Abstract
We reconstruct the phylogenetic relationships within the bacterial genus Pseudonocardia to evaluate two models explaining how and why Pseudonocardia bacteria colonize the microbial communities on the integument of fungus-gardening ant species (Attini, Formicidae). The traditional Coevolution-Codivergence model views the integument-colonizing Pseudonocardia as mutualistic microbes that are largely vertically transmitted between ant generations and that supply antibiotics that specifically suppress the garden pathogen Escovopsis. The more recent Acquisition model views Pseudonocardia as part of a larger integumental microbe community that frequently colonizes the ant integument from environmental sources (e.g., soil, plant material). Under this latter model, ant-associated Pseudonocardia may have diverse ecological roles on the ant integument (possibly ranging from pathogenic, to commensal, to mutualistic) and are not necessarily related to Escovopsis suppression. We test distinct predictions of these two models regarding the phylogenetic proximity of ant-associated and environmental Pseudonocardia. We amassed 16S-rRNA gene sequence information for 87 attine-associated and 238 environmental Pseudonocardia, aligned the sequences with the help of RNA secondary structure modeling, and reconstructed phylogenetic relationships using a maximum-likelihood approach. We present 16S-rRNA secondary structure models of representative Pseudonocardia species to improve sequence alignments and identify sequencing errors. Our phylogenetic analyses reveal close affinities and even identical sequence matches between environmental Pseudonocardia and ant-associated Pseudonocardia, as well as nesting of environmental Pseudonocardia in subgroups that were previously thought to be specialized to associate only with attine ants. The great majority of ant associated Pseudonocardia are closely related to autotrophic Pseudonocardia and are placed in a large subgroup of Pseudonocardia that is known essentially only from cultured isolates (rather than cloned 16S sequences). The preponderance of the known ant-associated Pseudonocardia in this latter clade of culturable lineages may not necessarily reflect abundance of these Pseudonocardia types on the ants, but isolation biases when screening for Pseudonocardia (e.g., preferential isolation of autotrophic Pseudonocardia with minimum-nutrient media). The accumulated phylogenetic patterns and the possibility of isolation biases in previous work further erode support for the traditional Coevolution-Codivergence model and calls for continued revision of our understanding how and why Pseudonocardia colonize the microbial communities on the integument of fungus-gardening ant species.
Keywords: Attine ant-microbe symbiosis, Mutualism, Antibiotic, Secondary rRNA structure
Introduction
Fungus-growing ants (Attini, Formicidae) cultivate a basidiomycete fungus in gardens that the ant nurture with dead or fresh plant substrate. The main food source for the ants is the cultivated fungus, which is thought to comprise the main living biomass in attine gardens (Mueller et al. 2005). The gardens are actually mini-ecosystems of competing, commensal, and mutualistic microbes, including a diverse assembly of filamentous fungi, yeasts, and bacteria (Bacci et al. 1995; Carreiro et al. 1997; Currie et al. 1999a; Santos et al. 2004; Mueller et al. 2005; Rodrigues et al. 2008; Rodrigues et al. 2009). The interaction network of these microbes is engineered in part by the physical manipulations and secretions of the ants (Santos et al. 2004; Mueller et al. 2005). Significant populations of microbes, including actinomycetes, also colonize the integument and the alimentary canal of the farming ants (the ant microbiome), as is probably true for most arthropods (Goodfellow and Williams 1983; Dattagupta et al. 2009 and references therein). A diversity of integumental actinomycete bacteria can be readily isolated from most attine ants with minimum-nutrient media favoring autotrophic bacterial growth, including bacteria in the genus Pseudonocardia (Cafaro and Currie 2005; Currie et al. 2006; Mueller et al. 2008; Fernández-Marín et al. 2009; Sen et al. 2009). As an extreme case, workers of the attine ant Mycocepurus smithii can be colonized by significant populations of several species of Pseudonocardia, one species of the closely related genus Amycolatopsis, and many other actinomycete and non-actinomycete bacteria (Sen et al. 2009).
