Abstract
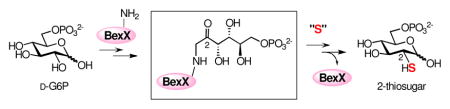
The first mechanistic insight into the 2-thiosugar production in an angucycline-type antibiotic, BE-7585A is reported. D-Glucose 6-phosphate was identified as the substrate for the putative thiosugar biosynthetic protein, BexX, by trapping the covalently bonded enzyme-substrate intermediate. The site-specific modification at K110 residue was determined by mutagenesis studies and LC-MS/MS analysis. A key intermediate carrying a keto functionality was confirmed to exist in the enzyme-substrate complex. These results suggest that the sulfur insertion mechanism in 2-thiosugar biosynthesis shares similarities with that for thiamin biosynthesis.
Highly modified sugars are commonly found in prokaryotes’ secondary metabolites.1,2 These sugars are vital components for the efficacy and specificity of many biologically active natural products,2 such that altering and/or exchanging these crucial sugar structures may enhance or vary the physiological characteristics of their parent molecules.2 Exploitation of the biosynthetic machineries is a powerful approach to generate new sugars,2,3 but requires a thorough understanding of the biosynthetic pathway of each target sugar including the genetic, enzymatic, and mechanistic information. Previous efforts have achieved notable advances in our understanding of the formation of deoxy-, amino- and branched-chain sugars.4 However, knowledge regarding the biosynthesis of thiosugars has been scarce due to the rareness of its natural occurrence. In fact, only a handful of thiosugar-containing natural products have been isolated thus far.5 Here, we report the first mechanistic investigation of the biosynthesis of a 2-thiosugar found in an angucycline-type antibiotic BE-7585A (1)6 produced by the soil bacteria, Amycolatopsis orientalis subsp. vinearia BA-07585. Notably, BE-7585A is one of the only two known natural products containing a 2-thiosugar moiety. Our results provide strong evidence supporting the intermediacy of a 2-keto-substrate–enzyme adduct and the involvement of a series of isomerization reactions as early steps in the pathway.
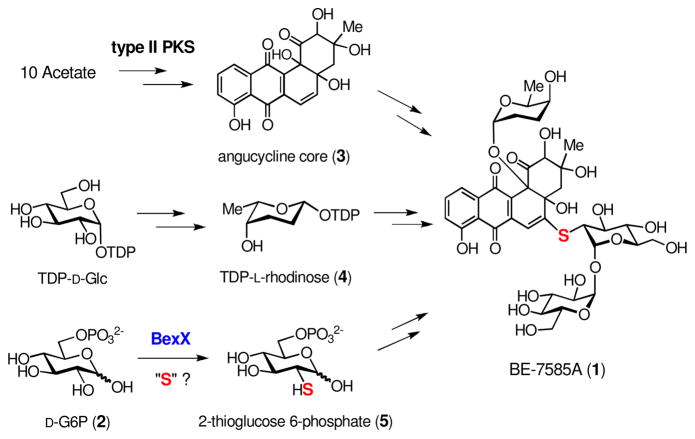
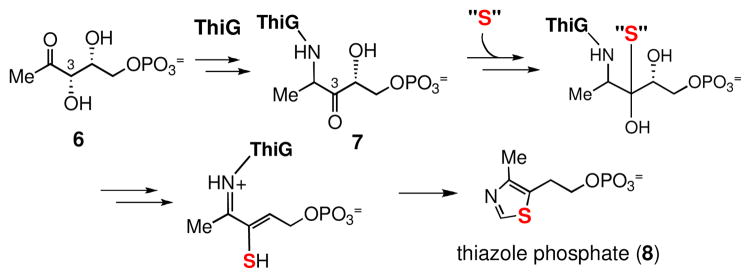
The biosynthetic gene cluster for BE-7585A (1) was recently identified by PCR-based screening of the cosmid library of A. orientalis, and a biosynthetic pathway for its formation was proposed (Scheme 1).5 Along with the genes encoding type II polyketide synthases (PKS) and rhodinose biosynthetic enzymes, a gene, bexX, whose translated sequence displays good similarity to thiazole synthases, ThiG (58% identity to ThiG from Stigmatella aurantiaca DW4/3-1;7 38% identity to ThiG from Bacillus subtilis subsp. subtilis str 1688) was also located in the gene cluster. Studies of thiamine biosynthesis in B. subtilis showed that the ThiG reaction is initiated by the Schiff base formation between Lys96 of ThiG and the 2-keto group of 1-deoxy-D-xylulose-5-phosphate (DXP, 6, Scheme 2).9 A sulfur carrier protein then delivers the sulfur atom to the keto moiety at C-3 of the DXP-lysine-ThiG adduct (7).10 Since ThiG is a key enzyme in the formation of thiazole phosphate (8) of thiamine, the newly identified bexX may also play a key role in the biosynthesis of 2-thiosugar in A. orientalis, and the mechanism of sulfur incorporation into the 2-thiosugar in BE-7585A could resemble that of ThiG reaction in thiamin biosynthesis.5
Scheme 1.
