure 4.
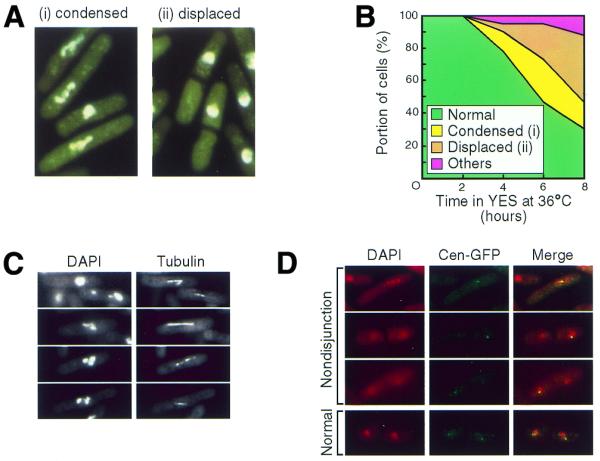
Defective chromosome segregation in the dhp1-1 mutant. (A) Nuclear structures of the dhp1-1 mutant at 36°C. dhp1-1 (MP102) cells grown at 36°C for 6 h were stained with DAPI to visualize the nucleus. Typical nuclear morphologies of defective chromosome segregation are indicated: (i) cells with condensed chromosomes and (ii) cells with displaced nuclei. See also (C) and (D). (B) Frequency of the dhp1-1 cells exhibiting defective chromosome segregation. The dhp1-1 cells grown at 26°C were cultured at 36°C and aliquots were fixed with 70% ethanol and stained with DAPI. At least 120 cells were analyzed at every interval. (C) Elongating mitotic spindles that failed to segregate chromosomes. The dhp1-1 cells cultured at 36°C for 6 h were subjected to immunofluorescence microscopy with the monoclonal anti-tubulin antibody YOL1/34. (D) Nondisjunction of sister chromatids in the dhp1-1 mutant. dhp1-1 cells (KP38) were cultured at 36°C for 6 h and the centromere DNA of chromosome I in the dhp1-1 mutant was visualized by using the Cen1–GFP system.
Next, we investigated whether loss of Dhp1p also causes abnormal chromosome segregation in the dhp1 null mutant. We constructed a dhp1 null mutant harboring the pREP81-dhp1+ plasmid, which carries the ORF for dhp1+ under the control of the weak nmt1 promoter Rep81, and grew the cells on thiamine-free plates. When Dhp1p expression was repressed by addition of thiamine, the cells ceased to grow (Fig. 5A) and exhibited chromosome segregation defects such as displacement of the nucleus (Fig. 5B), which was also observed in the dhp1-1 mutant at 36°C. Thus, we concluded that the dhp1+ gene is required for proper chromosome segregation.
Figure 5.
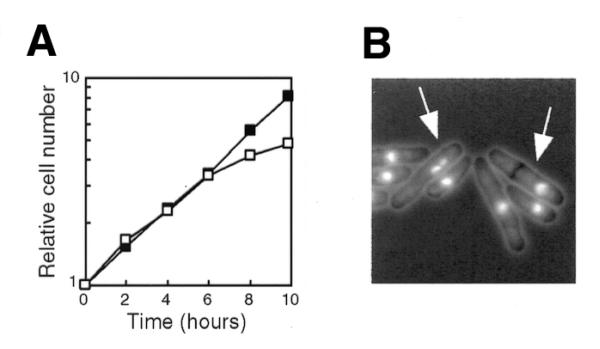
Depletion of Dhp1p from cells. (A) Growth of the Δdhp1 cells after shut-off of Dhp1p. Δdhp1 cells harboring pREP81-dhp1+ were exponentially grown in thiamine-free MM (+Ade) and further incubated with (open squares) or without (closed squares) 5 µg/ml thiamine at 30°C. (B) Nuclear structure of Dhp1p-depleted cells. The Δdhp1 cells harboring pREP81-dhp1+ were incubated for 10 h in the presence of thiamine and then fixed and stained with DAPI. Arrows indicate cells with defective chromosome segregation.