At the time of the discovery of actinomycete growth on the attine integument 10 years ago, it was thought that the ants promote the growth of antibiotic- secreting actinomycetes on their integument as a specific defense to suppress the garden parasite Escovopsis (Hypocreales, Ascomycota) (Currie et al. 1999a, b; Currie 2001). The almost simultaneous discovery of the parasite Escovopsis and the integumental actinomycete growth led to the immediate speculation that Escovopsis and the growth must somehow be ecologically linked. Specifically, Currie et al. (1999b) and Currie (2001) argued that attine ants sustain growth of the actinomycete Pseudonocardia as mutualists on their integument to specifically combat Escovopsis parasites; that the mutualistic Pseudonocardia have been engaged in a co-evolutionary arms race with Escovopsis since the origin of the attine ant-fungus mutualism 50 million years ago; and that Escovopsis largely failed to evolve effective resistance against Pseudonocardia because of some unknown disadvantage in the coevolutionary arms race (see also recent reviews discussing the Escovopsis-Pseudonocardia arms race; Suen and Currie 2008; Poulsen et al. 2009).
The original report and a string of follow-up studies seemed to support this ant-Escovopsis-Pseudonocardia co-evolution model, including (a) derived cuticular structures that seem designed to facilitate actinomycete colonization of the worker integument (Currie et al. 2006); (b) glandular structures underlying areas of significant actinomycete accumulation on the integument, suggesting that the ants may manipulate microbial growth through secretions (Currie et al. 2006); (c) greater susceptibility of gardens to Escovopsis infection if the integumental accretions are scraped off from workers (a procedure also likely to stress or injure ants) (Currie et al. 2003); (d) apparent clade-to-clade correspondences between ant, Escovopsis, and Pseudonocardia phylogenies, supporting the possibility of tight co-diversification in an ancient ant-Pseudonocardia association (Cafaro and Currie 2005; Poulsen et al. 2009); (e) marginal (but not strong) inhibition of Escovopsis by a peptide secreted in high concentrations by a Pseudonocardia isolate in vitro (Oh et al. 2009); (f) a growth-enhancing effect on the fungal cultivar by an unidentified actinomycete isolated from an ant integument (Currie et al. 1999b); and (g) specific antibiosis of a single Pseudonocardia isolate against Escovopsis but not against 17 non-Escovopsis test fungi (Currie et al. 1999b). Taken together, the accumulating evidence seemed to support ant-Escovopsis-Pseudonocardia co-evolution and the possibility of a 50-million-year-old Pseudonocardia-Escovopsis arms race.
The image of co-evolved, pesticide-secreting, integumental microbes of attine ants seemed so convincing that the National Science Teacher Association (NSTA) and several science museums in the USA began promoting the attine-Pseudonocardia-Escovopsis association as an indisputable example of coevolution (Diamond 2006). To visualize the evidence of co-evolved, specific antibiosis against Escovopsis, a hands-on exercise developed by NSTA asks biology students to measure growth of two garden pathogens, Escovopsis and Trichoderma, growing under the influence of Pseudonocardia secretions. Because the growth tracks on paper show that Pseudonocardia strongly inhibits Escovopsis but not Trichoderma (a finding originally reported in Currie et al. 1999b), Pseudonocardia apparently exhibits specialized antibiosis against Escovopsis, because, if Pseudonocardia “developed a substance that would kill only [Escovopsis], then that would support the idea that the two organisms had coevolved” (Diamond 2006, page 95; see www.nsta.org/pdfs/virus/Virus-Activity3.pdf).
