Abstract
Clathrin is a major vesicle coat protein involved in receptor-mediated endocytosis. In yeast and higher eukaryotes, clathrin is recruited to the plasma membrane during the early stage of endocytosis along with clathrin-associated adaptors. As coated pits undergo maturation, a burst of actin polymerization accompanies and helps drive vesicle internalization. Here, we investigate the dynamics of clathrin relative to the early endocytic patch protein Sla2p. We find that clathrin is recruited to the cortex prior to Sla2p. In the absence of clathrin, normal numbers of Sla2p patches form, but many do not internalize or are dramatically delayed in completion of endocytosis. Patches that do internalize receive Sla1p late, which is followed by Abp1, which appears near the end of Sla2p lifetime. In addition, clathrin mutants develop actin comet tails, suggesting an important function in actin patch organization/dynamics. Similar to its mammalian counterparts, the light chain (LC) subunit of yeast clathrin interacts directly with the coiled-coil domain of Sla2p. A mutant of Sla2p that no longer interacts with LC (sla2Δ376-573) results in delayed progression of endocytic patches and aberrant actin dynamics. These data demonstrate an important role for clathrin in organization and progression of early endocytic patches to the late stages of endocytosis.
Keywords: clathrin light chain, endocytosis, HIP1/R, Sla2p, TIRFM
Clathrin, a major vesicle coat protein, functions in receptor-mediated endocytosis (RME) as well as in sorting and retention of cargo at the trans-Golgi network (TGN) and endosomal system. Clathrin triskelions are recruited to membranes by cargo/lipid-binding adaptors, resulting in formation of the polygonal lattices of clathrin-coated pits (CCPs) and clathrin-coated vesicles (CCVs). Three clathrin heavy chains (HCs) comprise the structural backbone of the triskelion and are each bound by a clathrin light chain (LC). The HC has an N-terminal globular domain (HC-TD), distal leg, proximal leg that binds LC and a trimerization region near the C-terminus (1–3). The HC-TD is important for clathrin’s interactions with membrane-associated factors, such as the adaptors AP-1 and AP-2, AP180s, amphi-physins, epsins and others, involved in sorting into CCPs and CCV formation (3). These proteins all contain variations of the LLDLD-type clathrin-box motif or the (W box) clathrin-binding motif PWXXW, which mediate their interaction with the HC-TD (4).
Biochemical studies have suggested several direct roles for LC, including uncoating of CCVs, prevention of lattice assembly or trimerization of HCs (3,5–9). But the only in vivo evidence for a direct LC function comes from work in yeast, where the LC (Clc1p) is required for HC (Chc1p) trimerization and stability. Due to this requirement, the phenotypes of clc1Δ cells mimic what is observed in HC-deficient (chc1Δ) yeast (10,11). These phenotypes include defects in TGN/endosomal protein sorting/retention and RME (10–18), as well as increased cell size, polyploidy, temperature sensitive growth and aberrant actin organization (11,19–21). Deletion of the LC gene in Dictyostelium also causes severe phenotypes, affecting development, cytokinesis and osmoregulation (22). Surprisingly, siRNA directed against clathrin light chains A and B in Hela cells does not reduce EGF and transferrin receptor endocytosis, whereas a similar depletion of HC has a significant effect on RME (23). Possibly, LCs are less critical in some animal cells, because the stoichiometry of LC to HC in liver is fivefold less than that found in neuronal cells (24). Alternatively, the phenotypic differences between higher and lower eukaryotes upon depletion of clathrin LC could reflect the degree of effectiveness of gene knockouts versus siRNA techniques.
Aside from LC’s role in clathrin HC trimerization in vivo, another proposed function for clathrin LC comes from studies showing that LC interacts with mammalian Hip1 and Hip1R and its yeast homologue, Sla2p (19,25–27). This family of endocytic proteins all share an AP180 N-terminal homology (ANTH) domain found in proteins that interact with membrane inositol phospholipids, a C-terminal actin-binding talin-like domain and a central coiled-coil domain, which binds to clathrin LC (19,25–32). Like sla2Δ yeast, Hela cells subjected to Hip1R RNAi or neurons derived from Hip1 (−/−) mice display reduced endocytosis (33,34). Furthermore, Hip1R RNAi cultures accumulate arrested endocytic structures containing actin cytoskeletal proteins, clathrin, endocytic cargo and other endocytic factors (33). This has led to the conclusion that the animal Sla2p homologues regulate the interaction between actin and the clathrin endocytic machinery.
Recently, we demonstrated for the first time the existence of plasma membrane-associated clathrin in yeast (35), which was debated for many years because clathrin mutants only show partial endocytic phenotypes (36). We also found that recruitment of yeast clathrin to the cortex involves the epsin and AP180 homologues Ent1/2p and Yap1801/2p (35). As the later stages of endocytosis follow, clathrin disappears from the cell surface coinciding with a burst of actin polymerization, which presumably provides the force for vesicle fission and/or internalization (35). This sequence of assembly of clathrin and actin resembles what has been observed in animal cells (37,38) and was also reported for yeast in another recent study (39).
Other early endocytic factors present at the cortex include Sla2p, the SH3 domain protein Sla1p, an Eps15 homology domain protein Pan1p, and the N-Wasp homologue Las17p (40). These early proteins, including clathrin, become immobile at the cortex upon LAT-A treatment or in sla2Δ cells, demonstrating important roles for Sla2p and the actin cytoskeleton during endocytic patch progression and vesicle internalization (35,40). However, the relationship between clathrin and Sla2p during the maturation and progression of endocytic cortical patches remains unclear.
In this report, we show that the coiled-coil region of Sla2p interacts directly with LC, as seen for Hip1/R and LC in mammalian cells (25–27). By live cell imaging of cortical patch dynamics, we find clathrin is recruited prior to Sla2p, which is then followed by Sla1p, demonstrating there are at least three stages of early endocytic factor assembly. Furthermore, clathrin depletion dramatically affects the organization and turnover of endocytic patches containing Sla2p, Sla1p and Abp1p, having a distinct effect on the dynamics of each protein. Finally, removal of the Sla2p region that binds to LC results in delayed progression of endocytic cortical patches, suggesting this interaction is critical to promote efficient vesicle internalization.
Results
Clathrin LC mediates HC interaction with Sla2p
Previously a two-hybrid screen in our lab found that clathrin LC interacts with Sla2p (19). The two-hybrid analysis also showed an interaction between the central coiled-coil domain of Sla2p and a fragment containing the clathrin HC proximal arm (655–1653), although this interaction was weaker than the interaction seen between LC and Sla2p (Figure 1A, upper panel).
Figure 1. Clathrin LC is important for Sla2p–HC interaction, but clathrin HC is not required for Sla2p-LC binding.

(A) Wild-type (WT) (YPJ96-4), clc1Δ (SL4410) and chc1Δ (SL4409) two-hybrid reporter strains were transformed with GAL4-binding domain (GBD) bait plasmids pGBDU (GBD empty), pKH19 (GBD-CLC1) or pKH47 (GBD-sla2-292-501) in combination with GAL4 activation domain (GAD) prey plasmids pGAD (GAD empty), pKH24 (GAD-chc1-655-1653) or p31-2 (GAD-sla2-292-520). Equal numbers of cells from transformants were spotted on complete synthetic medium lacking leucine and uracil (C-LEU-URA) and medium lacking adenine, leucine and uracil (C-ADE-LEU-URA) and grown for 5 days to score for ADE2 reporter expression. (B) Bacterially purified GST fusions prebound to glutathione sepharose beads were incubated with yeast lysates from WT (SL1629) or clc1Δ (SL1628) to pull down clathrin HC. GST-Bmh1p was used as a control for nonspecific binding. (C) Same as (B), except GST fusions were incubated with yeast lysates from WT (SL1463 + pKH2) or chc1Δ (SL1927) to pull down clathrin LC. (B and C) Blots were probed with indicated antibodies. Ten microliters of cell lysates were run as input controls (lanes 1 and 6).
To determine whether Sla2p interacts directly with the HC or the LC, we generated deletions of CLC1 or CHC1 in the two-hybrid reporter strain so the interactions of baits and preys of Sla2p with HC (655-1653) or LC could be assayed in the absence of genomically encoded LC or HC, respectively. In the clc1Δ reporter strain, the interaction between HC and Sla2p (292-501) was ablated (Figure 1A, middle panel). This indicates that the Sla2p coiled-coil domain interaction with HC is probably mediated through LC. In contrast, the interaction between LC and Sla2p (292-520) still occurred in the absence of CHC1 (Figure 1A, lower panel). In addition, the known dimerization of Sla2p via the central coiled-coil domain (41,42) does not require the presence of either clathrin subunit (Figure 1A).
To further confirm that the HC interaction with the Sla2p coiled-coil domain is indirect, we incubated a bacterially purified GST-Sla2p fusion (289-583) with wild-type (WT), clc1Δ and chc1Δ yeast extracts. GST-Sla2p (289-583) was able to pull down clathrin HC from a WT yeast extract (Figure 1B, lane 5). However, GST-Sla2p (289-583) did not pull down HC in an extract derived from a clc1Δ strain (Figure 1B, lane 9), once again showing the dependence of the Sla2p coiled-coil/HC interaction on clathrin LC. GST-Clc1p could not pull down HC from a WT extract (Figure 1B, lane 4) but readily pulled down HC from a clc1Δ strain (Figure 1B, lane 8), showing that the HC in WT cells is prebound with LC. Consistent with the two-hybrid analysis, GST-Sla2p (289-583) associated with LC from a yeast extract in the presence or absence of HC (Figure 1C, lanes 5 and 9). Taken together, the two-hybrid and GST pull down data demonstrate a dependence on LC for an interaction between the Sla2p coiled-coil domain and the clathrin HC, while HC is not required for the interaction between Sla2p and LC.