Isolation of a multicopy suppressor gene for the dhp1-1 mutation
To search for factors that interact with dhp1+, we screened for multicopy suppressors of the dhp1-1 mutation. The genomic library of S.pombe was introduced into the dhp1-1 mutant and viable cells were selected at 35°C. Among the 1.3 × 105 clones screened, 11 clones were obtained. Nucleotide sequencing of the clones revealed that two genes were responsible for the suppression of temperature-sensitivity of the dhp1-1 mutant. One of them, designated din1+ (encoding the Dhp1p-interacting protein; DDBJ accession no. AB045607), was found to be identical to the uncharacterized ORF SPAC19D5.06c in the S.pombe chromosome I cosmid c19D5 (EMBL accession no. Z99531). The other is under investigation in our laboratory. A database search for homology revealed that Din1p had partial homology with the deduced amino acid sequence of S.cerevisiae YGL246c/RAI1 (50), human DOM3Z (51) and Caenorhabditis elegans dom-3 (52), suggesting that Din1p is conserved among eukaryotes.
When the plasmid containing the din1+ gene (pREP41-din1+) was introduced into the dhp1-1 cells, the temperature-sensitivity was abrogated at 36°C, although the growth rate was slightly lower than that of wild-type cells growing at the same temperature (Fig. 6A). In addition, there was no significant loss of viability or any defective chromosome segregation in dhp1-1 cells carrying pREP41-din1+ at 36°C (data not shown). Furthermore, we found that the reduction in Dhp1-1p levels was considerably suppressed by pREP41-din1+ at 36°C (Fig. 6B). Dhp1-1p was still present in dhp1-1 cells carrying pREP41-din1+ even after 6 h at 36°C, while it could not be detected under the same conditions in dhp1-1 cells containing the vector alone. These data indicate that the din1+ gene product can suppress the temperature-sensitivity of dhp1-1 by preventing the loss of Dhp1-1p. During the preparation of this manuscript, Rai1p was reported to interact with, and stabilize the exoribonuclease activity of, Rat1p in S.cerevisiae (50), a result that is consistent with our observations in S.pombe.
Figure 6.
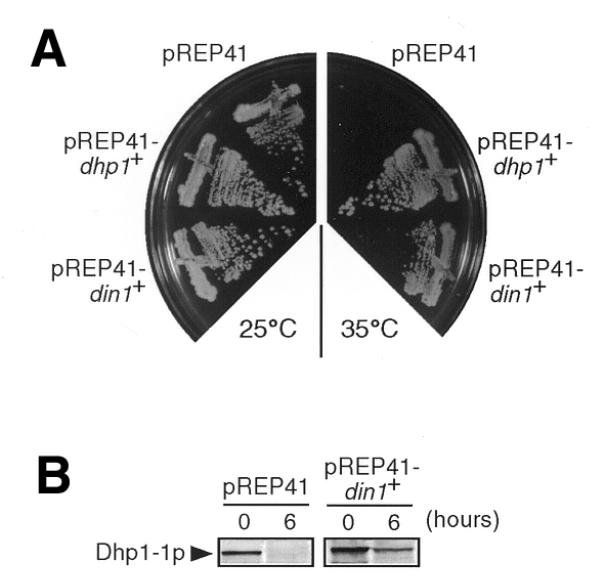
Suppression of the dhp1-1 mutation by the din1+ gene. (A) Abrogation of the temperature-sensitivity of dhp1-1 by din1+. The dhp1-1 mutant (MP102) harboring pREP41, pREP41-dhp1+ or pREP41-din1+ was streaked onto MM (+Ade) plates and incubated at 25°C for 5 days or at 35°C for 2 days. (B) Rescue of the reduction in the Dhp1-1p level by Din1p overproduction at the restrictive temperature. The dhp1-1 cells harboring pREP41 or pREP41-din1+ were cultured in MM (+Ade) at 36°C. Soluble cell lysates prepared from aliquots were separated by 8.3% SDS–PAGE and subjected to western blotting with anti-Dhp1p antiserum.
