Summary
Clathrin-coated vesicle formation is responsible for membrane traffic to and from the endocytic pathway during receptor-mediated endocytosis and organelle biogenesis, influencing how cells relate to their environment. Generating these vesicles involves self-assembly of clathrin molecules into a latticed coat on membranes that recruits receptors and organizes protein machinery necessary for budding. Here we define a molecular mechanism regulating clathrin lattice formation by obtaining structural information from co-crystals of clathrin subunits. Low resolution X-ray diffraction data (7.9–9.0Å) was analyzed using a combination of molecular replacement with an energyminimized model, and non-crystallographic symmetry averaging. Resulting topological information revealed two conformations of the regulatory clathrin light chain bound to clathrin heavy chain. Based on protein domain positions, mutagenesis and biochemical assays, we identify an electrostatic interaction between the clathrin subunits that allows the observed conformational variation in clathrin light chains to alter the conformation of the clathrin heavy chain and thereby regulate assembly.
Introduction
Clathrin-mediated membrane traffic influences cell signaling and metabolic homeostasis by regulating receptor transport during endocytosis and organelle biogenesis. Receptor sorting in these pathways results from self-assembly of the triskelion-shaped clathrin protein into a polyhedral lattice that coats transport vesicles and concentrates vesicle cargo (Brodsky et al., 2001; Ungewickell and Hinrichsen, 2007). The triskelion is formed from three clathrin heavy chain (CHC) subunits, with associated clathrin light chain (CLC) subunits that prevent spontaneous CHC self-assembly at physiological pH (Ungewickell and Ungewickell, 1991; Liu et al., 1995; Ybe et al., 1998). During clathrin-coated vesicle (CCV) formation in cells, adaptor molecules induce the basic clathrin self-assembly reaction, as well as target clathrin to membranes and select vesicle cargo (Greene et al., 2000). Thus cellular clathrin assembly must be a balance between negative CLC and positive adaptor regulation. It is therefore puzzling that siRNA depletion of CLC from tissue culture cells or in vivo perturbation of CLC’s ability to regulate clathrin assembly have little effect on endocytosis, one of the major pathways mediated by CCVs (Chen and Brodsky, 2005; Huang et al., 2004; Poupon et al., 2008; Wang et al., 2006; Wilbur et al., 2008). To address this conundrum, we investigated CLC regulation of lattice formation through crystallographic and biochemical analysis.
There are two isoforms of CLC in vertebrates (LCa and LCb) with 60% sequence identity and each has neuronal splicing variants (Brodsky et al., 2001). The differential and tissue-specific functions of the CLCs have yet to be defined. Their longest shared sequence, 22 residues near the N-termini, is the binding site for huntingtin-interacting protein 1 (HIP1) and its related protein (HIP1R). This “consensus” sequence also comprises an acidic patch (EED) responsible for regulation of the pH sensitivity of clathrin self-assembly (Chen and Brodsky, 2005; Legendre-Guillemin et al., 2005; Ybe et al., 1998). CHCs are divided into domains starting at the N-terminal domain (residues 1 to 330 in mammalian CHC), which is followed by the ankle (~331 to 838), distal leg (~839 to 1073), knee (~1074 to 1197), proximal leg (~1198 to 1575) and trimerization domain (~1576 to 1675) (Fotin et al., 2004). The combination of the knee, proximal leg, and trimerization domain constitute the hub (1074–1675), which is a self-assembling fragment (Liu et al., 1995). A 2.9 Å resolution structure of the proximal leg revealed CHC repeats (CHCRs) comprising 10 helices of 10–12 residues each connected by loops, creating two helical and two loop faces to the triskelion leg (Ybe et al., 1999). Mapping CLC position along CHC by electron microscopy and immunolabeling has generated conflicting evidence for a bent or an extended CLC conformation, suggesting possible conformational lability (Kirchhausen and Toyoda, 1993; Nathke et al., 1992). A reconstructed image of the assembled clathrin lattice between 7.9 and 12 Å resolution based on cryo-electron microscopy (cryoEM) showed the backbone of the central third of the CLC as an alpha helix bound to the loop face of the CHC (Fotin et al., 2004), as predicted by mutagenesis and modeling studies (Chen et al., 2002). How the N and C-terminal thirds of CLC interact with CHC was not evident from these studies.
Here we define the relative topological disposition of the entire CLC (bovine LCb, neuronal isoform) bound to the CHC hub by analysis of low resolution X-ray diffraction data from the crystallized complex, resolving, the CLC termini. Comparison of this disassembled fragment to assembled clathrin (Fotin et al., 2004), suggested that conformational changes in the CLC N-termini negatively regulate lattice assembly by changing CHC knee conformation. This mechanism for CLC control of assembly was verified by mutagenesis and biochemical assays and is consistent with cellular assays that indicate CLC depletion is still permissive for clathrin function.
Results
Structure determination for the clathrin hub-LCb complex at low resolution
Purified hub-LCb complex was crystallized by hanging drop vapor diffusion and formed tetragonal crystals in the spacegroup I4122, or P42212. Despite significant efforts to improve the resolution, the crystals only diffracted to 7.9Å (I4122 spacegroup). Few crystal structures have been solved at this low resolution, but even with associated limitations on revealing atomic detail, establishing the distribution of electron density can provide insight into protein topology. Crystal data was phased by molecular replacement using a model that combined multiple sources of information. An all atom model of clathrin was created based on revised sequence homology alignments for CHCRs and structurally fitted to the cryoEM structure. Loops that varied significantly between the model and the cryoEM structure were refined using a previously described loop modeling protocol and the modeled structure was energy minimized (Jacobson et al., 2004). The final molecular replacement search probe was a trimer of CHC residues 1280–1630 with all side chain atoms but no information from CLC or the CHC N-terminus (residues 1074–1279). Initial maps generated from the 7.9 Å (I4122) data clearly showed density that could be assigned to residues N-terminal to 1280 of CHC and to CLC. Individual helices were clearly defined, even outside the search model. Additional modeled CHC regions were built into maps generated by non-crystallographic symmetry averaging and refined against crystal data using a modified simulated annealing protocol with a low parameter to data ratio. Overall >90% of CLC was built, as polyalanine helices and the entire structure was refined to Rwork and Rfree of 0.42 each (Figure 1, Table 1). The previously determined 2.9 Å resolution structure of the proximal leg region (Ybe et al., 1999) fit very well into the refined electron density of the clathrin hub from the 7.9 Å (I4122) hub-LCb co-crystal (Figure 1B). Diffraction data to 9 Å resolution from a second hub-LCb crystal (P42212 spacegroup) was analyzed very conservatively, for comparison with the 7.9 Å (I4122) structure. The complete CHC structure was built into this 9 Å (P42212) data and refined using only a few rounds of standard rigid body refinement (Figure S1A and S1B).