Clathrin light chain and Sla2p directly interact through the Sla2p coiled-coil domain
The interaction of LC with the Sla2p coiled-coil domain in the two-hybrid analysis and pull down from yeast extracts could be indirect. Therefore, we tested for direct interactions by incubating a bacterially expressed 6xHis–Clc1p fusion with several bacterially expressed GST-Sla2p truncation and deletion fusions. GST-Sla2p (292-968) and GST-Sla2p (289-583) interacted with 6xHis-Clc1p (Figure 2A, lanes 4 and 5), showing that the smaller fragment containing the Sla2p coiled-coil domain is sufficient to bind LC. Next, to determine whether any region outside of the coiled-coil domain bound the LC, pull downs with a GST–Sla2p fusion deleting amino acids 376–573 was compared with a GST-Sla2p full-length fusion. The GST-Sla2p-Δ376–573 did not interact with 6xHis-Clc1p, unlike the full-length GST-Sla2p (Figure 2A, lanes 6 and 7). Therefore, Sla2p and LC interact directly, and the central region of Sla2p containing the coiled-coil domain is necessary and sufficient for binding to LC.
Figure 2. Clathrin LC and Sla2p interact directly in a region containing the coiled-coil domain of Sla2p, but the HC terminal domain does not interact with Sla2p.

(A) Bacterially expressed GST fusions prebound to glutathione sepharose beads were used to pull down bacterially expressed 6xHis-Clc1p. GST-Bmh1p was used as a control for nonspecific binding. Blots were probed with indicated antibodies. (B) Bacterially expressed GST fusions prebound to glutathione sepharose beads were incubated with WT (SL1629) yeast extract to pull down clathrin HC. (C) YPJ96-4 was transformed with GBD bait plasmids pTMN9 (GBD-SLA2), pKH49 (GBD-sla2-292-968), pKH47 (GBD-sla2-292-501), pTMN38 (GBD-ENT1) or pGBDU (GBD empty) in combination with GAD prey plasmids pGAD (GAD empty) and pTMN8 (GAD-chc1-1-500). Equal numbers of cells from transformants were spotted on complete synthetic medium lacking leucine and uracil (C-LEU-URA) and medium lacking adenine, leucine and uracil (C-ADE-LEU-URA) and grown for 3 days. (D) Coomassie Blue-stained gel of fractions (31–47) from a Sephacryl S-1000 column derived from CCV purification of WT (SL1463) yeast. (E) Western blots of yeast extracts from WT + pSLA2 2μ (SL1463 + YEplac195-SLA2), WT (SL1463) and sla2Δ (HR1965) and fraction number 35 from S-1000 CCV preparation shown in D concentrated 50-fold. Blots were probed with αSla2p and αClc1p antibodies.
Clathrin HC terminal domain does not bind Sla2p
Many proteins containing the clathrin-box motif related to LLDLD are known to interact with the HC-TD. Hip1, but not Hip1R, interacts with mammalian HC via this type of motif (25,43–45). Two related sequences exist in Sla2p [amino acids 152–156 (ILDLM) and 573–577 (ILDAI)]. To examine whether Sla2p lacking the LC-binding region interacts with the HC-TD, we tested whether GST-Sla2p-Δ376–573 was capable of interacting with HC from a yeast extract. As a positive control, GST-LLDLD was used to pull down HC from a WT extract (Figure 2B, lane 4). The Sla2p coiled-coil domain deletion construct interacted very weakly with clathrin HC in the presence (Figure 2B, lane 5) or absence of endogenous LC (data not shown). This weak interaction left open the possibility of other indirect interactions between Sla2p and HC or that Sla2p binds to HC-TD through the LLDLD-like motifs or other sequences.
To test whether the HC-TD interacts with Sla2p, we tested two-hybrid analysis of Sla2p (292-501), Sla2p (292-968) and full length Sla2p baits with a HC-TD (Chc1p-1-500) prey. None of the Sla2p baits interacted with the TD prey; however, an Ent1p bait, which contains the C-terminal clathrin box motif LIDL* did bind the TD, verifying that the HC-TD prey was functional (Figure 2C). Because neither the potential clathrin-box motifs nor other Sla2p sequences interact with the HC-TD using two-hybrid analysis, the weak HC interaction with Sla2p seen in Figure 2B may occur indirectly through proteins other than the LC.
Sla2p is not associated with CCVs
Because Hip1/R are associated with mammalian CCVs (25,43,45), we examined whether Sla2p is a stable component of yeast CCVs. A 100 000 × g pellet was generated from cell lysates by differential centrifugation and fractionated over a Sephacryl S-1000 column (46); however, Sla2p could not be detected in CCV fractions, even after peak clathrin-containing fractions were concentrated 50-fold (Figure 2D,E). Therefore, Sla2p is not a stable component of yeast CCVs or endocytic CCVs are short-lived due to rapid uncoating. Alternatively, Sla2p association with clathrin LC may only occur prior to vesicle budding.
Clathrin is recruited to cortical patches prior to Sla2p
Previous studies showed that Sla2p or clathrin are recruited to the cortex during endocytosis prior to the burst of Abp1p/actin accumulation, indicating that they are both early endocytic patch components (35,40). To determine whether clathrin and Sla2p share similar cortical lifetimes, we co-expressed GFP-LC and Sla2p-mRFP in cells. Using wide-field fluorescence microscopy, we found that patches of cortical clathrin and Sla2p only partially overlapped, similar to what we saw previously with Sla2p-GFP with HC-mRFP (35) (Figure 3A). To be sure, we were selectively observing cortical clathrin, and that the signal was not from internal patches of clathrin on Golgi/endosomal structures, we used total internal reflection fluorescence microscopy (TIRFM) (35). Again, we saw some clathrin patches overlapping with, while others were independent of, Sla2p (Figure 3B). Time-lapse images from the TIRF microscopy found that 90% of Sla2p patches are recruited to sites already containing clathrin (n = 38). Therefore, most of the Sla2p-independent clathrin patches represent early clathrin patches that have not yet received Sla2p. The clathrin patches ranged in lifetime from 30 seconds to several minutes with an average lifetime of 53 ± 16 seconds, whereas Sla2p patches had shorter lifetimes of 29 ± 12 seconds (Figure 3D,E; n = 34 patch events). On average, Sla2p was recruited 25 ± 16 seconds after the initial recruitment of clathrin, while both proteins disappeared from the cortex simultaneously (Figure 3D,E).
Figure 3. Clathrin is recruited prior to Sla2p during endocytic patch formation in wild-type cells.
GFP-CLC1 SLA2-mRFP strain SL5121 was viewed by wide-field (A) and TIRF microscopy (B). Note many cortical patches of clathrin do not contain Sla2p, consistent with earlier recruitment of clathrin. (C) Time-lapse video of a representative cortical clathrin patch at 3-second intervals visualized using wide-field imaging. The bottom line is a reference for the starting point of the clathrin patch when it is still at the cortex. The clathrin patch moves upwards in the video approximately 200 nm, illustrating its movement away from the cell surface. (D) Time-lapse videos of patches with GFP-LC and Sla2p-mRFP shown at 6.9-second intervals using TIRFM. Note clathrin recruitment prior to Sla2p and simultaneous disappearance of Sla2p and clathrin. In videos with longer Sla2p/clathrin lifetimes, fluctuations in signal intensities were observed, possibly due to photobleaching. The video in the top panel in (D) was started after clathrin already appeared. Clathrin patch lifetimes cannot be calculated from such examples, but these events clearly demonstrate earlier recruitment of clathrin. (E) Bar graph showing the average patch lifetimes of Sla2p-mRFP and GFP-LC using TIRFM. Lifetimes of Sla2p and LC were significantly different (p< 0.001). Scale bar = 5 μm. Time scales (C & D) are in seconds.
The lifetime of Sla2p patches observed using TIRFM was shorter than originally reported using wide-field microscopy (40), because Sla2p would not be visible as it moved 200 nm away from the cell surface. TIRFM should only penetrate approximately 60 nm deep into the cell, assuming an average cell wall thickness of approximately 90 nm (47). Using wide-field microscopy, we found that clathrin also undergoes approximately 200 nm of inward movement from the cortex just before its disappearance (Figure 3C; n = 12 patch events), like Sla1p, Pan1p and Sla2p (40). These data show that clathrin is an early endo-cytic patch protein, which is recruited to the cortex prior to Sla2p, and disappears from the cortex simultaneously with Sla2p after 200 nm of inward movement.
Clathrin is required for normal progression of Sla2p patches and cortical actin dynamics
As clathrin is recruited prior to Sla2p, we examined whether the absence of HC or LC would affect Sla2p-GFP recruitment and/or dynamics using wide-field microscopy. As a marker for cortical actin, we examined Abp1-mRFP. As described by Kaksonen et al. (40), in WT cells Sla2p was recruited to the cortex prior to Abp1p, and as the Sla2p signal declined, the Abp1p signal intensified and disappeared soon after (Supplemental Movie S1). Sla2p patches in our WT cells had a lifetime of 38 ± 9 seconds by wide-field imaging (as compared with 29 seconds by TIRFM) and temporally overlapped with Abp1 patches, which displayed lifetimes of 13 ± 3 seconds (Figure 4A1,B1; n = 42 events).
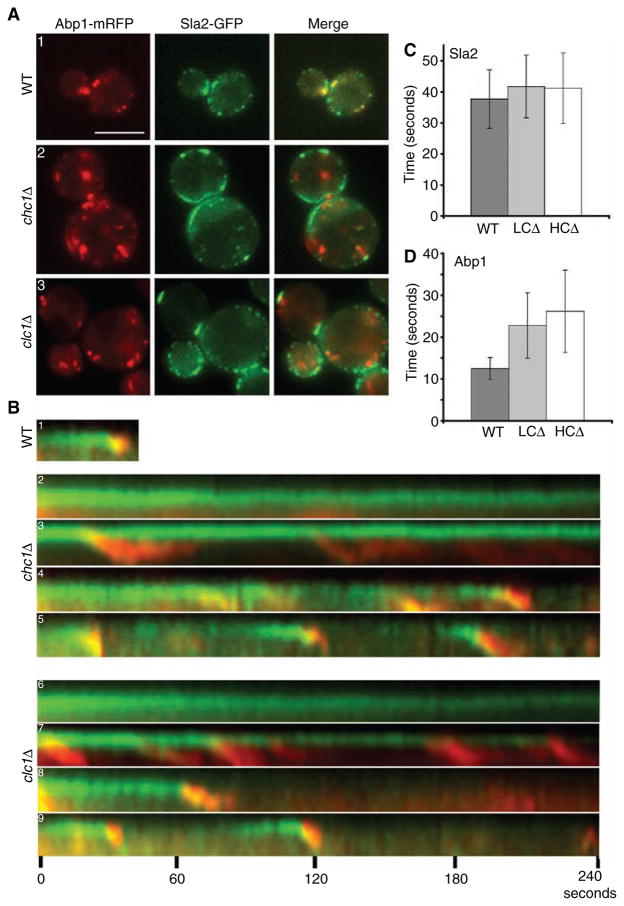
Figure 4. Clathrin is required for progression of Sla2p patches and normal actin dynamics.