To investigate the function of the din1+ gene, we performed a gene disruption experiment of din1+ and successfully obtained haploid cells lacking the din1+ gene. The Δdin1 mutant is viable even at the high temperature of 35°C (data not shown), indicating that the din1+ gene is not essential for cell growth.
DISCUSSION
In the present study, we have characterized the fission yeast dhp1+ gene, which encodes a putative exoribonuclease. Dhp1p has sequence similarity to Rat1p, and both of them are essential for cell viability (1,4,6,20). Since expression of dhp1+ rescued the temperature-sensitivity of a rat1 mutant, dhp1+ is not only structurally, but also functionally homologous to RAT1 (20). Rat1p is a nuclear protein that has 5′→3′ exoribonuclease activity (1). A series of analyses of rat1 mutants revealed that Rat1p is involved in RNA trafficking and processing (4,17,27,28). The functional complementation of the rat1 mutant by dhp1+ expression indicates that Dhp1p is a putative 5′→3′ exoribonuclease. We previously identified a mouse ortholog of dhp1+/RAT1, termed Dhm1, and showed that expression of the Dhm1 cDNA in fission yeast could suppress the temperature-sensitivity of the dhp1-1 allele as well as the lethality of the Δdhp1 allele (23). These data suggest that the functions of exoribonucleases of Rat1p/Dhp1p members are well conserved throughout eukaryotic evolution.
We found that Dhp1p, like Rat1p, is required for a certain type of RNA metabolism. Poly(A)+ RNA accumulated in the nucleus of dhp1-1 mutant at the restrictive temperature; this is also a characteristic of the budding yeast rat1-1 mutant (4). This result is consistent with the idea that Dhp1p is a functional homolog of Rat1p and that it possesses 5′→3′ exoribonuclease activity and suggests that Dhp1p and Rat1p are required for the nuclear export of mRNA. However, another possibility is that the accumulated poly(A)+ RNA corresponds to non-mRNA species that were aberrantly produced owing to defective 5′→3′ exoribonuclease activity. Recently, it was reported that aberrantly polyadenylated snoRNAs accumulate in budding yeast exosome mutants (41) that were originally isolated as mutants defective in mRNA export (33). The exosome was identified as a protein complex with 3′→5′ exoribonuclease activity required for RNA processing in budding yeast (30,31). The exoribonuclease activity of Rat1p is required for the processing of a variety of RNA species, suggesting that this may also be the case for Dhp1p. In support of the latter possibility, the cytoplasmic signal for poly(A)+ RNA appeared not to be significantly reduced, irrespective of the level of nuclear accumulation of poly(A)+ RNA in the dhp1-1 mutant. Further research to identify the nature of the accumulated RNA species in the nucleus has to be done to address this issue in more detail.
Characterization of the dhp1-1 mutant revealed involvement of the dhp1+ gene in chromosome segregation. At the restrictive temperature, aberrant segregation of chromosomes was observed in the dhp1-1 mutant but this has not been reported in rat1 mutants. Some cells in the mutant cell population contained condensed chromosomes, which frequently consisted of three blocks of each chromosome that were pulled by the mitotic spindle. However, the sister chromatids appeared not to be fully separated, resulting in unequal or defective chromosome segregation. Analyses of chromosomal behavior using the Cen1–GFP system confirmed that dhp1-1 was unable to separate sister chromatids efficiently. In addition, similar mitotic defects were observed in the dhp1 null mutant, demonstrating that these phenotypes are not specific to the dhp1-1 mutant. Thus, we conclude that dhp1+ is required for the normal execution of chromosome segregation either directly or indirectly.