Scheme 2.
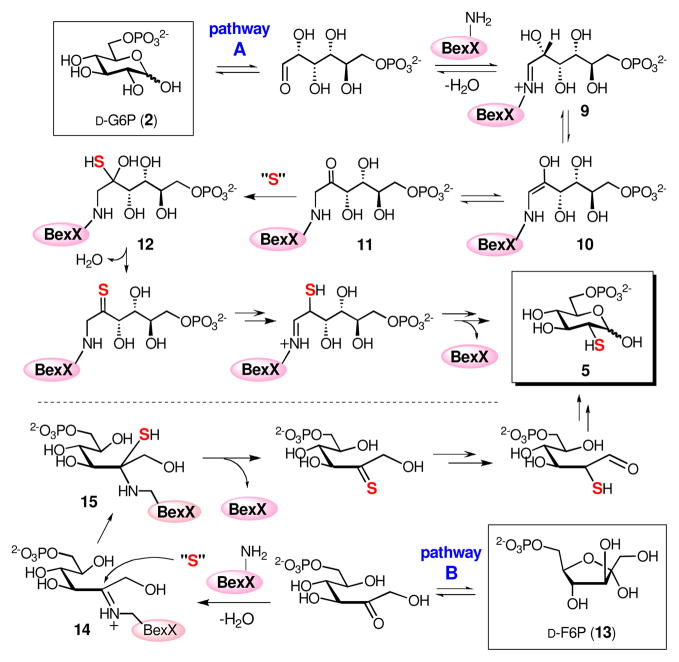
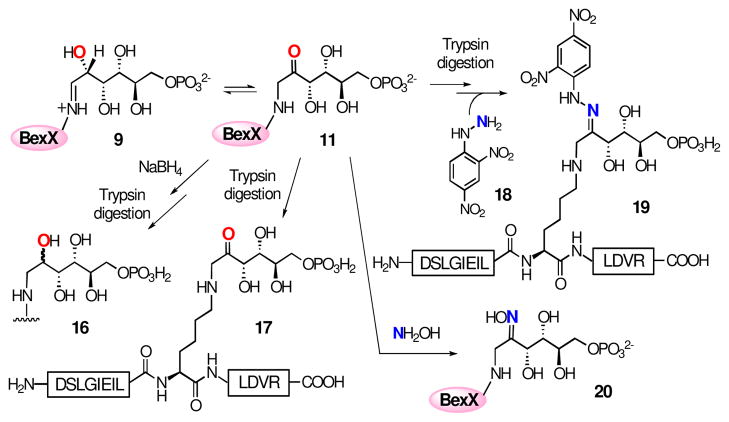
On the basis of the structure of 2-thiosugar in BE-7585A (1), D-glucose 6-phosphate (D-G6P, 2) is a postulated substrate for BexX. A possible biosynthetic pathway for 2-thiosugar is shown in Scheme 3 (pathway A). An active-site lysine residue of BexX may first form a Schiff base with the linear form of 2 at C-1. The resultant imine intermediate 9 facilitates abstraction of proton at C-2 to give an enamine intermediate 10. The enamine 10 is then tautomerized to C-2 ketone 11, which can be attacked by an activated sulfur donor to incorporate a sulfur atom at C-2. Alternatively, the hexose monophosphate substrate may be D-fructose 6-phosphate (D-F6P, 13), which could form a Schiff base with the active-site lysine at C-2 (14, Scheme 3, pathway B) to directly activate the target position as seen in glucosamine biosynthesis.11
Scheme 3.