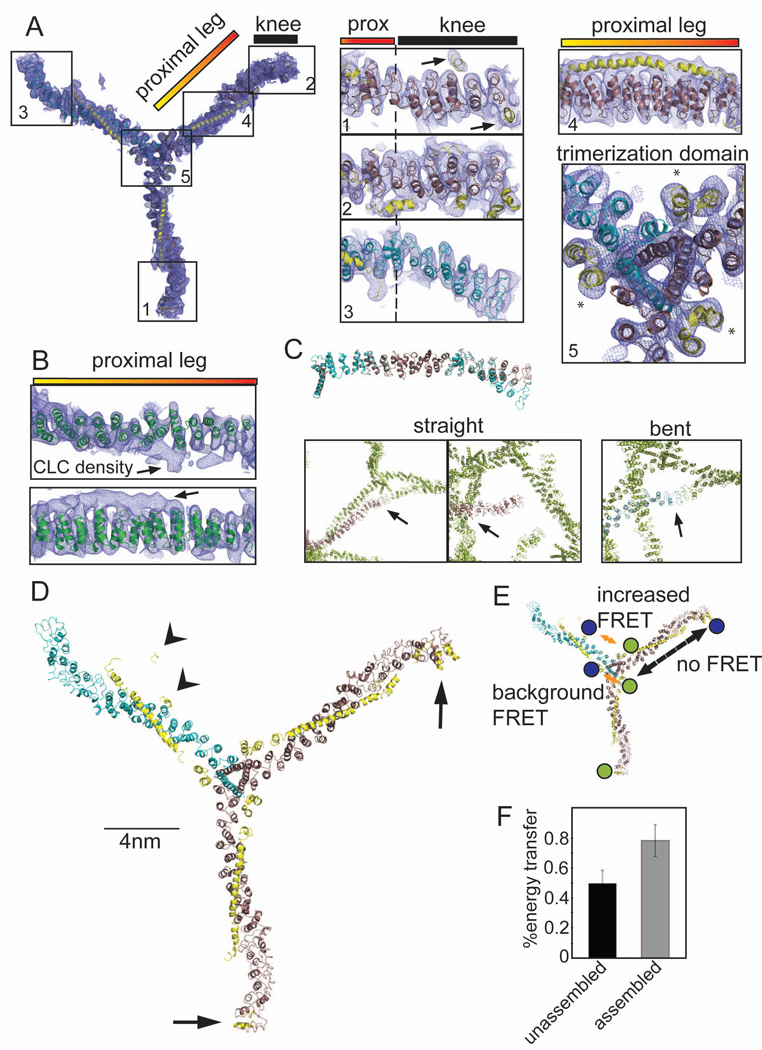
Figure 1. 7.9 Å resolution electron density of the clathrin hub-LCb complex and conformational variability of clathrin light chains.
(A) Model of the hub-LCb complex determined by X-ray crystallography with 2Fo-Fc electron density (0.06e/A3, 1.2σ, blue mesh). Details are shown in numbered boxes indicating the regions. Regions 1 and 2 show CHC straight knee conformations (brown ribbons) and region 3 shows the CHC bent knee conformation (blue ribbons), N-terminal to the proximal (prox) leg. CLC residues are shown as yellow ribbons and highlighted by arrows in region 1. Asterisks in region 5 (trimerization domain) indicate helices from the C-terminus of CLC that make contacts with two different CHCs. (B) The high resolution structure of the clathrin heavy chain proximal leg (1B89, green ribbon, Ybe et al., 1999) fitted into the 2Fo-Fc electron density from the proximal leg region. Arrows point to density corresponding to CLC. (C) Straight (brown ribbons) or bent (blue ribbons) legs from the 7.9 Å, spacegroup I4122 structure were aligned with each other in PYMOL. Boxes show contacts between crystal symmetry-mates (green) surrounding the N-terminus of straight or bent legs, colored as above. Arrows point to areas of symmetry-mate contact closest to the N-terminus of each leg. (D) Structural model of the clathrin hub-LCb complex. CHC helices are in brown (straight conformation) and blue (bent conformation) and CLC is in yellow. CLC extends from the trimerization domain to the knee region on two legs with straight knees (arrows) while on the bent leg CLC forms a more compact structure in the N-terminal region (arrowheads). (E) Diagram indicating the possible FRET interactions and the locations of the fluorescent dyes in blue and green. Orange arrows indicate distances favorable for FRET between two clathrin light chains (background at the trimerization domain or between termini of clathrin light chains). Dashed black arrow indicates a distance unfavorable for FRET. (F) CLC labeled randomly with two different fluorophores was bound to clathrin hub and these complexes were mixed with complexes of hub with unlabeled CLC to prevent intermolecular energy transfer (as determined in Figure S1D). FRET was measured in either the unassembled or the assembled state. Each bar represents the mean ±SEM (n=3).
Table 1.