The chromosomal morphology of dhp1-1 resembles that of dis mutants, which were isolated as being defective in the separation of the sister chromatids (43). They fell into at least three complementation groups, and the corresponding genes were determined. The dis1+ gene encodes a microtubule-associating protein (53), while dis2+ is a type I protein phosphatase (54,55). Interestingly, dis3+ encodes a fission yeast ortholog of a component of the exosome (30,31,42). Although exoribonuclease activity of Dis3p or Dhp1p has not been experimentally determined, processing of some RNA species may be directly involved in the proper execution of chromosome segregation. However, it still remains unclear whether particular RNA species, and not just proteins, have a role to play in chromosome segregation. As mentioned above, it is possible that mRNA export might be defective in the dhp1-1 mutant. Thus, defective chromosome segregation might be a secondary effect of defective mRNA export, which could lead to the depletion of proteins involved in the mitotic apparatus. In addition, it is also possible that Dhp1p may be directly involved in chromosome segregation by its distinct activity from that of exoribonuclease. Further study will be needed to elucidate the role of Dhp1p in chromosome segregation.
Finally, we isolated a novel gene, which genetically interacts with dhp1+ in fission yeast. It was isolated as a multicopy suppressor of dhp1-1 and named din1+. Overproduction of Din1p rescued the temperature-sensitivity and defective chromosome segregation phenotype of dhp1-1, and prevented the reduction in the amount of the mutant Dhp1-1 protein. However, the lethality of the dhp1 deletion mutant was not rescued by overproducing Din1p (data not shown). These results indicate that Din1p overexpression rescued the temperature-sensitivity of dhp1-1 by maintaining nearly normal levels of mutant Dhp1-1p in the cells. Din1p is likely to interact with, and stabilize, Dhp1p although one cannot rule out the possibility that overproduction of Din1p affects the level of dhp1-1 mRNA or efficiency of Dhp1-1p translation. The C-terminal domain of Dhp1p may be required for efficient interaction with Din1p. The idea that Din1p physically interacts with Dhp1p is supported by very recent work in S.cerevisiae. Rai1p, an ortholog of Din1p, was found to bind to Rat1p, an ortholog of Dhp1p (50). Gene disruption of din1+ revealed that, unlike dhp1+, it is not essential for cell viability, indicating that din1+ is dispensable for the essential function of dhp1+. The rai1 null mutant is also viable in S.cerevisiae, although the null mutation allele of rai1 confers slow growth on the cells. These data suggest that Din1p may be an auxiliary protein that stabilizes Dhp1p and/or facilitates its activity. Furthermore, the roles of Din1p may be compensated by other proteins that are functionally redundant. It is possible that Dhp1p may form an exosome-like complex with Din1p and other proteins. This idea would gain further credence if other factors that interact with dhp1+ were identified.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr M.Yanagida for the S.pombe strain and to Drs Y.Watanabe and T.Takeda for the S.pombe genomic and cDNA libraries. The work was supported by Grants-in-Aid for Scientific Research (B) and Scientific Research on Priority Areas (B) to H.I. from the Ministry of Education, Science, Sports and Culture of Japan.