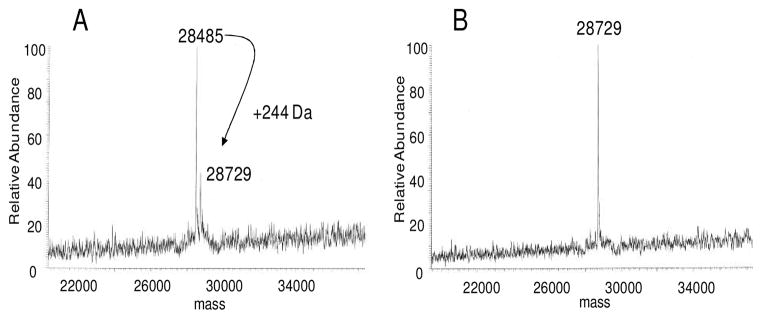
In order to verify the predicted function of BexX, the corresponding protein, BexX, was heterologously expressed in Escherichia coli and purified as a C-terminal His6-tagged protein (see Figure S1).12 Interestingly, ESI-MS of the isolated protein displayed two peaks. The dominant peak corresponds to the expected His6-tagged BexX (calcd 28488, obsd 28485), and the minor peak shows a mass increase of ~240 Da than the parent peak. Upon NaBH4 reduction, the second peak of the reduced enzyme became more apparent (obsd 28729) (Figure 1A), and the mass increase (244 Da) is consistent with a reduced dehydration adduct between BexX and a hexose monophosphate substrate (e.g., the reduced 9 or 14). It is thus reasonable to assume that part of the purified BexX contains a trapped reaction intermediate, and the binding of the substrate-enzyme complex is likely an imine linkage, which can be stabilized by hydride reduction.
Figure 1.
(A) ESI-MS of C-His6-tagged BexX treated with NaBH4. (B) ESI-MS of C-His6-tagged BexX incubated with D-G6P (2) prior to NaBH4 treatment. Calculated molecular weights of C-His6-BexX (268 aa) and C-His6-BexX–D-G6P (reduced) are 28488, and 28732, respectively.
To test this hypothesis, the purified BexX was incubated with the putative substrate D-G6P (2) and treated with NaBH4 prior to MS analysis. As shown in Figure 1B, the addition of 2 resulted in complete conversion of the parent protein peak to the enzyme-substrate-modified peak. In contrast, no change of peak distribution was discernible when D-F6P (13), D-glucose, or DXP (6) was used instead of D-G6P (see Figure S2).12 Thus, the observed covalent modification of BexX is clearly D-G6P (2) specific. These results indicate that D-G6P is the substrate for BexX, and formation of an imine intermediate (9) with an active-site lysine residue is the initial step of the BexX reaction.
Sequence alignment revealed that the catalytic lysine residue (Lys96) in ThiG of Bacillus subtilis has a counterpart, Lys110, in BexX (see Figure S4).12 To investigate whether Lys110 plays an active role in BexX catalysis, the K110A mutant was constructed, heterologously expressed in E. coli, and purified as a C-terminal His6-tagged protein. ESI-MS of the purified mutant protein exhibited only one peak whose mass matches the calculated molecular mass of K110A (calcd 28431, obsd 28430) without any modification. No change was noted even after treatment with D-G6P (2) and NaBH4 (see Figure S3).12 This finding strongly implicates Lys110 as the site where the adduct with D-G6P is formed.