Crystallographic data collection and refinement for clathrin hub-LCb complex
| 7.9 Å Native data (high resolution shell) |
9 Å Native data (high resolution shell) |
|
|---|---|---|
| Data collection | ||
| Space group | I4122 | P42212 |
| Cell dimensions (Å) | a=b=228.5 c=710.3 | a=b=229.7 c=512.2 |
| Wavelength (Å) | 1.1 | 1.1 |
| Resolution (Å) | 250–7.94 (8.55–7.94) | 500–9 (9.23–9.00) |
| Rsym (%) | 7.5 (88.2) | 17.7 (60.5) |
| I/σI | 4.7 (2.8) | 5.3 (1.5) |
| Completeness (%) | 99 (97.2) | 78.7 (78.1) |
| Redundancy | 9.0 (9.7) | 3.1 (3.2) |
| Total # Obs. Unique | 10,700 (2,079) | 8,530 (838) |
| Reflections | ||
| Refinement | Rigid Body Simulated Annealing |
Standard Rigid Body |
| Resolution (Å) | 100–7.9 | 500–9 |
| # of Reflections used in refinement |
10,208 (test set 566) |
8,097 (test set 431) |
| Rwork | 0.42 | 0.48 |
| Rfree (5%) | 0.42 | 0.47 |
| Average B factor (Å2) | 295.203 | 131.8 |
Many tests were used to determine the validity of structural features of the hub-LCb complex established from the 7.9 Å (I4122) X-ray diffraction data, while a minimal refinement procedure was used for the 9 Å (P42212) data (Figure S1 and S2). For the structure generated from the 7.9 Å (I4122) data, decoy poly-alanine helices misplaced into solvent density always returned negative density after refinement, verifying the refinement protocol (Figure S2D). Only helices built into density greater than 0.9 σ reliably improved the refinement and only when near the protein envelope. Based on these tests, a strict cut-off of 1.2 σ was use for placing helices. Test models of the clathrin hub in a variety of conformations were assessed for correspondence to the molecular replacement solution after simple rigid body refinement and could be clearly identified as matching the electron density or not, supporting further the validity of the model refinement.
The final model derived from the 7.9 Å (I4122) crystal localized CLC density relative to CHC density (Figure 1), in molecular detail comparable to a cryoEM map. The residue numbers of most CHC helices could be assigned by extension from the search model, but numbering the predicted CLC helices was not possible. Nonetheless, the relative disposition of the two subunits revealed conformational differences between the clathrin legs that were evident in models generated from both the 7.9 Å (I4122) and the 9.0 Å (P42212) crystals (Figure 1; Figure S1A–S1C).
Identification of conformational changes in clathrin
In both crystals there is one hub-LCb complex per asymmetric unit and two different conformations of hub legs. One crystal contact in each of the structures might reflect assembly-like interactions but most contacts were compatible with hub in a disassembled state. Comparison to the cryoEM lattice structure (Fotin et al., 2004) revealed that the “pucker” of the unassembled CHC hub is similar to that seen for assembled triskelia (Movie S1) and that the proximal leg regions are similarly rigid and straight (Figure 1A, 1C, and 1D; Figure S1A). In contrast, the knee regions of legs in both determined structures displayed either a “bent” conformation or a “straight” conformation (Figure 1A, 1C 1D; Movie S1; Figure S1A–C). The legs with bent knees from both determined structures aligned well with the structure of CHC from cryoEM reconstructions (Fotin et al., 2004) (Figure S1C). The two straight knee conformations in the 7.9 Å (I4122) crystal have different contacts suggesting contacts are not significantly influencing the observed conformations (Figure 1C). In the 7.9 Å (I4122) structure, the straight knees are associated with extended CLCs (Figure 1A, and 1D; Movie S1) with assigned CLC density extending from the CHC trimerization domain along the proximal leg to the crease of the CHC knee where the CLC N-termini are bound. For one of these extended CLCs, continuous density assignable to most of the CLC was resolved. The bent knee displayed by the third leg has no detectable CLC density in the knee region (Figure 1A and 1D), suggesting a more compact structure for the associated CLC. Density assigned to this compact CLC is present at the trimerization domain and spans about half of the proximal leg (Figure 1D; Movie S1), with the CLC N-terminus bent away from the CHC knee region. It is unclear whether this exact positioning is physiological, though the release of the N-terminus from the knee region appears to be critical for regulating assembly of clathrin. In both structures, density assignable to the C-terminus of each CLC interacts with helices derived from two different CHCs at the hub vertex (Figure 1A, trimerization domain), explaining the biochemical and genetic evidence that CLCs contribute stability to the trimerization domain (Chen et al., 2002; Huang et al., 1997; Pishvaee et al., 1997; Wang et al., 2006; Ybe et al., 2007).
Clathrin light chain switches conformations during hub assembly
Correspondence of the hub leg with a bent knee conformation to legs in the assembled clathrin lattice (Figure S1C) suggested that bent knees with the compact CLC conformation are compatible with assembly, while straight knees with extended CLC are characteristic of disassembled hub. To test whether CLCs switch conformations upon assembly, the proximity of their N and C termini was assessed in disassembled and assembled hub-CLC complexes using Förster resonance energy transfer (FRET). To attach fluorescent probes near the CLC termini, the neuronal isoform of human LCb, was engineered to have a single cysteine at either terminus, by introducing two mutations (S9C and C190S). Then labeling with a mixture of Alexa 555 and Alexa 647 yielded CLCs with a different fluorophore at either terminus or labeled with the same fluorophore (Figure 1E). Labeled CLCs were bound to CHC hubs and their FRET measured under disassembled conditions. The extended conformation of CLC is too distant for significant FRET between coupled fluorophores, but a background level of FRET (efficiency of 49% ± 8%) results from different fluorophores at the C-termini of CLCs juxtaposed at the hub trimerization domains and from some CLCs stochastically adopting the compact conformation (Figure 1E and 1F). Hubs with labeled CLCs were then mixed with an excess of hubs with unlabeled CLCs, and FRET was measured following induction of assembly. The FRET signal was significantly increased (efficiency of 78% ± 10%) when hub-CLC complexes were assembled (Figure 1F), indicating increased proximity of the CLC termini. The excess of hubs with unlabeled CLC eliminates the possibility that increased FRET upon assembly results from interactions between triskelia. This was established empirically by titration of unlabeled hubs into the assembly reaction (Figure S1D). These FRET data confirm that CLC switches to a more compact conformation when clathrin assembles.