DDBJ/EMBL/GenBank accession no. AB045607
References
- 1.Kenna M., Stevens,A., McCammon,M. and Douglas,M.G. (1993) An essential yeast gene with homology to the exonuclease-encoding XRN1/KEM1 gene also encodes a protein with exoribonuclease activity. Mol. Cell. Biol., 13, 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens A. (1980) Purification and characterization of a Saccharomyces cerevisiae exoribonuclease which yields 5′-mononucleotides by a 5′→3′ mode of hydrolysis. J. Biol. Chem., 255, 3080–3085. [PubMed] [Google Scholar]
- 3.Stevens A. and Poole,T.L. (1995) 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J. Biol. Chem., 270, 16063–16069. [DOI] [PubMed] [Google Scholar]
- 4.Amberg D.C., Goldstein,A.L. and Cole,C.N. (1992) Isolation and characterization of RAT1: an essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes Dev., 6, 1173–1189. [DOI] [PubMed] [Google Scholar]
- 5.Aldrich T.L., Di Segni,G., McConaughy,B.L., Keen,N.J., Whelen,S. and Hall,B.D. (1993) Structure of the yeast TAP1 protein: dependence of transcription activation on the DNA context of the target gene. Mol. Cell. Biol., 13, 3434–3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Di Segni G., McConaughy,B.L., Shapiro,R.A., Aldrich,T.L. and Hall,B.D. (1993) TAP1, a yeast gene that activates the expression of a tRNA gene with a defective internal promoter. Mol. Cell. Biol., 13, 3424–3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larimer F.W. and Stevens,A. (1990) Disruption of the gene XRN1, coding for a 5′→3′ exoribonuclease, restricts yeast cell growth. Gene, 95, 85–90. [DOI] [PubMed] [Google Scholar]
- 8.Dykstra C.C., Kitada,K., Clark,A.B., Hamatake,R.K. and Sugino,A. (1991) Cloning and characterization of DST2, the gene for DNA strand transfer protein β from Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 2583–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson A.W. and Kolodner,R.D. (1991) Strand exchange protein 1 from Saccharomyces cerevisiae. A novel multifunctional protein that contains DNA strand exchange and exonuclease activities. J. Biol. Chem., 266, 14046–14054. [PubMed] [Google Scholar]
- 10.Kim J., Ljungdahl,P.O. and Fink,G.R. (1990) kem mutations affect nuclear fusion in Saccharomyces cerevisiae. Genetics, 126, 799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Z. and Gilbert,W. (1994) The yeast KEM1 gene encodes a nuclease specific for G4 tetraplex DNA: implication of in vivo functions for this novel DNA structure. Cell, 77, 1083–1092. [DOI] [PubMed] [Google Scholar]
- 12.Kipling D., Tambini,C. and Kearsey,S.E. (1991) rar mutations which increase artificial chromosome stability in Saccharomyces cerevisiae identify transcription and recombination proteins. Nucleic Acids Res., 19, 1385–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heyer W.D., Johnson,A.W., Reinhart,U. and Kolodner,R.D. (1995) Regulation and intracellular localization of Saccharomyces cerevisiae strand exchange protein 1 (Sep1/Xrn1/Kem1), a multifunctional exonuclease. Mol. Cell. Biol., 15, 2728–2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu C.L. and Stevens,A. (1993) Yeast cells lacking 5′→3′ exoribonuclease 1 contain mRNA species that are poly(A) deficient and partially lack the 5′ cap structure. Mol. Cell. Biol., 13, 4826–4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muhlrad D., Decker,C.J. and Parker,R. (1994) Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes Dev., 8, 855–866. [DOI] [PubMed] [Google Scholar]
- 16.Muhlrad D., Decker,C.J. and Parker,R. (1995) Turnover mechanisms of the stable yeast PGK1 mRNA. Mol. Cell. Biol., 15, 2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henry Y., Wood,H., Morrissey,J.P., Petfalski,E., Kearsey,S. and Tollervey,D. (1994) The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J., 13, 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stevens A., Hsu,C.L., Isham,K.R. and Larimer,F.W. (1991) Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J. Bacteriol ., 173, 7024–7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson A.W. (1997) Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol., 17, 6122–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugano S., Shobuike,T., Takeda,T., Sugino,A. and Ikeda,H. (1994) Molecular analysis of the dhp1+ gene of Schizosaccharomyces pombe: an essential gene that has homology to the DST2 and RAT1 genes of Saccharomyces cerevisiae. Mol. Gen. Genet., 243, 1–8. [DOI] [PubMed] [Google Scholar]