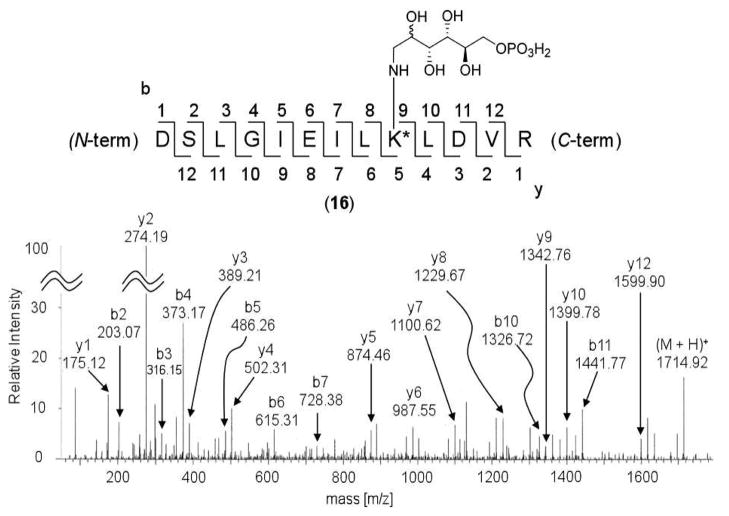
To ensure that the modification indeed occurs at Lys110, the wild-type BexX–D-G6P complex was reduced by NaBH4 and then subjected to proteolysis with trypsin. The tryptic digested peptide fragments were analyzed by LC-MS/MS, and the peptide fragment D102–R114 [calcd m/z 1714.89 (M + H+, z = 1), obsd 1714.92] contains a D-G6P adduct (16, Figure 2, also see Figures S5).12 Further analysis of the observed b and y ions revealed the attachment of the hexose phosphate at K110 (Figure 2). These results unambiguously demonstrated that the sugar modification indeed occurs at a specific lysine residue, i.e., K110, consistent with the proposed mechanism shown in Scheme 3.
Figure 2.
LC-MS/MS of the identified peptide (D102–R114) with a D-G6P coupled to the active site lysine (K110) (16). Calculated mass of 16 is m/z 1714.89 (M + H+, z = 1), obsd 1714.92.
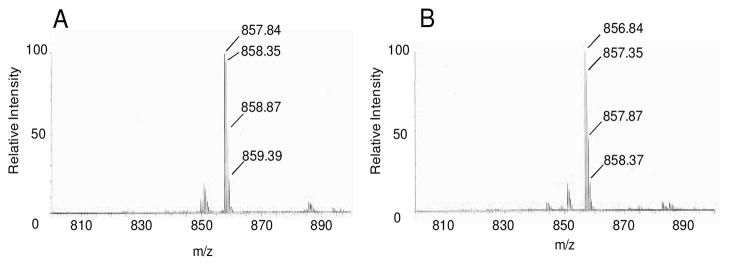
Interestingly, during the studies of the trypsin digested BexX-fragments, we noticed that the BexX-D-G6P adduct could be detected even without adding any reducing reagent (NaBH4) in the reaction mixture [calcd m/z 856.94 (M + 2H+, z = 2), obsd 856.84] (Figure 3B, also see Figure S6).12 This observation was surprising because it is unlikely that the proposed imine intermediate (9), without prior reduction, could survive the trypsin digestion as well as the conditions used for the LC-MS/MS analysis. This prompted us to reconsider the chemical nature of the trapped intermediate in the active site of BexX. The initially formed iminium intermediate 9 is in equilibrium with 10 and 11 (Scheme 3). A likely candidate for the stable covalent BexX–D-G6P adduct is compound 11 which is not readily hydrolyzable in solution. Accordingly, the tryptic fragment 16 detected in the LC-MS/MS analysis could result from hydride reduction of the imine moiety in 9 and/or the 2-keto group in 11 [calcd m/z 857.95 (M + 2H+, z = 2), obsd 857.84] (Figure 3A). However, without prior reduction, the modified D102-R114 fragment shown in Figure 3B likely has a structure of 17 (Scheme 4).