Electrostatic assembly regulation by clathrin light chain involves a loop at the heavy chain knee
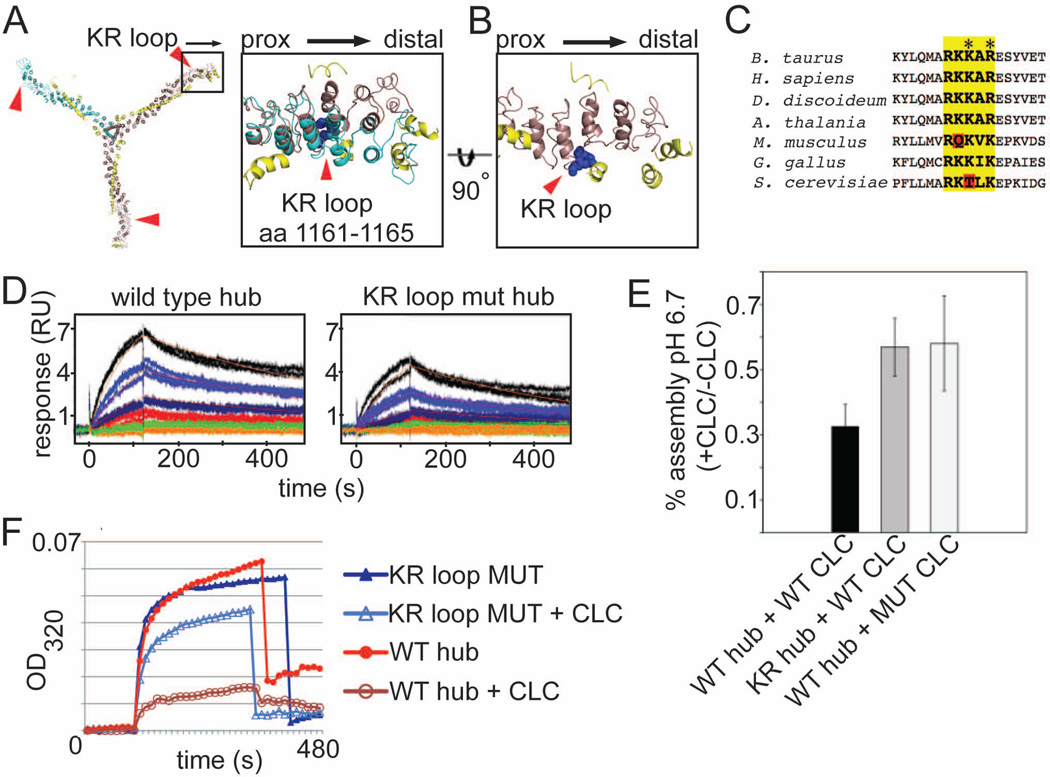
In the extended CLC conformation, the N-terminal CLC residues interact with CHC residues in the crease of the knee, near a loop comprised of basic residues (R1161KKAR1165 or KR loop) (Figure 2A–2C). This positioning and its evolutionary conservation (Figure 2C) suggested the possibility that the KR loop might be the complementary half of an interaction with the N-terminal EED sequence of CLC that controls pH sensitivity of clathrin assembly. The KR loop is also the hinge point for the straight and bent CHC knee (Figure 2A and 2B), situating that loop at a site where CHC knee conformation would be affected by CLC binding. To test the role of the KR loop in assembly regulation, recombinant CHC hub fragments were mutated at K1163 and R1165 to glutamate and aspartate, respectively. Both wild-type and KR-mutant CHC hub bound CLC with similar affinity revealing a slight difference in on rate, with no difference in off rate (Figure 2D). Both hub forms were fully saturated with CLC and the complexes tested for assembly properties. When CLC was bound to wild-type hub, it inhibited assembly at pH 6.7, as reported (Ybe et al, 1998). When CLC was bound to the KR-mutant hub, its ability to inhibit hub assembly at pH 6.7 was significantly impaired (Figure 2E and 2F). Thus the KR loop is implicated in assembly control. The partial susceptibility of the KR-mutant to CLC regulation suggests either that additional minor CLC-CHC interactions contribute to assembly regulation or that the remaining positively charged residues in the KR loop can still interact weakly with the EED sequence. These positively charged residues were retained in the mutant to minimize perturbation of the CHC structure. Mutation of the E20ED22 sequence to KKK (Figure 2E) or QQN (Ybe et al., 1998) in LCb impaired CLC control of assembly similarly to the KR loop mutation. Attempts to regain regulation by combining the charge switch mutant of CHC (KR-mutant) with that of CLC (EED to KKK) led to inconclusive results, likely due to failure to recapitulate local pKa’s required for their electrostatic interaction. These mutagenesis experiments suggest the KR loop on CHC is the binding partner of the regulatory EED sequence of CLC and support a central role for their interaction in assembly regulation. Localization of this electrostatic switch to CLC-knee interactions and not to the assembly interface of the lattice suggests negative regulation of clathrin assembly by CLCs results from their effect on CHC knee bending.
Figure 2. The KR loop in the clathrin heavy chain knee participates in electrostatic regulation of assembly by clathrin light chain.