- 21.Szankasi P. and Smith,G.R. (1996) Requirement of S. pombe exonuclease II, a homologue of S. cerevisiae Sep1, for normal mitotic growth and viability. Curr. Genet., 30, 284–293. [DOI] [PubMed] [Google Scholar]
- 22.Bashkirov V.I., Scherthan,H., Solinger,J.A., Buerstedde,J.M. and Heyer,W.D. (1997) A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol., 136, 761–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shobuike T., Sugano,S., Yamashita,T. and Ikeda,H. (1995) Characterization of cDNA encoding mouse homolog of fission yeast dhp1+ gene: structural and functional conservation. Nucleic Acids Res., 23, 357–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shobuike T., Sugano,S., Yamashita,T. and Ikeda,H. (1997) Cloning and characterization of mouse Dhm2 cDNA, a functional homolog of budding yeast SEP1. Gene, 191, 161–166. [DOI] [PubMed] [Google Scholar]
- 25.Sato Y., Shimamoto,A., Shobuike,T., Sugimoto,M., Ikeda,H., Kuroda,S. and Furuichi,Y. (1998) Cloning and characterization of human Sep1 (hSEP1) gene and cytoplasmic localization of its product. DNA Res., 5, 241–246. [DOI] [PubMed] [Google Scholar]
- 26.Zhang M., Yu,L., Xin,Y., Hu,P., Fu,Q., Yu,C. and Zhao,S. (1999) Cloning and mapping of the XRN2 gene to human chromosome 20p11.1-p11.2. Genomics, 59, 252–254. [DOI] [PubMed] [Google Scholar]
- 27.Petfalski E., Dandekar,T., Henry,Y. and Tollervey,D. (1998) Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol., 18, 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qu L.H., Henras,A., Lu,Y.J., Zhou,H., Zhou,W.X., Zhu,Y.Q., Zhao,J., Henry,Y., Caizergues-Ferrer,M. and Bachellerie,J.P. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villa T., Ceradini,F., Presutti,C. and Bozzoni,I. (1998) Processing of the intron-encoded U18 small nucleolar RNA in the yeast Saccharomyces cerevisiae relies on both exo- and endonucleolytic activities. Mol. Cell. Biol., 18, 3376–3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allmang C., Petfalski,E., Podtelejnikov,A., Mann,M., Tollervey,D. and Mitchell,P. (1999) The yeast exosome and human PM-Scl are related complexes of 3′→5′ exonucleases. Genes Dev., 13, 2148–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitchell P., Petfalski,E., Shevchenko,A., Mann,M. and Tollervey,D. (1997) The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′→5′ exoribonucleases. Cell, 91, 457–466. [DOI] [PubMed] [Google Scholar]
- 32.van Hoof A. and Parker,R. (1999) The exosome: a proteasome for RNA? Cell, 99, 347–350. [DOI] [PubMed] [Google Scholar]
- 33.Kadowaki T., Chen,S., Hitomi,M., Jacobs,E., Kumagai,C., Liang,S., Schneiter,R., Singleton,D., Wisniewska,J. and Tartakoff,A.M. (1994) Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J. Cell Biol., 126, 649–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Allmang C., Mitchell,P., Petfalski,E. and Tollervey,D. (2000) Degradation of ribosomal RNA precursors by the exosome. Nucleic Acids Res., 28, 1684–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Allmang C., Kufel,J., Chanfreau,G., Mitchell,P., Petfalski,E. and Tollervey,D. (1999) Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J., 18, 5399–5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Briggs M.W., Burkard,K.T. and Butler,J.S. (1998) Rrp6p, the yeast homologue of the human PM-Scl 100-kDa autoantigen, is essential for efficient 5.8 S rRNA 3′ end formation. J. Biol. Chem., 273, 13255–13263. [DOI] [PubMed] [Google Scholar]
- 37.de la Cruz J., Kressler,D., Tollervey,D. and Linder,P. (1998) Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J., 17, 1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell P., Petfalski,E. and Tollervey,D. (1996) The 3′ end of yeast 5.8S rRNA is generated by an exonuclease processing mechanism. Genes Dev., 10, 502–513. [DOI] [PubMed] [Google Scholar]