Figure 3.
LC-MS of trypsin-digested BexX-D-G6P with (A) or without (B) NaBH4 treatment (positive mode analysis). Calculated m/z values of 16 and its unreduced form (17) are 857.95 (M + 2H+, z = 2) and 856.94 (M + 2H+, z = 2), respectively.
Scheme 4.
To test this hypothesis, the trypsin-digested BexX–D-G6P sample was treated with a carbonyl-specific labeling reagent, 2,4-dinitrophenylhydrazine (DNPH, 18), and the resulting mixture was analyzed by LC-MS (Figure 4A). The observed m/z 946.96 ion (M + 2H+, z = 2) is consistent with the modified peptide, D102–R114, coupled with D-G6P and DNPH (19, Scheme 4). In a separate experiment, the undigested BexX–D-G6P complex was treated with another carbonyl-reactive reagent, NH2OH, and analyzed by ESI-MS. The observed ion of 28742 (Figure 4B) matches the calculated molecular mass of BexX–D-G6P–NH2OH imine adduct (20, Scheme 4). These results strongly support the presence of a keto intermediate (11) in the protein–substrate complex.
Figure 4.
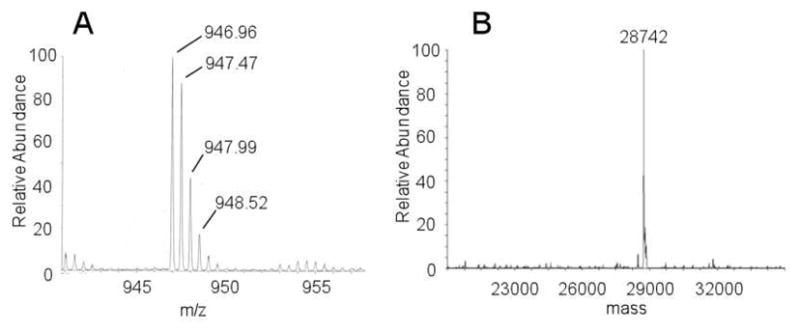
(A) LC-MS of the modified peptide (19) derived from trypsin-digested BexX–D-G6P after DNPH treatment. Calculated m/z value of 19 is 946.95 (M + 2H+, z = 2). (B) ESI-MS of C-His6-BexX–D-G6P ketone intermediate trapped by NH2OH (20). Calculated molecular weight of 20 is 28745.
In summary, a key enzyme involved in the biosynthesis of 2-thiosugar in BE-7585A, BexX, was expressed and purified. Its physiological substrate was verified to be D-G6P (2). A stable protein–substrate adduct was observed in the as-isolated BexX by ESI-MS, and the covalent modification was demonstrated to be substrate-specific (i.e., only with D-G6P) and site-specific (at K110). Most significantly, the sugar substrate in the covalent D-G6P-enzyme adduct exists at least partially in its 2-keto form (11), which could be directly trapped by carbonyl-directing reagents, DNPH (18) and NH2OH. These results provide the first mechanistic insight into the 2-thiosugar formation reaction, which closely resembles that of the ThiG-catalyzed thiazole formation in thiamin biosynthesis.13 The initial steps must involve several isomerization steps to generate the 2-keto intermediate (9 → 10 → 11), priming the sugar substrate to accept the thiol group from a nucleophilic sulfur donor. A potential sulfur carrier protein such as ThiS in thiamin biosynthesis is not found in the BE-7585A biosynthetic cluster. However, a pathway-independent sulfur carrier protein or an endogenous cystein desulfurase harboring a reactive thiocarboxylate or a cystein persulfide may serve as the sulfur donor by attacking the 2-keto intermediate 11 to introduce the sulfur atom.14,15 A. orientalis genome mining is in progress in order to find this sulfur transferring protein.