(A1–A3). WT ABP1-mRFP SLA2-GFP (SL5185), chc1Δ ABP1-mRFP SLA2-GFP (SL5240) and clc1Δ ABP1-mRFP SLA2-GFP (SL5226) cells were viewed using wide-field microscopy. (B1–B9) Kymographs generated from single pixel wide lines placed through cortical patches from cells described in (A). Images were collected at two-second intervals and viewed up to 4 min. Note different types of patch dynamics in clathrin mutants: (B2 and B6) immobile Sla2p patches without actin assembly; (B3 and B7) Sla2p patches with actin comet tails and repeated patch internalization from the same site; (B4 and B8) Sla2p and Abp1p patches with extended lifetimes that do mature and internalize; (B5 and B9) relatively normal Sla2p lifetimes with extended Abp1p lifetimes. (C and D) Bar graphs showing average patch lifetimes of Sla2p-GFP and Abp1p-mRFP in WT and clathrin mutants for relatively normal internalization events (n = 30). (C) Patch lifetimes of Sla2p in WT and clathrin mutants (B5 and B9) were not significantly different (p > 0.05). (D) Patch lifetimes of Abp1p were slowed almost 2-fold in clathrin mutants (p < 0.001) (B5 and B9) Scale bar = 5 μm.
To our surprise, and in contrast to results we published previously (19), we found strong cortical recruitment of Sla2p-GFP in clathrin HC (chc1Δ) and LC (clc1Δ) mutant cells (Figure 4A). We suspect we did not observe cortical Sla2p in clathrin HC-depleted cells previously because of technical problems with the immunofluorescence techniques and antibody reagents available at the time. Clathrin mutants have a severely thickened cell wall, and fixation and penetration of antibodies may have been inefficient.
We observed at least four different types of Sla2p patch structures and dynamics in chc1Δ and clc1Δ cells by wide-field live cell imaging (Figure 4A2,A3,4B). The first class had immobile Sla2p patches that were usually large and/or clustered in zones along the cell surface. These Sla2p patches showed highly elongated lifetimes extending 4 min or longer (Figure 4B2,B6). In addition, the patches never received cortical actin during the 4-min time course, demonstrating a complete failure in recruitment of cortical actin. The second class of Sla2p patches seen in clathrin mutants were also large and immobile but developed long actin-containing structures, resembling actin comet tails (Figure 4B3,B7). These large immobile Sla2p patches randomly released smaller Sla2p patches at their tips in intervals of approximately 20 seconds to several minutes (Supplemental Movie S2). As each smaller Sla2p patch budded off, it was accompanied by a cortical actin patch, which eventually elongated into an actin comet tail, waving into the cytosol for up to 1 min. The actin comet tail often appeared to hold onto the smaller Sla2p patch up to 1500 nm away from the cortex until they both dissipated. The actin comet tail structures formed by Abp1-mRFP were also confirmed using Alexa-phalloidin staining (data not shown). The third class seen in clathrin mutants had Sla2p patches that were more normal in size and that completely budded off from the cortex along with cortical actin (Figure 4B4,B8). However, these Sla2p patches persisted at the cortex for about 1–4 min before recruitment of Abp1. Finally, in the fourth class, Sla2p patches were similar to those seen in WT cells (Figure 4B5,B9). The Sla2p signal had an average lifetime of about 41 ± 10 seconds (chc1Δ and clc1Δ cells), although the actin/Abp1 fluorescence persisted for 26 ± 10 seconds in chc1Δ and 23 ± 8 seconds in clc1Δ (Figure 4C,D; n = 30 each), almost twice as long as seen in WT cells. All four classes of Sla2p patch events were well represented in the cell population, and no differences were observed between chc1Δ and clc1Δ cells in the frequency of these events. These data demonstrate an important role for clathrin in the organization and normal turnover of endocytic cortical patches.
Actin comet tails and cortical patches in clathrin mutants contain endocytic membrane
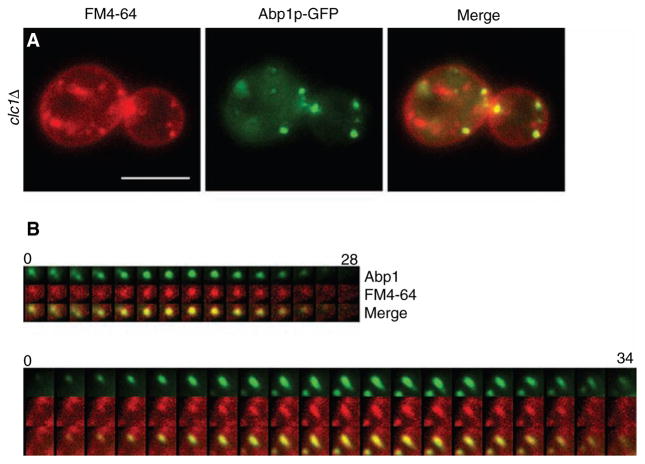
The aberrant actin structures observed in the clathrin mutants could represent completely failed or elongated endocytic events. To test whether the clathrin mutants internalized membrane into these actin structures, clc1Δ cells were stained with the lipophillic dye FM4-64 for 2 min, washed and resuspended in fresh medium and viewed after 20 min incubation. The wash helped deplete the strong band of FM4-64 from the plasma membrane, allowing better visualization of the dye at de novo cortical patches. Not only did clc1Δ cells take up the FM4-64 into newly forming cortical patches (Figure 5A,B; top panel), FM4-64 was also present in actin comet tails (Figure 5B; bottom panel). These data show that clathrin mutants do internalize plasma membrane at cortical actin patches and both normal looking and aberrant actin structures contain invaginating membrane.
Figure 5. Actin comet tails and patches in clathrin mutants contain endocytic membrane.
(A) clc1Δ ABP1-GFP (SL5101) cells stained with 40 μM FM4-64 and incubated for 20 min before viewing using wide-field microscopy. Note patches of Abp1p associated with FM4-64 seen as yellow in the merge. (B) Two examples of time-lapse videos showing concentration of FM4-64 into de novo actin patches (top panel) and actin comet tails (bottom panel) from clc1Δ cells. Note first three frames in the top left corner of B), top panel show a residual disappearing Abp1p patch different than the de novo patch that appears in the middle of frame number 3. Scale bar = 5 μm. Time scales in (B) are in seconds.
Sla1p, but not Sla2p, shows reduced cortical recruitment and patch lifetime in clathrin mutants
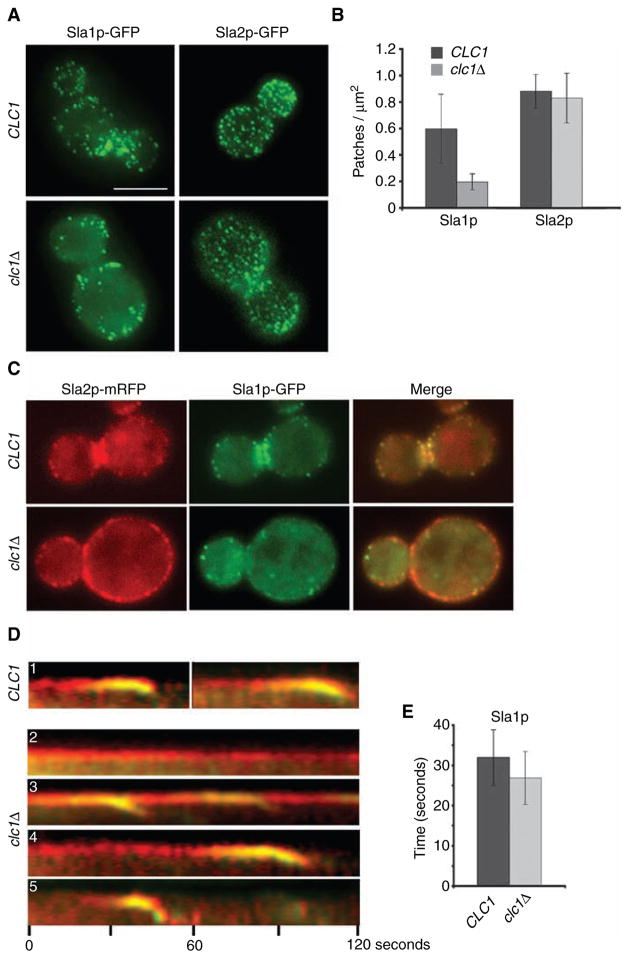
A recent report showed that clathrin mutants have a reduced number of Sla1p and Las17p patches per cell (39). To determine whether there is a similar effect on Sla2p, the number of Sla2p and Sla1p patches/μm2 were compared in our WT and clc1Δ strains. Similar to the 69% reduction reported by Kaksonen et al. (39), Sla1p patches decreased from 0.6 ± 0.26 patches/μm2 in WT to 0.2 ± 0.06 patches/μm2 in our clc1Δ cells (Figure 6A–B). In contrast, the number of Sla2p patches did not significantly change between WT (0.88 ± 0.12) and clc1Δ (0.83 ± 0.19) (Figure 6A–B).
Figure 6. Sla1p, but not Sla2p, shows decreased patch lifetime and cortical recruitment in clathrin mutants.