- 39.Zanchin N.I. and Goldfarb,D.S. (1999) The exosome subunit Rrp43p is required for the efficient maturation of 5.8S, 18S and 25S rRNA. Nucleic Acids Res., 27, 1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson J.S.J. and Parker,R. (1998) The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J., 17, 1497–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van Hoof A., Lennertz,P. and Parker,R. (2000) Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol., 20, 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kinoshita N., Goebl,M. and Yanagida,M. (1991) The fission yeast dis3+ gene encodes a 110-kDa essential protein implicated in mitotic control. Mol. Cell. Biol., 11, 5839–5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohkura H., Adachi,Y., Kinoshita,N., Niwa,O., Toda,T. and Yanagida,M. (1988) Cold-sensitive and caffeine-supersensitive mutants of the Schizosaccharomyces pombe dis genes implicated in sister chromatid separation during mitosis. EMBO J., 7, 1465–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alfa C., Fantes,P., Hyams,J., Mcleod,M. and Warbrick,E. (1993) Experiment with Fission Yeast: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Basi G., Schmid,E. and Maundrell,K. (1993) TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene, 123, 131–136. [DOI] [PubMed] [Google Scholar]
- 46.Beach D., Piper,M. and Nurse,P. (1982) Construction of a Schizosaccharomyces pombe gene bank in a yeast bacterial shuttle vector and its use to isolate genes by complementation. Mol. Gen. Genet., 187, 326–329. [DOI] [PubMed] [Google Scholar]
- 47.Nabeshima K., Nakagawa,T., Straight,A.F., Murray,A., Chikashige,Y., Yamashita,Y.M., Hiraoka,Y. and Yanagida,M. (1998) Dynamics of centromeres during metaphase–anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol. Biol. Cell, 9, 3211–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatebayashi K., Kato,J. and Ikeda,H. (1998) Isolation of a Schizosaccharomyces pombe rad21ts mutant that is aberrant in chromosome segregation, microtubule function, DNA repair and sensitive to hydroxyurea: possible involvement of Rad21 in ubiquitin-mediated proteolysis. Genetics, 148, 49–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Azad A.K., Tani,T., Shiki,N., Tsuneyoshi,S., Urushiyama,S. and Ohshima,Y. (1997) Isolation and molecular characterization of mRNA transport mutants in Schizosaccharomyces pombe. Mol. Biol. Cell, 8, 825–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xue Y., Bai,X., Lee,I., Kallstrom,G., Ho,J., Brown,J., Stevens,A. and Johnson,A.W. (2000) Saccharomyces cerevisiae RAI1 (YGL246c) is homologous to human DOM3Z and encodes a protein that binds the nuclear exoribonuclease Rat1p. Mol. Cell. Biol., 20, 4006–4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang Z., Shen,L., Dangel,A.W., Wu,L.C. and Yu,C.Y. (1998) Four ubiquitously expressed genes, RD (D6S45)-SKI2W (SKIV2L)-DOM3Z-RP1 (D6S60E), are present between complement component genes factor B and C4 in the class III region of the HLA. Genomics, 53, 338–347. [DOI] [PubMed] [Google Scholar]
- 52.Paulsen J.E., Capowski,E.E. and Strome,S. (1995) Phenotypic and molecular analysis of mes-3, a maternal-effect gene required for proliferation and viability of the germ line in C. elegans. Genetics, 141, 1383–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nabeshima K., Kurooka,H., Takeuchi,M., Kinoshita,K., Nakaseko,Y. and Yanagida,M. (1995) p93dis1, which is required for sister chromatid separation, is a novel microtubule and spindle pole body-associating protein phosphorylated at the Cdc2 target sites. Genes Dev., 9, 1572–1585. [DOI] [PubMed] [Google Scholar]
- 54.Kinoshita N., Ohkura,H. and Yanagida,M. (1990) Distinct, essential roles of type 1 and 2A protein phosphatases in the control of the fission yeast cell division cycle. Cell, 63, 405–415. [DOI] [PubMed] [Google Scholar]
- 55.Ohkura H., Kinoshita,N., Miyatani,S., Toda,T. and Yanagida,M. (1989) The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell, 57, 997–1007. [DOI] [PubMed] [Google Scholar]