Supplementary Material
Acknowledgments
This work was supported in part by the National Institutes of Health Grants (GM035906).
Footnotes
Supporting Information Available. Experimental details, ESI-MS spectra, LC-MS spectra and protein sequence analysis. This material is available free of charge via the Internet at http://pubs.acs.org.
References and Notes
- 1.(a) Weymouth-Wilson AC. Nat Prod Rep. 1997;14:99–110. doi: 10.1039/np9971400099. [DOI] [PubMed] [Google Scholar]; (b) Kren V, Martinkova L. Curr Med Chem. 2001;8:1303–1328. doi: 10.2174/0929867013372193. [DOI] [PubMed] [Google Scholar]; (c) Thorson JS, Hosted J, Jiang J, Biggins JB, Ahlert J. Curr Org Chem. 2001;5:139–167. [Google Scholar]
- 2.(a) Melancon CE, III, Thibodeaux CJ, Liu H-w. ACS Chem Biol. 2006;8:499–504. doi: 10.1021/cb600365q. [DOI] [PubMed] [Google Scholar]; (b) Mendez C, Luzhetskyy A, Bechthold A, Salas JA. Curr Top Med Chem. 2008;8:710–724. doi: 10.2174/156802608784221532. [DOI] [PubMed] [Google Scholar]; (c) Williams GJ, Gantt RW, Thorson JS. Curr Opin Chem Biol. 2008;12:556–564. doi: 10.1016/j.cbpa.2008.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Solenberg PJ, Matsushima P, Stack DR, Wilkie SC, Thompson RC, Baltz RH. Chem Biol. 1997;4:195–202. doi: 10.1016/s1074-5521(97)90288-x. [DOI] [PubMed] [Google Scholar]; (b) Madduri K, Kennedy J, Rivola G, Inventi-Solari A, Filippini S, Zanuso G, Colombo AL, Gewain KM, Occi JL, MacNeil DJ, Hutchinson CR. Nat Biotech. 1998;16:69–74. doi: 10.1038/nbt0198-69. [DOI] [PubMed] [Google Scholar]; (c) Yamase H, Zhao L, Liu H-w. J Am Chem Soc. 2000;122:12397–12398. [Google Scholar]; (d) Mendez C, Salas JA. Trends Biotechnol. 2001;19:449–456. doi: 10.1016/s0167-7799(01)01765-6. [DOI] [PubMed] [Google Scholar]; (e) Griffith BR, Langenhan JM, Thorson JS. Curr Opin Biotechnol. 2005;16:622–630. doi: 10.1016/j.copbio.2005.10.002. [DOI] [PubMed] [Google Scholar]; (f) Melancon CE, III, Yu W-l, Liu H-w. J Am Chem Soc. 2005;127:12240–12241. doi: 10.1021/ja053835o. [DOI] [PubMed] [Google Scholar]; (g) Melancon CE, III, Liu H-w. J Am Chem Soc. 2007;129:4896–4898. doi: 10.1021/ja068254t. [DOI] [PMC free article] [PubMed] [Google Scholar]; (h) Mendez C, Luzhetskyy A, Bechthold A, Salas JA. Curr Top Med Chem. 2008;8:710. doi: 10.2174/156802608784221532. [DOI] [PubMed] [Google Scholar]
- 4.(a) He X, Agnihotri G, Liu H-w. Chem Rev. 2000;100:4615–4661. doi: 10.1021/cr9902998. [DOI] [PubMed] [Google Scholar]; (b) He XM, Liu H-w. Curr Opin Chem Biol. 2002;6:590–597. doi: 10.1016/s1367-5931(02)00367-8. [DOI] [PubMed] [Google Scholar]; (c) He XM, Liu H-w. Annu Rev Biochem. 2002;71:701–754. doi: 10.1146/annurev.biochem.71.110601.135339. [DOI] [PubMed] [Google Scholar]; (d) Thibodeaux CJ, Melancon CE, III, Liu H-w. Nature. 