(A) Expanded boxed region shows the position of the KR loop (blue spheres, residues 1161–1165 in CHC) relative to CLC. The straight knee conformation (brown) associated with the extended form of CLC (yellow) and the bent knee conformation (cyan) associated with the compact form of CLC (not shown) are overlaid. The KR loop is indicated by red arrowheads. Major deviations between the overlaid structures occur distal to the KR loop. (B) Expanded box region of a, rotated 90° (straight knee only). (C) Alignment of various CHC protein sequences around the KR loop (bold, yellow highlight). Red highlights show differences that alter the charge distribution. Residues mutated to alter the KR loop charge indicated by asterisks (K1163E and R1165D). (D) Surface plasmon resonance (SPR) analysis of clathrin light chain interactions with wild type clathrin hub or KR loop mutant (mut) hub. Human neuronal LCb was immobilized on the SPR chip surface and wild type or KR loop mutant hub flowed over at different concentrations. The fitted binding affinity for wild type was 25 nM while KR loop mutant was 59.1 nM. The difference in affinity for CLC was due to a slowed on-rate for the KR loop mutant likely due to decreased electrostatic attraction. (E) Purified hubs (wild-type, WT or mutant, KR) were saturated with CLC (wild type neuronal LCb (WT CLC) or neuronal LCb with E20ED22 mutated to K20KK22 (MUT CLC)) or left unoccupied and assayed for assembly at pH 6.7 by light scattering. The ratio of assembly signal for hub with light chain to assembly signal for hub without light chain for is plotted for each reaction (mean±SEM). (F) Representative kinetic assembly data for WT or KR loop mutant (MUT) hub with or without CLC bound, as indicated. Assembly is detected photometrically by an increase in light scatter at OD320.
Modeling clathrin light chain regulation of assembly
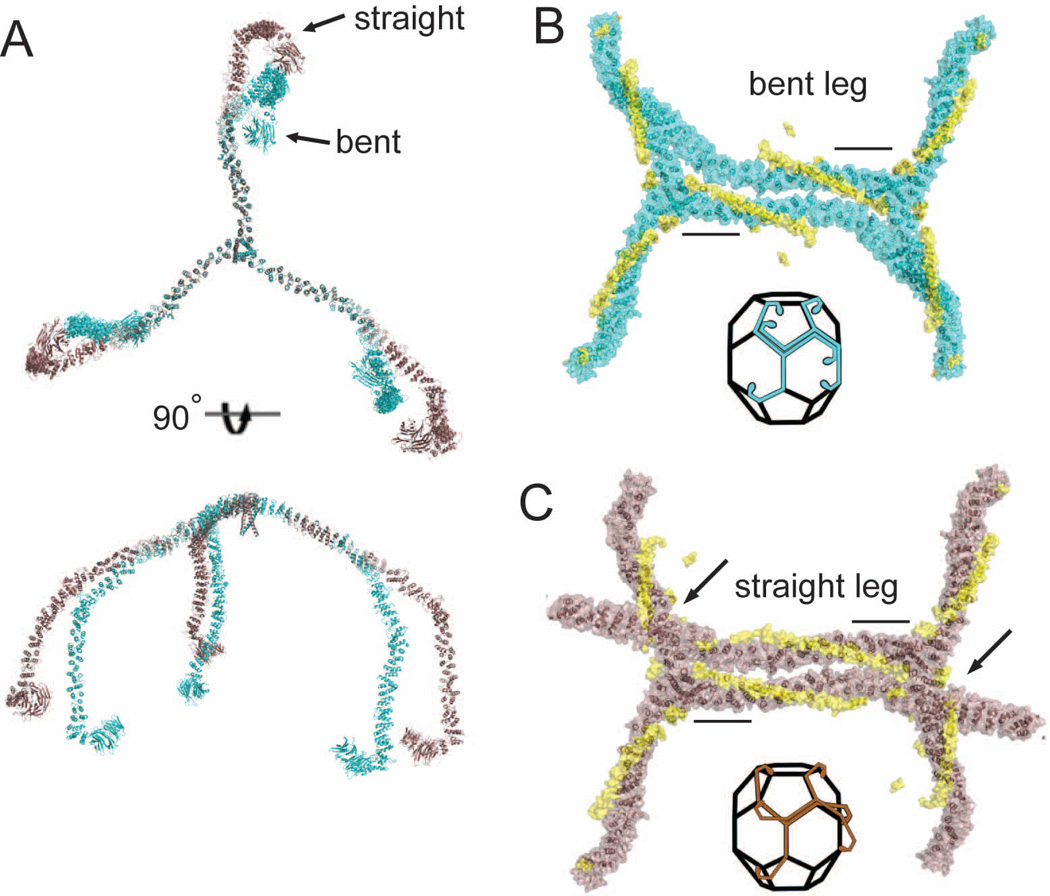
The consistent angle of orientation of triskelion legs is lost when clathrin light chains are removed, supporting a role for CLC in controlling knee bending (Schmid et al., 1982; Ungewickell, 1983). To visualize CLC-induced changes in CHC bending and how they relate to the mechanism of clathrin assembly, complete clathrin triskelia with straight or bent knees were modeled by structural alignment (Figure 3A), revealing an obvious difference in knee angle. Triskelion hubs with one full bent or straight leg modeled were then aligned with the hub regions in an assembled lattice (at both a hexagon (Figure 3B and 3C) and pentagon edge). This alignment revealed that the straight knee conformation would clash with adjacent triskelia, and be refractory to assembly (Figure 3C). Critical assembly interactions involving the distal leg and the ankle domains have been predicted (Fotin et al., 2004; Wakeham et al., 2003) and both would be misaligned in the extended CLC-straight CHC knee conformation (Figure 3C). The compact CLC conformation, in which CLC is dissociated from the knee, is permissive for assembly by allowing the CHC knee to bend and accommodate to either hexagonal or pentagonal angles.
Figure 3. Modeling clathrin light chain-induced conformational changes in the triskelion knee and comparison to the assembled clathrin lattice.