(A) WT SLA1-GFP (SL5318), clc1Δ SLA1-GFP (SL5317), WT SLA2-GFP (SL4781) and clc1Δ SLA2-GFP (SL4782) were viewed using wide-field microscopy. Z stacks were collected at an interval of 0.25 μm, and the maximum intensity projection is shown in all panels, except for clc1Δ SLA2-GFP, as the presence of large Sla2p-GFP patches led to the exclusion of weak intensity Sla2p-GFP patches in the projection image. In this case, a single plane from the z-axis focused on the cell surface is shown. (B) Quantification of Sla1p-GFP and Sla2p-GFP patches/μm2 in WT and clc1Δ cells from (A). Note the difference between WT and clc1Δ for Sla1p-GFP (p < 0.001) but not Sla2p-GFP (p > 0.2) (n ≈ 17 cells per strain). (C–D) WT SLA1-GFP SLA2-mRFP (SL5318) and clc1Δ SLA1-GFP SLA2-mRFP (SL5317) were viewed using wide-field microscopy, and time-lapse images were collected every 2 seconds. Note different types of patch dynamics in clathrin mutants: (D1) Sla2p and Sla1p in WT cells; (D2) immobile Sla2p patches without Sla1p; (D3) Sla2p patches recruiting Sla1p and repeated patch internalization from the same site; (D4) Sla2p patches with extended lifetimes that acquire Sla1p and internalize and (D5) relatively normal Sla2p lifetime with shortened Sla1p lifetime. (E) Bar graph showing difference in patch lifetimes of Sla1p-GFP between WT (SL5318, n = 30) and clc1Δ (SL5317, n = 37) (p < 0.005). Scale bar = 5 μm.
The greater number of Sla2p than Sla1p patches per cell in the clathrin mutant suggested that many Sla2p patches would not contain Sla1p. To confirm this, we co-expressed Sla2p-mRFP and Sla1p-GFP in a clc1Δ strain and found that there were Sla2p patches all around the cortex, but they only occasionally overlapped with Sla1p patches (Figure 6C). Time-lapse videos of the overlapping patches revealed that Sla2p was recruited to the cortex prior to Sla1p in clc1Δ cells (Figure 6D). As seen for Sla2p/Abp1p (see Figure 4), at least four distinct classes of Sla2p/Sla1p events were observed in the clathrin mutant (Figure 6D). Some Sla2p patches remained immobile and never received Sla1p (Figure 6D2), while others were immobile and periodically shed patches containing Sla1p (D3). Longer-lived Sla2p patches that eventually internalized also recruited Sla1p but towards the end of their lifetime (D4); likewise, the Sla2p patches with normal lifetimes also recruited Sla1p (D5). Overall, Sla1p was only recruited to sites containing pre-existing Sla2p, and all sites where the Sla2p patch matured and internalized received Sla1p prior to inward movement.
The previous study of patch dynamics in clathrin mutants also found that Sla1p patch lifetime was accelerated (39). In our clc1Δ cells, Sla1p-GFP patch lifetime also decreased from 32 ± 7 seconds in WT to 27 ± 7 seconds (Figure 6E; n = 30 events), and nearly all of the Sla1p patches were mobile and internalized. In contrast, as described above (Figure 4), many of the Sla2p patches were large and immobilized at the cell surface, and even in patches that completed internalization, the Sla2p lifetimes were slower or at best similar to WT. Furthermore, even in WT cells, the lifetime of Sla1p (32 ± 7 seconds; Figure 6E) was significantly shorter than Sla2p (38 ± 9 seconds; Figure 4C) (p < 0.01, n = 30 patch events), which is also reflected by the fact that there are more patches with Sla2p than Sla1p (Figure 6B). Therefore, Sla2p is recruited to the cortex prior to Sla1p in both WT and clathrin mutant cells.
The Sla2 coiled-coil domain is important for normal turnover of clathrin, Sla2p and Abp1p
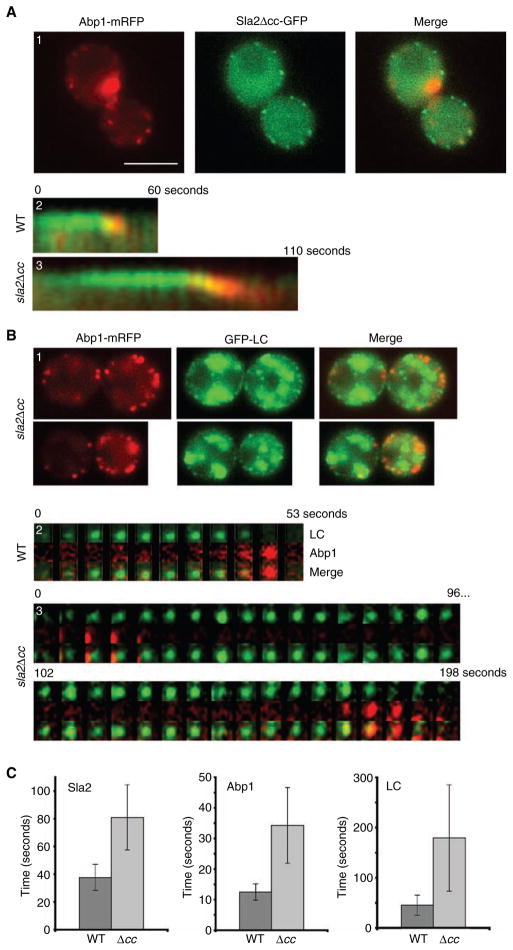
As the Sla2Δ coiled-coil (cc) mutant protein does not interact with clathrin LC, we tested the effects of this mutation on the dynamics of clathrin, Sla2p and Abp1p. To examine Sla2p/Abp1p dynamics in the sla2Δcc mutant, we co-expressed Abp1p-mRFP with Sla2pΔcc-GFP as the sole source of each protein. Unlike WT, sla2Δcc cells displayed elongated patch lifetimes of 79 ± 23 seconds for Sla2pΔcc-GFP and 32 ± 12 seconds for Abp1p-mRFP (Figure 7A1–A3,C; n = 30 events). This increase in patch lifetime for Sla2pΔcc and Abp1p is consistent with the reduced rate of alpha factor uptake by Ste2p in sla2Δcc cells (41).
Figure 7. Sla2Δcc has increased Abp1, Sla2p and clathrin cortical patch lifetime.
(A) sla2Δcc-GFP ABP1-mRFP (SL5186) (A1 and A3) and SLA2-GFP ABP1-mRFP (SL5185) (A2) cells were visualized using wide-field microscopy. (A1) Single frame and (A2–A3) kymographs generated from single pixel lines through individual cortical patches collected from time-lapse videos at 2-second intervals. Note elongated patch lifetimes of cells expressing Sla2pΔcc and Abp1p. (B) sla2Δcc ABP1-mRFP GFP-CLC1 (SL5198) cells imaged using wide-field (B1) and TIRFM time-lapse at 6-second intervals (B3). (B2) WT ABP1-mRFP GFP-CLC1 (SL5090) cells imaged using TIRFM at 5.3-second intervals. (C) Bar graphs showing the average patch lifetimes for Sla2p-GFP, Abp1p-mRFP and GFP-LC in WT and sla2Δcc cells. Note dramatic increase in patch lifetimes in the sla2Δcc mutant (p < 0.001 for all). Scale bar = 5 μm.
To observe the effect of sla2Δcc on cortical clathrin, we measured the lifespan of GFP-LC along with Abp1p-mRFP by TIRFM. Consistent with the elongation of Sla2p and Abp1p lifespan, GFP-LC lifetime increased from 49 ± 22 seconds (n = 61) to 180 ± 106 seconds (n = 13) (Figure 7B1–B3,C). Although the clathrin lifespan was increased in the sla2Δcc mutant, Abp1p was still recruited to clathrin patches, with 91% (n = 58) of Abp1p patches forming at sites containing GFP-LC. These results show that the Sla2p coiled-coil domain is not required for its own recruitment or that of Abp1p and clathrin; however, loss of this Sla2p domain affects the normal dynamics of both early and late endocytic factors.
Discussion
Physical interactions between Sla2p and the clathrin subunits
In this study, we show that the interaction between yeast clathrin LC and the Sla2p coiled-coil domain is direct and occurs independently of clathrin HC. This interaction is conserved, as mammalian LCs interact directly with the coiled-coil domain of the Sla2p homologues, Hip1 and Hip1R (HIP12) (25–27). In addition to the LC interaction, Hip1, but not Hip1R, also interacts with the HC-TD through its clathrin box motif (25,43,45). We did detect a weak interaction between yeast HC and Sla2pΔcc, which suggested Sla2p might interact with HC-TD through its two sequences related to the LLDLD-like clathrin box motif. But, by two hybrid analysis, we found TD does not interact with these motifs under conditions where Ent1p bound strongly. This suggests that Sla2p is more similar to Hip1R, which lacks clathrin-binding motifs and an AP-2-binding site (25). However, we cannot completely rule out a TD–Sla2p interaction, because HIP1 also does not bind mammalian TD by two-hybrid analysis (26). Still, the weak association between HC and Sla2p-Δcc could be mediated by multivalent interactions between endocytic factors that can bridge Sla2p and clathrin HC even in the absence of LC-Sla2p binding. For example, Sla2p binds to Sla1p, which interacts directly with Pan1p (48,49). Pan1p EH domains, in turn, bind Ent1/2p and Yap1801/1802p, which have clathrin TD-binding motifs (50,51).