2007;446:1008–1016. doi: 10.1038/nature05814. [DOI] [PubMed] [Google Scholar]; (e) Thibodeaux CJ, Melancon CE, III, Liu H-w. Angew Chem Int Ed. 2008;47:9814–9859. doi: 10.1002/anie.200801204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sasaki E, Ogasawara Y, Liu H-w. J Am Chem Soc. 2010;132:7405–7417. doi: 10.1021/ja1014037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.(a) Okabe T, Suda H, Sato F, Okanishi M. Banyu Pharmaceutical Co., Ltd; Japan: 02-16894 A. Jpn Kokai Tokkyo Koho JP. 1990; (b) Etoh H, Iguchi M, Nagasawa T, Tani Y, Yamada H, Fukami H. Agric Biol Chem. 1987;51:1819–1824. [Google Scholar]
- 7.Ronning CM, Nierman WC. In: Myxobacteria: Multicellularity and Differentiation. Whitworth DE, editor. ASM Press; Washington, DC: 2008. pp. 285–298. [Google Scholar]
- 8.Park JH, Dorrestein PC, Zhai H, Kinsland C, McLafferty FW, Begley TP. Biochemistry. 2003;42:12430–12438. doi: 10.1021/bi034902z. [DOI] [PubMed] [Google Scholar]
- 9.Dorrestein PC, Zhai H, Taylor SV, McLafferty FW, Begley TP. J Am Chem Soc. 2004;126:3091–3096. doi: 10.1021/ja039616p. [DOI] [PubMed] [Google Scholar]
- 10.Dorrestein PC, Zhai H, McLafferty FW, Begley TP. Chem Biol. 2004;11:1373–1381. doi: 10.1016/j.chembiol.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 11.Mouilleron S, Badet-Denisot MA, Golinelli-Pimpaneau B. J Biol Chem. 2006;281:4404–4412. doi: 10.1074/jbc.M511689200. [DOI] [PubMed] [Google Scholar]
- 12.See Supporting Information for details.
- 13.Begley TP. Nat Prod Rep. 2006;23:15–25. doi: 10.1039/b207131m. [DOI] [PubMed] [Google Scholar]
- 14.(a) Begley TP, Xi J, Kinsland C, Taylor S, McLafferty F. Curr Opin Chem Biol. 1999;3:623–629. doi: 10.1016/s1367-5931(99)00018-6. [DOI] [PubMed] [Google Scholar]; (b) Fontecave M, Ollagnier-de-Choudens S, Mulliez E. Chem Rev. 2003;103:2149–2166. doi: 10.1021/cr020427j. [DOI] [PubMed] [Google Scholar]; (c) Mueller EG. Nat Chem Biol. 2006;2:185–194. doi: 10.1038/nchembio779. [DOI] [PubMed] [Google Scholar]; (d) Kessler D. FEMS Microbiol Rev. 2006;30:825–840. doi: 10.1111/j.1574-6976.2006.00036.x. [DOI] [PubMed] [Google Scholar]; (e) Booker SJ, Cicchillo RM, Grove TL. Curr Opin Chem Biol. 2007;11:543–552. doi: 10.1016/j.cbpa.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; (f) Jurgenson CT, Begley TP, Ealick SE. Annu Rev Biochem. 2009;78:569–603. doi: 10.1146/annurev.biochem.78.072407.102340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.It was demonstrated that bisulfide could be used as a substitute for the sulfur carrier protein, ThiS-thiocarboxylate, in thiamin biosynthesis.8 However, this is not the case for BexX, since release of the expected thiosugar product 5 from 11 was not observed in the presence of high concentration of bisulfide (up to 20 mM).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.