(A) Clathrin heavy chain triskelia with all straight knees or all bent knees were modeled by alignment of distal legs (residues 1–1130) from the cryoEM structure of Fotin et al. (2004) with the hub structures determined here. Straight conformation (brown) and bent conformation (cyan) of knee regions are shifted ~30° starting from the KR loop hinge point. (B and C) Alignment of two modeled hubs, both with a single full length bent knee leg (B) or straight knee leg (C), with two assembled triskelia in the cryoEM structure (Fotin et al., 2004)(1XI4). Black bars indicate regions used to align cryoEM and hub crystal structures and the clathrin light chains are shown in yellow in their relative positions in the hub-LCb complex. Arrows indicated point of steric clashes in C. Insets indicate the geometric alignment of the bent or straight leg triskelia as modeled in (A) along edges of the clathrin basket.
Discussion
In this study we propose and validate a model for how CLCs negatively regulate clathrin assembly. The model is based on structural features determined from low resolution diffraction data obtained from two different crystals of hub-LCb complexes. We used a combination of molecular replacement and non-crystallographic symmetry averaging to refine X-ray data at 7.9 and 9.0 Å resolution, and found that it was possible to clearly assign density to each clathrin subunit. Determined density maps revealed similar protein topology from the two different data sets with different space groups and crystal contacts, generating confidence in our structural analysis. As expected this approach did not reveal structural details at atomic resolution. However, coordinated conformational differences in CLC and CHC were clearly observed, suggesting that CLC’s negative regulation of clathrin assembly is due to its ability to alter CHC knee conformation. This hypothesis was validated biochemically by mapping assembly control residues to the CHC knee and the CLC N-terminus, locating an electrostatic interaction known to regulate assembly. Additionally, conformational changes of CLC during clathrin assembly were verified by FRET during assembly reactions. Together the structural and functional data described here suggest that electrostatic interactions control whether the CLC is bound to the CHC knee. If CLC is bound, the knee is straight and assembly is inhibited. If the CLC is retracted from the knee, in a more compact state, assembly is favored. This model explains the pH sensitivity of CLC regulation of clathrin assembly in vitro and provides a starting point for understanding cellular control of clathrin assembly.
The model established here predicts that absence of CLC creates triskelia with greater knee flexibility that should be able to assemble. Compatible with this prediction, previous biochemical analysis has shown that clathrin can assemble without CLC in vitro and will assemble from pieces when the distal leg is completely severed from the knee (Greene et al., 2000; Ungewickell et al., 1982). Our model thereby explains clathrin’s ability to assemble and function in tissue culture cells depleted of CLC (Huang et al., 2004; Poupon, et al. 2008). Proteomic analysis of CCVs from different tissues suggests that CLC expression levels are stoichiometric with CHC only in brain (Girard et al., 2005). Fewer CLCs per triskelion in non-neuronal tissue would decrease the influence of CLCs on assembly and render various assembly-promoting adaptors more influential. Thus, we propose that the ability of CLC to control clathrin assembly is more effective in neurons where clathrin assembly has to be finely coordinated for synaptic vesicle protein recapture and is additionally controlled by neuron-specific versions of adaptors (Brodsky et al., 2001). This is consistent with the observation that in non-neuronal tissue the main detectable phenotype of CLC disruption is related to the Hip-binding function of CLCs and consequent regulation of the actin cytoskeleton, rather than an effect on CHC assembly (Poupon et al., 2008; Chen and Brodsky, 2005; Wilbur et al., 2008). It is interesting to note the predicted phosphorylation sites for LCb (Hill et al., 1988) are located in the knee-binding region, so phosphorylation might be another mechanism to regulate knee flexibility. The Hip proteins also bind the N-terminal region of CLC and promote assembly of clathrin in vitro (Chen and Brodsky, 2005; Legendre-Guillemin et al., 2005). These potential regulatory features suggest that knee bending might be gradually manipulated such that CLC conformation could influence the hexagon-pentagon switch during lattice curvature, a proposed function of knee flexibility (Musacchio et al., 1999). In conclusion we have localized full length CLC relative to the CHC and defined the mechanism for CLC regulation of clathrin lattice assembly. This should provide a framework for establishing how other modes of regulation occur in clathrin mediated-endocytosis, particularly in neurons, and generate new concepts for understanding regulation of self-assembling systems in general.
Experimental Procedures
Protein production and assembly assays
Bovine clathrin heavy chain residues 1074–1675 (hub) were expressed and purified as described (Liu et al., 1995). Bovine neuronal clathrin light chain b (LCb) used in crystallography was expressed and purified as described (Ybe et al., 1998). Human neuronal LCb was expressed and purified as a his-tagged fusion protein for assembly assays, FRET assays and SPR. His-tagged LCb behaved identically to untagged bovine LCb (data not shown). Assembly assays were performed as described (Ybe et al., 1998).
Crystal growth and data collection
Crystals were grown in 200mM citrate, 16–22% glycerol, 2% trifluoroethanol. Data was collected at the Advanced Light Source beamline 8.3.1. To collect low resolution data, two separate data sets were collected. The highest resolution reflections were collected using standard procedures. To gain adequate separation of the very low resolution reflections in the second data set, the detector was moved to the maximum distance from the crystal. With no further modifications the majority of the expected low resolution reflections could be accurately measured.
Data for the I4122 spacegroup was originally processed to 8.3 Å with an I/sigma I of approximately 4 using DENZO. To improve the resolution, maps were calculated using phases calculated from a molecular replacement solution (described below). Data was then processed to higher resolution in 0.1 Å or 0.2 Å steps, using HKL2000, despite worsening processing statistics and maps recalculated. This iterative process continued until visual inspection of the maps showed no improvement.
Molecular replacement model development
To generate an accurate model for molecular replacement, comparative models of the clathrin heavy chain repeats were generated. Structural alignment to the cryoEM structure (pdb code 1×14), loop modeling and energy minimization were used to build a complete model of CHC. This all atom model was truncated to a trimer of residues 1280–1630 and used successfully to find a molecular replacement solution where more simplistic search models had failed.