Clathrin is recruited prior to Sla2p at endocytic patches
The direct interaction between LC and Sla2p suggested that these proteins function together at the cortex during endocytosis. To better understand this interaction, we followed their dynamics simultaneously by live cell imaging. Previously, it was shown that both clathrin and Sla2p are early endocytic patch factors, relative to the late-stage components, such as Abp1p or Arp2/3 complex proteins, present during actin recruitment and vesicle internalization (35,40,52). Here, using TIRF microscopy, we found that clathrin is recruited to the cortex on average 25 seconds prior to Sla2p, suggesting a role for clathrin during the earliest stages of endocytosis. This is consistent with findings that clathrin is also recruited prior to Las17p (39). In addition, both clathrin and Sla2p disappear from the cortex at the same time, after the initial recruitment of Abp1p. By wide-field microscopy, we found that clathrin undergoes inward movement from the cortex about 200 nm before its disappearance. This 200 nm inward movement [also seen by Kaksonen et al. (39)] is identical to that of Sla2p, Sla1p and Pan1p during the end of their patch lifetimes, but is a much shorter distance than the 500–1000 nm movement reported for the later Abp1p/actin-containing patches as they move into the cell (40).
It is not clear whether the slight inward movement of patches containing Sla2p, Sla1p, Pan1p and clathrin represent completely budded CCVs or deeply invaginated CCPs at the plasma membrane. Although Hip1/R are highly abundant on mammalian CCVs, we could not detect Sla2p on purified yeast CCVS. If we assume Sla2p exits/uncoats along with clathrin, this result argues that endo-cytic CCVs may not exist in yeast, or only do so for several seconds, preventing purification in significant amounts relative to TGN/endosomal derived CCVs. Whether or not true endocytic CCVs exist in yeast, it is clear that Sla1p/Sla2p/Pan1p and clathrin uncoat/disassemble prior to the actin-associated long-range movement.
Clathrin’s roles in cortical patch dynamics and endocytosis
Whether clathrin plays a direct role during endocytosis in yeast was debated for some time (36) due to the partial alpha factor uptake defect of clathrin mutants (17). However, our recent studies clearly showed clathrin is a dynamic component of the endocytic machinery in yeast (35). Thus, the partial endocytic defect of clathrin mutants might be manifested by a similar partial effect on all endo-cytic sites, or, alternatively, only a subset of endocytic sites is still relatively functional. Our identification of at least four types of Sla2p patch populations and aberrant actin structures in cells lacking HC or LC helps to clarify this issue. Some Sla2p patches in the clathrin mutants were abnormally large and immobile, whereas others completed internalization, albeit often with slower kinetics. There were also major defects in cortical patch mobility and unusual patterns of assembly and disassembly of actin in clathrin mutants, including actin comet tails pluming inward from some Sla2p patches. This could be caused by improper activation, arrangement or recruitment of the Arp2/3 activators Las17p, Pan1p, Abp1p and Myo3/5p (53–57). Nevertheless, even in endocytic patch events with relatively normal Sla2p timing, Abp1 patch lifetimes were delayed approximately twofold in clathrin mutants, and both Abp1p and Sla2p seemed to move deeper into the cell and persisted together longer than normal (compare Figure 4B1–B5,4B9). Even though these internalization events seem abnormal, the delay in Abp1/actin release may not actually contribute to the alpha factor endocytosis defects observed. The extended Abp1 phase could occur late after sequestration of alpha factor and vesicle pinching off. Therefore, the slowed maturation of Sla2p patches and the fewer sites that complete internalization probably account for the partial endocytic defect seen in clathrin mutants. It is still possible that without clathrin, fluid phase endocytosis or another endocytic pathway is up-regulated, as was seen in dominant negative dynamin-expressing animal cells (58). However, these pathways would have to be actin independent, as nearly all clathrin patches progress to the actin/Abp1 phase, and mutations or drugs that directly perturb actin function have strong endocytic phenotypes (35,39,40).
Sla1p and Sla2p dynamics are distinct
Recent studies of Kaksonen et al. (39) examining Sla1p dynamics in clathrin-deficient cells were remarkably different from what we observed for Sla2p. They also observed that Abp1p lifetimes are slowed in the clathrin mutants, but they found that Sla1p patches are fewer in number and their lifetimes are shorter than normal. They suggested that the accelerated Sla1p dynamics accounted for the 56–73% reduction in the number of endocytic sites, which in turn caused the partial internalization defect seen in the absence of clathrin. Our examination of both Sla1p and Sla2p leads to a different interpretation of these findings.
We also found that the number of cortical Sla1p patches/μm2 is greatly decreased in clc1Δ, but the number of Sla2p patches is essentially the same in WT and the clathrin mutant. One possibility that could account for this difference is that Sla2p can bind lipid via its ANTH domain, which may allow for normal cortical recruitment in the absence of clathrin. Similar results have also been seen with clathrin adaptors in animal cells. RNAi directed against clathrin HC showed no effect on the number of endocytic patches present at the plasma membrane, based on localization studies of AP-2, AP180/CALM, Eps15 and epsin (59,60). Therefore, adaptor recruitment to the plasma membrane does not require clathrin.
Not only do Sla2p and Sla1p differ in that the lifetime of Sla1p patches is shortened, but nearly all Sla1p patches acquire Abp1/actin and are internalized in clathrin mutants. In contrast, many Sla2p patches are large and immobile. Interestingly, the Sla2p patches that do internalize in clathrin mutants always receive Sla1p (and presumably Abp1p) prior to their internalization. This suggests that Sla1p is only recruited to those Sla2p patches that are competent or primed to complete endocytosis. Alternatively, recruitment of Sla1p could be less efficient without clathrin, but once recruited, it could trigger the rapid movement seen. The more rapid patch lifetime of Sla1p supports the idea that other steps needed for rapid internalization have been completed prior to Sla1p arrival at endocytic sites. Importantly, this sequential assembly appears to be a normal event, as we found that Sla1p is recruited after Sla2p even in WT cells. Taken together, our results demonstrate that without clathrin there is not decrease in the number of potential endocytic sites; rather, there is a reduction in the number of competent endocytic sites that can recruit Sla1p and Abp1p to complete internalization.
Role of the clathrin–Sla2p interaction
Sla2p-deficient cells accumulate immobile cortical patches containing Sla1p, Pan1p, Las17p, clathrin, as well as AP180 and epsin adaptors, and they often develop wide actin comet tails spread over a large domain of the cell surface (35,40,48). These large patches are unable to progress to endocytic pits and vesicles. This suggests that Sla2p plays a central regulatory role to promote maturation of the early patch stage to the invagination step. In contrast, the actin comet tails that develop in the clathrin mutants are very narrow and extend only as a single patch, rather than from a broad region of the cell surface. These actin comet tails are invaginating membrane as they are labeled with the lipophillic dye FM4-64. Furthermore, many of the sites marked by Sla2p do eventually acquire Sla1p and Abp1, which is a prerequisite for completion of endocytosis. Therefore, clathrin may act as a scaffold to properly establish the early endocytic patches. If all other factors are available, endocytic sites can still assemble and mature, although the process is less efficient.
What might then be the function of the clathrin LC–Sla2p interaction? The sla2Δcc mutation completely disrupts LC binding in vitro and strongly reduces the interaction with clathrin HC in a yeast lysate. The near-doubling in patch lifetime for Abp1p, clathrin and Sla2p in the coiled-coil mutant indicates that in the absence of LC–Sla2p association, overall patch progression to and during the mobile late phase is delayed. Sla2p–LC interaction could promote patch mobility by serving as a signal for clathrin triskelions to rearrange or release and reassemble during membrane curvature and vesicle internalization. This model is supported by the ability of the Sla2p homologues, Hip1 and Hip1R, to promote clathrin assembly into clathrin cages, while perturbation of LC–Hip1/R interaction reduces lattice assembly (26,27). Alternatively, LC itself could stimulate Sla2p-mediated progression of endocytic patches to the mobile actin phase. Note, we cannot rule out that the coiled-coil region of Sla2p, which is also a dimerization domain, has other interactions that contribute to the observed phenotypes. However, taken together, our data and the previous in vitro assembly studies (26,27) suggest that the Sla2p–LC interaction is a critical step regulating the progression of endocytic events in both yeast and animal cells.
In conclusion, we have shown that clathrin is an early endocytic factor, arriving at endocytic sites prior to Sla2p, which is followed by recruitment of Sla1p, establishing at least three temporal stages of early endocytic patch assembly. Our previous studies found that clathrin recruitment is dependent on the epsin and AP180 adaptors (35); hence, these proteins are probably recruited to early patches before or along with clathrin. Both clathrin and Sla2p uncoat/disappear at the onset of intense Abp1 recruitment, which is when endocytic vesicles complete their internalization and move away from the cell surface. We propose that clathrin provides an early scaffolding or regulatory function for proper endocytic patch formation and efficient priming of Sla2p-containing patches, a necessary step for recruitment of Sla1p and, ultimately, Abp1p/Actin. Furthermore, our data support a direct in vivo role of clathrin LC–Sla2p interaction for endocytic vesicle formation. Future studies will be necessary to determine how LC binding regulates Sla2p’s role in this process.
Methods and Materials
Strains, media and genetic methods
Strains used in this study are listed in Table 1. YEPD and synthetic dropout media were prepared as described in (61). Yeast mating, sporulation and tetrad analysis were performed as described in (62). Yeast were transformed using the method as described in Gietz et al. (63), and cells were cultured at 30 °C.
Table 1.