Phasing and Refinement of Crystal Data
Crystal data was phased by molecular replacement using Phaser (McCoy et al., 2007), built with COOT (Emsley and Cowtan, 2004), and refined with CNS (Brunger, 2007; Brunger et al., 1998) or REFMAC (Murshudov et al., 1997).
The following strict guidelines for building into maps were followed to help prevent over-interpretation of the low resolution information. All new structural elements were built into 2Fo-Fc map density scaled to 0.06 e/Å3 (1.2 σ) or higher with positive Fo-Fc density associated with it. Novel regions from CLC were built as poly-alanine helices due to the low resolution of the maps. Since the CLC was not a continuous chain, the numbering of newly built residues is arbitrary. Building and numbering of clathrin heavy chain was based upon the refined all atom model used for molecular replacement.
To improve the maps prior to model building for the 7.9 Å (I4122) data, two steps were taken. First one round of rigid body refinement was performed using REFMAC, with each leg within the molecular replacement search model defining a rigid body. This was performed to be sure the position of each leg was correct within the trimer. NCS averaging of maps (FOM weighted phases with observed amplitudes) generated from the first round of rigid body refinement, across all three legs, caused density near the N-terminus of CHC to disappear, while density for the helical region of clathrin light chain became more defined (Figure S2B). NCS averaging all possible combinations of two legs led to determination that one randomly assigned pair of legs retained density at the N-terminus (Figure S2B). This suggests that these two legs are very similar in conformation while the third (chain C) is different.
Adjustments to the model were initially followed by simple rigid body refinements with two domains for each leg and the domain boundary at the flexible knee joint. Further rigid body refinements after model adjustments occured with increasingly smaller domains strongly restrained by the NCS. Details of the domain boundaries and NCS restraint parameters can be found in the supplementary information.
Subsequent refinement consisted of NCS restrained rigid body refinement alternating with NCS restrained domain B factor refinement. Final rounds of refinement, used to help minimize model bias, were achieved through simulated annealing with individual helices as rigid body groups while maintaining NCS restraints. The NCS restraints were gradually decreased through the last rounds of simulated annealing. Overall less than 2000 parameters were refined against greater than 10,000 reflections. Maps calculations for refinement steps were either with CNS 1.2 including default optimization of bulk solvent mask scaling parameters or with REFMAC with solvent scaling parameters, ionic radius and shrink equal to 2.0. This was necessary for calculation of high quality maps similar to previously determined low resolution structures (DeLaBarre and Brunger, 2003, 2006). The 9 Å (P42212) data was phased by molecular replacement and minimally refined with standard rigid body refinements.
Testing for model bias with decoy building
The intentional addition of helices outside but near 2Fo-Fc map density for the 7.9 Å (I4122) was used as a test for building with minimal model bias (Figure S2D). Helices placed in density weaker than 0.9 σ consistently appeared in negative density upon refinement and produced higher Rwork or Rfree values. The density, especially that of the N-termini of the clathrin light chains, represents the position of the domain relative to other domains of both CLC and CHC.
Modeling complete triskelia by combination of new crystallographic data with cryoEM data
Structural alignments between elements from the crystal structure reported here and the cryoEM structure by Fotin et al. (pdb 1XI4) were used to model full triskelia. Full legs were built by aligning the distal leg (residues 1–1130 of the cryo-EM lattice) with residues 1074–1130 from the crystal structure, which are N-terminal to the hinge point in the knee region (red arrowheads, Fig. 2A–2C). Three straight legs or three bent legs were trimerized by alignment with the C-terminal part of the hub. Crystal structures and data have been deposited to the Protein Databank with codes 3LVG and 3LVH.
Förster resonance energy transfer
FRET assays were performed by labeling a double mutant of LCb (S9C and C190S) with a mixture of Alexa 555 and Alexa 647 fluorescent dyes and measuring steady state energy transfer under assembly or disassembly conditions.
Surface plasmon resonance
For SPR analysis LCb was covalently coupled to the surface of a CM5 flow cell (Biacore) and clathrin hub was flowed over the surface. Data was collected on a Biacore T100 biosensor and processed with Scrubber (University of Utah, Center for Biomolecular Interactions).
Acknowledgements
We thank Dr. Luke Rice and Dr. Pascal Egea for technical assistance with X-ray crystallography and discussions. This work was supported by NIH grants GM038093 to F.M.B and S10RR023443 to R.J.F. and a grant for interdisciplinary research to M.P.J. and F.M.B. from the UCSF School of Pharmacy.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Brodsky FM, Chen CY, Knuehl C, Towler MC, Wakeham DE. Biological basket weaving: formation and function of clathrin-coated vesicles. Annu Rev Cell Dev Biol. 2001;17:517–568. doi: 10.1146/annurev.cellbio.17.1.517. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Chen CY, Brodsky FM. Huntingtin-interacting protein 1 (Hip1) and Hip1-related protein (Hip1R) bind the conserved sequence of clathrin light chains and thereby influence clathrin assembly in vitro and actin distribution in vivo. J Biol Chem. 2005;280:6109–6117. doi: 10.1074/jbc.M408454200. [DOI] [PubMed] [Google Scholar]
- Chen CY, Reese ML, Hwang PK, Ota N, Agard D, Brodsky FM. Clathrin light and heavy chain interface: alpha-helix binding superhelix loops via critical tryptophans. Embo J. 2002;21:6072–6082. doi: 10.1093/emboj/cdf594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Considerations for the refinement of low-resolution crystal structures. Acta Crystallogr D Biol Crystallogr. 2006;62:923–932. doi: 10.1107/S0907444906012650. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- Girard M, Allaire PD, McPherson PS, Blondeau F. Non-stoichiometric relationship between clathrin heavy and light chains revealed by quantitative comparative proteomics of clathrin-coated vesicles from brain and liver. Mol Cell Proteomics. 2005;4:1145–1154. doi: 10.1074/mcp.M500043-MCP200. [DOI] [PubMed] [Google Scholar]
- Greene B, Liu SH, Wilde A, Brodsky FM. Complete reconstitution of clathrin basket formation with recombinant protein fragments: adaptor control of clathrin self-assembly. Traffic. 2000;1:69–75. doi: 10.1034/j.1600-0854.2000.010110.x. [DOI] [PubMed] [Google Scholar]
- Hill BL, Drickamer K, Brodsky FM, Parham P. Identification of the phosphorylation sites of clathrin light chain LCb. J Biol Chem. 1988;263:5499–5501. [PubMed] [Google Scholar]
- Huang F, Khvorova A, Marshall W, Sorkin A. Analysis of clathrin-mediated endocytosis of epidermal growth factor receptor by RNA interference. J Biol Chem. 2004;279:16657–16661. doi: 10.1074/jbc.C400046200. [DOI] [PubMed] [Google Scholar]
- Huang KM, Gullberg L, Nelson KK, Stefan CJ, Blumer K, Lemmon SK. Novel functions of clathrin light chains: clathrin heavy chain trimerization is defective in light chain-deficient yeast. J Cell Sci. 1997;110(Pt 7):899–910. doi: 10.1242/jcs.110.7.899. [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Toyoda T. Immunoelectron microscopic evidence for the extended conformation of light chains in clathrin trimers. J Biol Chem. 1993;268:10268–10273. [PubMed] [Google Scholar]
- Legendre-Guillemin V, Metzler M, Lemaire JF, Philie J, Gan L, Hayden MR, McPherson PS. Huntingtin interacting protein 1 (HIP1) regulates clathrin assembly through direct binding to the regulatory region of the clathrin light chain. J Biol Chem. 2005;280:6101–6108. doi: 10.1074/jbc.M408430200. [DOI] [PubMed] [Google Scholar]
- Liu SH, Wong ML, Craik CS, Brodsky FM. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell. 1995;83:257–267. doi: 10.1016/0092-8674(95)90167-1. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of Applied Crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Smith CJ, Roseman AM, Harrison SC, Kirchhausen T, Pearse BM. Functional organization of clathrin in coats: combining electron cryomicroscopy and X-ray crystallography. Mol Cell. 1999;3:761–770. doi: 10.1016/s1097-2765(01)80008-3. [DOI] [PubMed] [Google Scholar]
- Nathke IS, Heuser J, Lupas A, Stock J, Turck CW, Brodsky FM. Folding and trimerization of clathrin subunits at the triskelion hub. Cell. 1992;68:899–910. doi: 10.1016/0092-8674(92)90033-9. [DOI] [PubMed] [Google Scholar]
- Pishvaee B, Munn A, Payne GS. A novel structural model for regulation of clathrin function. Embo J. 1997;16:2227–2239. doi: 10.1093/emboj/16.9.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poupon V, Girard M, Legendre-Guillemin V, Thomas S, Bourbonniere L, Philie J, Bright NA, McPherson PS. Clathrin light chains function in mannose phosphate receptor trafficking via regulation of actin assembly. Proc Natl Acad Sci U S A. 2008;105:168–173. doi: 10.1073/pnas.0707269105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid SL, Matsumoto AK, Rothman JE. A domain of clathrin that forms coats. Proc Natl Acad Sci U S A. 1982;79:91–95. doi: 10.1073/pnas.79.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E. Biochemical and immunological studies on clathrin light chains and their binding sites on clathrin triskelions. Embo J. 1983;2:1401–1408. doi: 10.1002/j.1460-2075.1983.tb01598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungewickell E, Unanue ER, Branton D. Functional and structural studies on clathrin triskelions and baskets. Cold Spring Harb Symp Quant Biol. 1982;46(Pt 2):723–731. doi: 10.1101/sqb.1982.046.01.069. [DOI] [PubMed] [Google Scholar]
- Ungewickell E, Ungewickell H. Bovine brain clathrin light chains impede heavy chain assembly in vitro. J Biol Chem. 1991;266:12710–12714. [PubMed] [Google Scholar]
- Ungewickell EJ, Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol. 2007;19:417–425. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Wakeham DE, Chen CY, Greene B, Hwang PK, Brodsky FM. Clathrin self-assembly involves coordinated weak interactions favorable for cellular regulation. Embo J. 2003;22:4980–4990. doi: 10.1093/emboj/cdg511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang Y, O'Halloran TJ. Clathrin light chain: importance of the conserved carboxy terminal domain to function in living cells. Traffic. 2006;7:824–832. doi: 10.1111/j.1600-0854.2006.00438.x. [DOI] [PubMed] [Google Scholar]
- Wilbur JD, Chen CY, Manalo V, Hwang PK, Fletterick RJ, Brodsky FM. Actin binding by Hip1 (huntingtin-interacting protein 1) and Hip1R (Hip1-related protein) is regulated by clathrin light chain. J Biol Chem. 2008;283:32870–32879. doi: 10.1074/jbc.M802863200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ybe JA, Brodsky FM, Hofmann K, Lin K, Liu SH, Chen L, Earnest TN, Fletterick RJ, Hwang PK. Clathrin self-assembly is mediated by a tandemly repeated superhelix. Nature. 1999;399:371–375. doi: 10.1038/20708. [DOI] [PubMed] [Google Scholar]
- Ybe JA, Greene B, Liu SH, Pley U, Parham P, Brodsky FM. Clathrin self-assembly is regulated by three light-chain residues controlling the formation of critical salt bridges. Embo J. 1998;17:1297–1303. doi: 10.1093/emboj/17.5.1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ybe JA, Perez-Miller S, Niu Q, Coates DA, Drazer MW, Clegg ME. Light chain C-terminal region reinforces the stability of clathrin heavy chain trimers. Traffic. 2007;8:1101–1110. doi: 10.1111/j.1600-0854.2007.00597.x. [DOI] [PubMed] [Google Scholar]