Saccharomyces cerevisiae strains
| Strain | Genotypea |
|---|---|
| HR1965 | MATa his4 leu2 ura3 his4 lys2 bar1 sla2Δ::LEU2d |
| HR3399 | MATa leu2 ura3 his3 sla2Δ::LEU2 sla2Δ376-573::TRP1d |
| SL1463 | MATα leu2 ura3-52 trp1 his3-Δ200 |
| SL1628 | MATa leu2 ura3-52 trp1 his3-Δ200 clc1Δ::HIS3 YEp24-CHC1 [2μ, URA3, CHC1] |
| SL1629 | MATa leu2 ura3-52 trp1 his3-Δ200 YEp24-CHC1 |
| SL1927 | MATα leu2 ura3-52 ade6 chc1Δ::LEU2 pKH2 [2μ, URA3, CLC1] |
| SL4409 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 gal4Δ ade2 gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ chc1Δ::TRP1b |
| SL4410 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 gal4Δ ade2 gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ clc1Δ::TRP1b |
| SL4781 | MATα leu2 ura3 his3-Δ200 trp1 sla2Δ::LEU2 p111-SLA2-GFP [CEN, LEU2, TRP1, SLA2-GFP] |
| SL4782 | MATa leu2 ura3 his3-Δ200 trp1 clc1Δ::KanMX6 sla2Δ::LEU2 p111-SLA2-GFP [CEN, LEU2, TRP1, SLA2-GFP] |
| SL5090 | MATα leu2 ura3-52 trp1 his3-Δ200 clc1Δ::HIS3 ABP1-mRFP::KanMX6 pTMN17 [CEN, TRP1, GFP-CLC1] |
| SL5101 | MATa leu2 ura3 his3 ABP1-GFP::HIS3 clc1Δ::KanMX6 pRS426 [2μ, URA3]c |
| SL5121 | MATα leu2-3,112 ura3-52 trp1 his3-Δ200 clc1Δ::HIS3 SLA2-mRFP::KanMX6 pTMN17 [CEN, TRP1, GFP-CLC1] |
| SL5185 | MATα leu2 ura3 trp1 his3-Δ200 sla2Δ::LEU2 ABP1-mRFP::KanMX6 p111-Sla2-GFP [CEN, LEU2, TRP1, SLA2-GFP] |
| SL5186 | MATα leu2 ura3 trp1 his3-Δ200 sla2Δ::LEU2 ABP1-mRFP::KanMX6 pTMN52 [CEN, LEU2, sla2Δcc-GFP::TRP1] |
| SL5198 | MATa leu2 ura3 his3 clc1Δ::HIS3 sla2Δ::HIS3 sla2Δcc::TRP1 ABP1-mRFP::KanMX6 pTMN37 [CEN, URA3, GFP-CLC1]d |
| SL5226 | MATa leu2 ura3 his3-Δ200 trp1 chc1Δ::LEU2 sla2Δ::LEU2 ABP1-mRFP::KanMX6 p111-Sla2-GFP [CEN, LEU2, SLA2-GFP::TRP1] |
| SL5240 | MATa leu2 ura3 his3-Δ200 trp1 clc1Δ::HIS3 sla2Δ::LEU2 ABP1-mRFP::KanMX6 p111-Sla2-GFP [CEN, LEU2, SLA2-GFP::TRP1] |
| SL5317 | MATα leu2 ura3-52 trp1 his3-Δ200 clc1Δ::HIS3 SLA2-mRFP::KanMX6 SLA1-GFP::TRP1 |
| SL5318 | MATα leu2 ura3-52 trp1 his3-Δ200 SLA2-mRFP::KanMX6 SLA1-GFP::TRP1 |
| YPJ96-4 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-901 gal4Δ ade2 gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZb |
Plasmids
Plasmids pGAD-C1 (GAD empty, LEU2, 2μ) and pGBDU-C1 (GBD empty, URA3, 2μ) are described in (64). p31-2 (GAD-sla2-292-520, LEU2, 2μ), pKH31 (GBD-CLC1, URA3, 2μ), pKH49 (GBD-sla2-292-968, URA3, 2μ), pKH47 (GBD-sla2-292-501, URA3, 2μ) and pKH24 (GAD-chc1-655-1653, LEU2, 2μ) are described in (19). p31-6 (GAD-sla2-289-583, LEU2, 2μ) and p31-10 (GAD-sla2-292-968, LEU2, 2μ) were isolated from the LC two-hybrid screen (19). pTMN17 [GFP-CLC1, TRP1, CEN] and pTMN37 [GFP-CLC1, URA3, CEN] are described in Newpher et al. (35). YEp24-CHC1 and pKH2 [CLC1, URA3, 2μ] were previously described in (11). p111-SLA2-GFP (CEN, TRP1, LEU2) was a gift from María Isabel Geli and Fatima Idrissi. Plasmids pTMN1 (GST-CLC1) and pTMN3 (6xHis-CLC1 for bacterial expression) were constructed by polymerase chain reaction (PCR) amplifying the Clc1p-coding sequence flanked by BamH1 and EcoR1 restriction sites and subcloning into pGEX-2T (Amersham Biosciences, Piscataway, NJ, USA) and pRSET-A (Invitrogen, Carlsbad, CA, USA), respectively.
In-frame fusions of SLA2 were generated by subcloning the SLA2 open--reading frame (ORF) on a BamH1/Sal1 fragment into pGEX-4T-1 (Amersham Biosciences) and pGBDU-C1, generating pTMN5 (GST-SLA2 for bacterial expression) and pTMN9 (GBD-SLA2, URA3, 2μ), respectively. The HC terminal domain two-hybrid prey was constructed by subcloning the CHC1 ORF encoding amino acids 1–500 on a BamH1/Sal1 fragment into pGAD-C1 generating pTMN8 (GAD-CHC1-TD, LEU2, 2μ). pTMN13 (GST-sla2-Δ376-573 for bacterial expression) was generated by cutting pWA30 (41) with Nsi1 and ligating the sla2-Δ376-573 fragment into Nsi1 cut pTMN5. pTMN38 (GBD-ENT1, URA3, 2μ) was generated using PCR amplification of the ENT1 ORF with primers to allow gap repair into pGBDU-C1. GST-sla2-289-583 and GST-sla2-292-968 bacterial expression plasmids were generated by subcloning EcoR1/Sal1 SLA2 fragments from p31-6 and p31-10 (19), respectively, into pGEX-4T1. pTMN52 was constructed by cutting p111-SLA2-GFP with Nsi1 and gap repairing off the chromosomal sla2Δ376-573 internal deletion in HR3399 (SL3671). YEplac195-SLA2, GST-BMH1 and GST-ETLLDLDF were generous gifts from Howard Riezman, Mike Yaffe and Linton Traub, respectively. All plasmids generated from PCR fragments were verified by sequencing.
Gene deletions and integrations
To generate deletions of CLC1 (SL4410) and CHC1 (SL4409) in the two-hybrid reporter strain, we replaced the complete coding sequences with the TRP1 gene. Plasmid pFA6a-TRP1 (65) was amplified with primers extended by DNA flanking the CLC1 or CHC1 ORFs, and the PCR were products transformed into YPJ96-4 (64). All knockouts were verified using colony PCR.
SLA2-mRFP::KanMX6 cells were generated by integrating a PCR product amplified from pFA6a-mRFP::KanMX6 using primers flanked by sequences surrounding the SLA2 stop codon. SLA1-GFP::TRP1 was generated amplifying a PCR product from pFA6a-GFP::TRP1 using primers flanked by sequences surrounding the SLA1 stop codon and then integrating into a clc1Δ::HIS3 SLA2-mRFP::KanMX6 double heterozygote. SLA1-GFP::TRP1 spore segregants were derived from this transformant.
Two-hybrid interaction analysis
For analysis of two-hybrid interactions, log phase cultures were diluted to 0.5 × 106 cells/mL for reporter strain (YPJ96-4) and 1.0 × 107 cells/mL for chc1Δ (SL4409) and clc1Δ (SL4410) reporters and spotted on C-LEU-URA plates to test general growth and C-ADE-LEU-URA plates to score induction of the GAL2-ADE2 reporter. Plates were grown 3–5 days at 30 °C.
Biochemical procedures
To purify GST fusion proteins from bacteria, we induced 500 mL of log phase cells for expression with 1 mM IPTG. After 5 h of induction, cells were lysed by sonication in PBS pH 8.0 containing 1% Triton-X-100 and a protease inhibitor cocktail made as described in (66). Lysates were spun at 20 000 × g for 30 min, and the supernatant was absorbed onto a column containing 500 μL of glutathione sepharose. Following three washes with bacterial lysis buffer, glutathione S-transferase (GST) fusions were eluted with 20 mM glutathione in 50 mM Tris pH 8.0 and centrifuged through VIVASCIENCE concentrators (Hannover, Germany). Concentrated samples were diluted to approximately 4 mg/mL in PBS with 50% glycerol and stored at −20 °C. Recombinant 6xHis-tagged fusion proteins were expressed and purified following the QIAexpressionist™ protocols from Qiagen® (Valencia, CA, USA) and stored as described above for GST fusions.
GST pull downs from yeast lysates were performed by prebinding bacterially purified GST fusions (200 nM) to 50 μL of glutathione sepharose beads for 1 h in 1 mL of HEKT buffer (20 mM Hepes pH 7.4, 1 mM ethylenediaminetetraa-cetic acid, 50 mM KCl, 0.5% Triton-X-100 + protease inhibitors). Yeast extracts were prepared using glass bead lysis of 2 × 108 cells in 1 mL HEKT buffer for 3 min in a Braun homogenizer. Lysates were spun for 10 min at 10 000 × g, and 1 mL of supernatant was incubated for 1 h at 4 °C with the prebound GST fusions. Following incubation, the beads were washed three times with 1 mL of HEKT buffer and eluted in 120 μL HEKT buffer plus 30 μL of ×5 SDS sample buffer and boiled 5 min at 95 °C. Thirty microlitres were analyzed using SDS–PAGE and immunoblot analysis. Clathrin HC was detected using mouse monoclonal antibodies to Chc1p (46), and the GST fusions were detected using rabbit anti-GST antibodies at 1:5000 (Santa Cruz Biotechnology, Santa Cruz, CA, USA). Clathrin LC was detected using rabbit polyclonal anti-Clc1p antibodies at 1:1000, a gift from Greg Payne. Goat anti-rabbit (Zymed/Invitrogen) and goat anti-mouse (Kirkegaard and Perry Laboratories, Gaitherburg, MD, USA) antibodies conjugated to horseradish peroxidase were used at 1:5000 and detected using enhanced chemiluminescence (Amersham).
For testing direct interactions, purified 6xHis-tagged Clc1p was added at 200 nM in 1 mL of HEKT buffer to GST fusions prebound to glutathione-agarose, and complexes were affinity purified as described above. Histidine-tagged Clc1p was detected with 6xHis mouse monoclonal antibodies at 1:1000 (Clonetech, Mountain View, CA, USA).
Clathrin-coated vesicles were purified from yeast as described in (46). Clathrin-rich fractions were concentrated 50-fold using VIVASPIN concentrators and analyzed using SDS–PAGE and immunoblotted with rabbit anti-Sla2p C-terminal antibodies at 1:1000, a gift from Howard Riezman.
For yeast extracts shown in Figure 2E, lysates were prepared by glass bead homogenation in 50 mM Tris–HCl pH 7.5, 1% Triton-X-100, 1% sodium deoxycholate, 0.1% SDS, 1 mM sodium pyrophosphate, 150 mM NaCl plus protease inhibitors at 2.0 × 108 cells/mL and were spun at 10 000 × g for 10 min. Supernatants were fractionated using SDS–PAGE and immunoblotted with anti-Sla2p C-terminal and rabbit anti-Clc1p antibodies.
Microscopy
Fluorescent fusion proteins were visualized in live cells by growing cultures overnight to mid-log phase in synthetic media. To track FM4-64 with Abp1-GFP in clc1Δ, cells were incubated with 40 μM FM4-64 for 2 min, washed in synthetic medium and viewed after 20 min of incubation at room temperature. Microscopy was performed using an Olympus fluorescence BX61 microscope equipped with Nomarski differential interference contrast (DIC) optics, a ×100 objective (numerical aperture 1.35), a Roper CoolSNAP HQ camera, Sutter Lambda 10+2 automated excitation and emission filter wheels and a 175 watt Xenon remote source lamp with liquid light guide. Images were acquired and processed using the Intelligent Imaging Innovations (Denver, CO, USA) SlideBook™ image analysis software and prepared with Adobe® Photoshop®7.
TIRFM was performed as described in Newpher et al. (35) for Abp1-mRFP/GFP-LC and Sla2p-mRFP/GFP-LC. For acquisition of GFP-LC and Abp1-mRFP in sla2ccΔ cells, a dual 488/543 excitation filter was used to acquire images simultaneously. In Figures 3A and 6B, the clathrin signal was intensified in Adobe® Photoshop®7 for better viewing of cortical clathrin.
Kymographs were generated from single pixel lines taken during 4-min time-lapse videos. Exposure times of 500 ms were used for all wide-field and TIRFM images, except for Sla2p-mRFP in Figure 3B,D, which used a 2000-ms exposure. Patch lifetimes of clathrin, Sla2p and Abp1p were displayed as the mean ± the standard deviation in seconds.
Patch numbers were quantified from Z stacks collected at 0.25-μm intervals. In most cases, total patches/cell were counted from maximum intensity projections of large budded and nonpolarized cells. In the case of SL4782, total patches per cell were counted from individual Z stacks, manually filtering out recounts of patches that bridged more than one section. The cell surface area was calculated by measuring the average diameter of the cell at 0 ° and 90 °. The total number of patches/cell was divided by the surface area to calculate patches/μm2.
Supplementary Material
Time-lapse video of WT cells expressing Abp1p-mRFP and Sla2p-GFP (SL5185). Images were captured every 2 seconds with 500-ms exposures for a duration of 4 min.
Time-lapse video of chc1Δ cells expressing Sla2p-GFP and Abp1p-mRFP (SL5226). Images were captured every 2 seconds with 500-ms exposures for a duration of 4 min.
Acknowledgments
We thank María Isabel Geli, Fatima Idrissi, Linton Traub, Mike Yaffe, Greg Payne and Howard Riezman for providing many strains, plasmids and antibodies. We thank Kenneth Henry, John Collette and Ji Suk Chang for many helpful discussions and Robin P. Smith for technical assistance with TIRF microscopy. TMN was supported by National Institutes of Health (NIH) training grant (T32 G08056-20). This work was funded by NIH grant (R01 G55796).
References
- 1.Pishvaee B, Munn A, Payne GS. A novel structural model for regulation of clathrin function. EMBO J. 1997;16:2227–2239. doi: 10.1093/emboj/16.9.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen CY, Reese ML, Hwang PK, Ota N, Agard D, Brodsky FM. Clathrin light and heavy chain interface: alpha-helix binding superhelix loops via critical tryptophans. EMBO J. 2002;21:6072–6082. doi: 10.1093/emboj/cdf594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- 4.McPherson PS, Ritter B. Peptide motifs: building the clathrin machinery. Mol Neurobiol. 2005;32:73–87. doi: 10.1385/MN:32:1:073. [DOI] [PubMed] [Google Scholar]
- 5.Brodsky FM, Hill BL, Acton SL, Nathke I, Wong DH, Ponnambalam S, Parham P. Clathrin light chains: arrays of protein motifs that regulate coated-vesicle dynamics. Trends Biochem Sci. 1991;16:208–213. doi: 10.1016/0968-0004(91)90087-c. [DOI] [PubMed] [Google Scholar]
- 6.DeLuca-Flaherty C, McKay DB, Parham P, Hill BL. Uncoating protein (hsc70) binds a conformationally labile domain of clathrin light chain LCa to stimulate ATP hydrolysis. Cell. 1990;62:875–887. doi: 10.1016/0092-8674(90)90263-e. [DOI] [PubMed] [Google Scholar]
- 7.Liu SH, Wong ML, Craik CS, Brodsky FM. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell. 1995;83:257–267. doi: 10.1016/0092-8674(95)90167-1. [DOI] [PubMed] [Google Scholar]
- 8.Ungewickell E, Ungewickell H. Bovine brain clathrin light chains impede heavy chain assembly in vitro. J Biol Chem. 1991;266:12710–12714. [PubMed] [Google Scholar]
- 9.Ybe JA, Ruppel N, Mishra S, VanHaaften E. Contribution of cysteines to clathrin trimerization domain stability and mapping of light chain binding. Traffic. 2003;4:850–856. doi: 10.1046/j.1600-0854.2003.0139.x. [DOI] [PubMed] [Google Scholar]
- 10.Chu DS, Pishvaee B, Payne GS. The light chain subunit is required for clathrin function in Saccharomyces cerevisiae. J Biol Chem. 1996;271:33123–33130. doi: 10.1074/jbc.271.51.33123. [DOI] [PubMed] [Google Scholar]
- 11.Huang KM, Gullberg L, Nelson KK, Stefan CJ, Blumer K, Lemmon SK. Novel functions of clathrin light chains: clathrin heavy chain trimerization is defective in light chain-deficient yeast. J Cell Sci. 1997;110:899–910. doi: 10.1242/jcs.110.7.899. [DOI] [PubMed] [Google Scholar]
- 12.Harsay E, Schekman R. A subset of yeast vacuolar protein sorting mutants is blocked in one branch of the exocytic pathway. J Cell Biol. 2002;156:271–285. doi: 10.1083/jcb.200109077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gurunathan S, David D, Gerst JE. Dynamin and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 2002;21:602–614. doi: 10.1093/emboj/21.4.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valdivia RH, Baggott D, Chuang JS, Schekman RW. The yeast clathrin adaptor protein complex 1 is required for the efficient retention of a subset of late Golgi membrane proteins. Dev Cell. 2002;2:283–294. doi: 10.1016/s1534-5807(02)00127-2. [DOI] [PubMed] [Google Scholar]
- 15.Gall WE, Geething NC, Hua Z, Ingram MF, Liu K, Chen SI, Graham TR. Drs2p-dependent formation of exocytic clathrin-coated vesicles in vivo. Curr Biol. 2002;12:1623–1627. doi: 10.1016/s0960-9822(02)01148-x. [DOI] [PubMed] [Google Scholar]
- 16.Seeger M, Payne GS. Selective and immediate effects of clathrin heavy chain mutations on Golgi membrane protein retention in Saccharomyces cerevisiae. J Cell Biol. 1992;118:531–540. doi: 10.1083/jcb.118.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan PK, Davis NG, Sprague GF, Payne GS. Clathrin facilitates the internalization of seven transmembrane segment receptors for mating pheromones in yeast. J Cell Biol. 1993;123:1707–1716. doi: 10.1083/jcb.123.6.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seeger M, Payne GS. A role for clathrin in the sorting of vacuolar proteins in the Golgi complex of yeast. EMBO J. 1992;11:2811–2818. doi: 10.1002/j.1460-2075.1992.tb05348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry KR, D’Hondt K, Chang J, Newpher T, Huang K, Hudson RT, Riezman H, Lemmon SK. Scd5p and clathrin function are important for cortical actin organization, endocytosis, and localization of Sla2p in yeast. Mol Biol Cell. 2002;13:2607–2625. doi: 10.1091/mbc.E02-01-0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lemmon SK, Freund C, Conley K, Jones EW. Genetic instability of clathrin-deficient strains of Saccharomyces cerevisiae. Genetics. 1990;124:27–38. doi: 10.1093/genetics/124.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lemmon SK, Jones EW. Clathrin requirement for normal growth of yeast. Science. 1987;238:504–509. doi: 10.1126/science.3116672. [DOI] [PubMed] [Google Scholar]
- 22.Wang J, Virta VC, Riddelle-Spencer K, O’Halloran TJ. Compromise of clathrin function and membrane association by clathrin light chain deletion. Traffic. 2003;4:891–901. doi: 10.1046/j.1600-0854.2003.00144.x. [DOI] [PubMed] [Google Scholar]
- 23.Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279:16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- 24.Girard M, Allaire PD, McPherson PS, Blondeau F. Non-stoichiometric relationship between clathrin heavy and light chains revealed by quantitative comparative proteomics of clathrin-coated vesicles from brain and liver. Mol Cell Proteomics. 2005;4:1145–1154. doi: 10.1074/mcp.M500043-MCP200. [DOI] [PubMed] [Google Scholar]
- 25.Legendre-Guillemin V, Metzler M, Charbonneau M, Gan L, Chopra V, Philie J, Hayden MR, McPherson PS. HIP1 and HIP12 display differential binding to F-actin, AP2, and clathrin. Identification of a novel interaction with clathrin light chain. J Biol Chem. 2002;277:19897–19904. doi: 10.1074/jbc.M112310200. [DOI] [PubMed] [Google Scholar]
- 26.Chen CY, Brodsky FM. Huntingtin-interacting protein 1 (Hip1) and Hip1-related protein (Hip1R) bind the conserved sequence of clathrin light chains and thereby influence clathrin assembly in vitro and actin distribution in vivo. J Biol Chem. 2005;280:6109–6117. doi: 10.1074/jbc.M408454200. [DOI] [PubMed] [Google Scholar]
- 27.Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, et al. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280:6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- 28.Legendre-Guillemin V, Wasiak S, Hussain NK, Angers A, McPherson PS. ENTH/ANTH proteins and clathrin-mediated membrane budding. J Cell Sci. 2004;117:9–18. doi: 10.1242/jcs.00928. [DOI] [PubMed] [Google Scholar]
- 29.Hyun TS, Rao DS, Saint-Dic D, Michael LE, Kumar PD, Bradley SV, Mizukami IF, Oravecz-Wilson KI, Ross TS. HIP1 and HIP1R stabilize receptor tyrosine kinases and bind 3-phosphoinositides via epsin N-terminal homology domains. J Biol Chem. 2004;279:14294–14306. doi: 10.1074/jbc.M312645200. [DOI] [PubMed] [Google Scholar]
- 30.Sun Y, Kaksonen M, Madden DT, Schekman R, Drubin DG. Interaction of Sla2p’s ANTH domain with PtdIns (4,5),P2 is important for actin-dependent endocytic internalization. Mol Biol Cell. 2005;16:717–730. doi: 10.1091/mbc.E04-08-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Engqvist-Goldstein AE, Kessels MM, Chopra VS, Hayden MR, Drubin DG. An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J Cell Biol. 1999;147:1503–1518. doi: 10.1083/jcb.147.7.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCann RO, Craig SW. The I/LWEQ module: a conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Engqvist-Goldstein AE, Zhang CX, Carreno S, Barroso C, Heuser JE, Drubin DG. RNAi-mediated Hip1R silencing results in stable association between the endocytic machinery and the actin assembly machinery. Mol Biol Cell. 2004;15:1666–1679. doi: 10.1091/mbc.E03-09-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Metzler M, Li B, Gan L, Georgiou J, Gutekunst CA, Wang Y, Torre E, Devon RS, Oh R, Legendre-Guillemin V, Rich M, Alvarez C, Gertsenstein M, McPherson PS, Nagy A, et al. Disruption of the endo-cytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 2003;22:3254–3266. doi: 10.1093/emboj/cdg334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Newpher TM, Smith RP, Lemmon V, Lemmon SK. In vivo dynamics of clathrin and its adaptor-dependent recruitment to the actin-based endocytic machinery in yeast. Dev Cell. 2005;9:87–98. doi: 10.1016/j.devcel.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 36.Baggett JJ, Wendland B. Clathrin function in yeast endocytosis. Traffic. 2001;2:297–302. doi: 10.1034/j.1600-0854.2001.002005297.x. [DOI] [PubMed] [Google Scholar]
- 37.Yarar D, Waterman-Storer CM, Schmid SL. A dynamic actin cytoske-leton functions at multiple stages of clathrin-mediated endocytosis. Mol Biol Cell. 2005;16:964–975. doi: 10.1091/mbc.E04-09-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Merrifield CJ, Feldman ME, Wan L, Almers W. Imaging actin and dynamin recruitment during invagination of single clathrin-coated pits. Nat Cell Biol. 2002;4:691–698. doi: 10.1038/ncb837. [DOI] [PubMed] [Google Scholar]
- 39.Kaksonen M, Toret CP, Drubin DG. A modular design for the clathrin-and actin-mediated endocytosis machinery. Cell. 2005;123:305–320. doi: 10.1016/j.cell.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 40.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 41.Wesp A, Hicke L, Palecek J, Lombardi R, Aust T, Munn AL, Riezman H. End4p/Sla2p interacts with actin-associated proteins for endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1997;8:2291–2306. doi: 10.1091/mbc.8.11.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang S, Cope MJ, Drubin DG. Sla2p is associated with the yeast cortical actin cytoskeleton via redundant localization signals. Mol Biol Cell. 1999;10:2265–2283. doi: 10.1091/mbc.10.7.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Metzler M, Legendre-Guillemin V, Gan L, Chopra V, Kwok A, McPherson PS, Hayden MR. HIP1 functions in clathrin-mediated endo-cytosis through binding to clathrin and adaptor protein 2. J Biol Chem. 2001;276:39271–39276. doi: 10.1074/jbc.C100401200. [DOI] [PubMed] [Google Scholar]
- 44.Engqvist-Goldstein AE, Warren RA, Kessels MM, Keen JH, Heuser J, Drubin DG. The actin-binding protein Hip1R associates with clathrin during early stages of endocytosis and promotes clathrin assembly in vitro. J Cell Biol. 2001;154:1209–1223. doi: 10.1083/jcb.200106089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mishra SK, Agostinelli NR, Brett TJ, Mizukami I, Ross TS, Traub LM. Clathrin- and AP-2-binding sites in HIP1 uncover a general assembly role for endocytic accessory proteins. J Biol Chem. 2001;276:46230–46236. doi: 10.1074/jbc.M108177200. [DOI] [PubMed] [Google Scholar]
- 46.Lemmon S, Lemmon VP, Jones EW. Characterization of yeast clathrin and anticlathrin heavy-chain monoclonal antibodies. J Cell Biochem. 1988;36:329–340. doi: 10.1002/jcb.240360403. [DOI] [PubMed] [Google Scholar]
- 47.Smith AE, Zhang Z, Thomas CR, Moxham KE, Middelberg AP. The mechanical properties of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2000;97:9871–9874. doi: 10.1073/pnas.97.18.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gourlay CW, Dewar H, Warren DT, Costa R, Satish N, Ayscough KR. An interaction between Sla1p and Sla2p plays a role in regulating actin dynamics and endocytosis in budding yeast. J Cell Sci. 2003;116:2551–2564. doi: 10.1242/jcs.00454. [DOI] [PubMed] [Google Scholar]
- 49.Tang HY, Xu J, Cai M. Pan1p, End3p, and S1a1p, three yeast proteins required for normal cortical actin cytoskeleton organization, associate with each other and play essential roles in cell wall morphogenesis. Mol Cell Biol. 2000;20:12–25. doi: 10.1128/mcb.20.1.12-25.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wendland B, Emr SD. Pan1p, yeast eps15, functions as a multivalent adaptor that coordinates protein-protein interactions essential for endocytosis. J Cell Biol. 1998;141:71–84. doi: 10.1083/jcb.141.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wendland B, Steece KE, Emr SD. Yeast epsins contain an essential N-terminal ENTH domain, bind clathrin and are required for endocyto-sis. EMBO J. 1999;18:4383–4393. doi: 10.1093/emboj/18.16.4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huckaba TM, Gay AC, Pantalena LF, Yang HC, Pon LA. Live cell imaging of the assembly, disassembly, and actin cable-dependent movement of endosomes and actin patches in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 2004;167:519–530. doi: 10.1083/jcb.200404173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duncan MC, Cope MJ, Goode BL, Wendland B, Drubin DG. Yeast Eps15-like endocytic protein, Pan1p, activates the Arp2/3 complex. Nat Cell Biol. 2001;3:687–690. doi: 10.1038/35083087. [DOI] [PubMed] [Google Scholar]
- 54.Lechler T, Shevchenko A, Li R. Direct involvement of yeast type I myosins in Cdc42-dependent actin polymerization. J Cell Biol. 2000;148:363–373. doi: 10.1083/jcb.148.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Evangelista M, Klebl BM, Tong AH, Webb BA, Leeuw T, Leberer E, Whiteway M, Tomas DY, Boone C. A role for myosin-I in actin assembly through interactions with Vrp1p, Bee1p, and the Arp2/3 complex. J Cell Biol. 2000;148:353–362. doi: 10.1083/jcb.148.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Winter D, Lechler T, Li R. Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr Biol. 1999;9:501–504. doi: 10.1016/s0960-9822(99)80218-8. [DOI] [PubMed] [Google Scholar]
- 57.Goode BL, Rodal AA, Barnes G, Drubin DG. Activation of the Arp2/3 complex by the actin filament binding protein Abp1p. J Cell Biol. 2001;153:627–634. doi: 10.1083/jcb.153.3.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Damke H, Baba T, van der Bliek AM, Schmid SL. Clathrin-independent pinocytosis is induced in cells overexpressing a temperature-sensitive mutant of dynamin. J Cell Biol. 1995;131:69–80. doi: 10.1083/jcb.131.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Motley A, Bright NA, Seaman MN, Robinson MS. Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol. 2003;162:909–918. doi: 10.1083/jcb.200305145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinrichsen L, Harborth J, Andrees L, Weber K, Ungewickell EJ. Effect of clathrin heavy chain- and alpha-adaptin-specific small inhibitory RNAs on endocytic accessory proteins and receptor trafficking in HeLa cells. J Biol Chem. 2003;278:45160–45170. doi: 10.1074/jbc.M307290200. [DOI] [PubMed] [Google Scholar]
- 61.Nelson KK, Lemmon SK. Suppressors of clathrin deficiency. overexpression of ubiquitin rescues lethal strains of clathrin-deficient Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:521–532. doi: 10.1128/mcb.13.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guthrie C, Fink GR, editors. Guide to Yeast Genetics and Molecular Biology. San Diego: Academic Press; 1991. [Google Scholar]
- 63.Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 64.James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 66.Stepp JD, Pellicena-Palle A, Hamilton S, Kirchhausen T, Lemmon SK. A late Golgi sorting function for Saccharomyces cerevisiae Apm1p, but not for Apm2p, a second yeast clathrin AP medium chain-related protein. Mol Biol Cell. 1995;6:41–58. doi: 10.1091/mbc.6.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse video of WT cells expressing Abp1p-mRFP and Sla2p-GFP (SL5185). Images were captured every 2 seconds with 500-ms exposures for a duration of 4 min.
Time-lapse video of chc1Δ cells expressing Sla2p-GFP and Abp1p-mRFP (SL5226). Images were captured every 2 seconds with 500-ms exposures for a duration of 4 min.