A series of recent studies have eroded much of the key evidence that was thought to support the original formulation of specific antibiosis and ant-Pseudonocardia- Escovopsis co-evolution. For example, Kost et al. (2007) and Sen et al. (2009) showed that Pseudonocardia from the integument of attine ants have generalized, broad-spectrum antibiotic activities, comparable to the activities of environmental (free-living) actinomycete bacteria, and that therefore the attine-associated Pseudonocardia are not antibioticially specialized and evolutionarily derived as postulated by Currie et al. (1999b). Moreover, antibiotics secreted in vitro by ant-associated Pseudonocardia strongly suppress or kill the cultivated fungi from the corresponding nests (Sen et al. 2009), contrary to the original report of a growth-enhancing effect of Pseudonocardia on the cultivated fungi (Currie et al. 1999b). Even more unfortunate was a great delay before other research groups could replicate some of the original experiments. Because of a regrettable oversight, the original report did not specify the microbial methods for isolating Pseudonocardia from attine ants (Currie et al. 1999b), and it was not until 2005, with the first publications of the exact methods to isolate Pseudonocardia from ants (Cafaro and Currie 2005), that other research groups began to evaluate the earlier findings. Reviewing the history of ant-actinomycete research in a recent commentary, Boomsma and Aanen (2009) concluded that there exists a need for more careful hypothesis testing to prevent oversimplified evolutionary conclusions, and that a key message from ten years of ant-Pseudonocardia research may be the reminder to scientists at large “to continuously re-evaluate what we know for a fact and what we merely infer”.
Following Boomsma and Aanen’s (2009) call for more careful hypothesis testing to avoid premature conclusions of adaptive and co-evolved antibiotic design (sensu Gould and Lewontin 1979), we expand here on a test of phylogenetic predictions that was first explored by Mueller et al. (2008), testing for close phylogenetic affinities between ant-associated and environmental Pseudonocardia lineages. This same test can now be applied to a much larger dataset, because four times the sequence information has become available at GenBank for Pseudonocardia. Our test specifically juxtaposes critical predictions of the traditional ant-Pseudonocardia-Escovopsis Coevolution-Codivergence model (Currie et al. 1999b; Poulsen et al. 2009) and predictions of the alternate Acquisition model (Kost et al. 2007; Mueller et al. 2008). These two models represent extreme viewpoints, but they make different predictions regarding the phylogenetic proximity of ant-associated and environmental Pseudonocardia.
Predictions of the traditional Coevolution-Codivergence model
The Coevolution-Codivergence model assumes that the diversity of attine ants associate only with a limited number of Pseudonocardia lineages, and that these few lineages have co-diverged with corresponding ant lineages during a long association spanning millions of years (Cafaro and Currie 2005; Suen and Currie 2008; Poulsen et al. 2007; Poulsen et al. 2009). These predictions rest on two main assumptions; first that each attine colony associates with only a single Pseudonocardia strain; and second, that Pseudonocardia lineages are largely vertically transmitted across many ant generations, punctuated by occasional horizontal transfer between ant lineages, and punctuated very rarely by de novo acquisition of Pseudonocardia from environmental sources (Cafaro and Currie 2005; Poulsen et al. 2007; Poulsen et al. 2009; Caldera et al. 2009). The traditional Coevolution-Codivergence model emphasizes rare acquisition from environmental sources, long-term vertical transmission of Pseudonocardia associates across many ant generations, and strong co-evolutionary interactions between Pseudonocardia and Escovopsis.
Prediction of the alternate Acquisition model
The Acquisition model assumes continual colonization of the attine integument by diverse microbes (including Pseudonocardia), microbial turnover on the integument, and regular loss (purging) of some integumental microbes. In essence, the Acquisition model assumes a microbiome on the ant integument that may parallel the complex surface microbiome of other arthropods (e.g., Dattagupta et al. 2009 and references therein). Specifically, the Acquisition model assumes that (i) a great diversity of microbes inhabits the ant integument; (ii) specific microbes colonize the ants predictably because the ants encounter these microbes inevitably in their particular environment or in their interactions with the gardening substrate; (iii) some of these integumental microbes may be transmitted through foundress queens between generations, but long-term vertical transmission across many ant generations is limited because of continual microbial turnover on the ant integument; (iv) not all Pseudonocardia strains on the ant integument are mutualists (some may be commensal, some may even be pathogenic); (v) antibiotic effects against Escovopsis may be incidental byproducts—but not necessarily co-evolved adaptations—of microbial evolution occurring before colonization of the ant integument or occurring in situ on the integument under microbe-microbe competition for limited resources (Kost et al. 2007; Mueller et al. 2008; Sen et al. 2009). The Acquisition model emphasizes frequent acquisition of Pseudonocardia from environmental sources, limited vertical transmission of Pseudonocardia associates across many ant generations, and low potential or absence of Pseudonocardia-Escovopsis co-evolution.
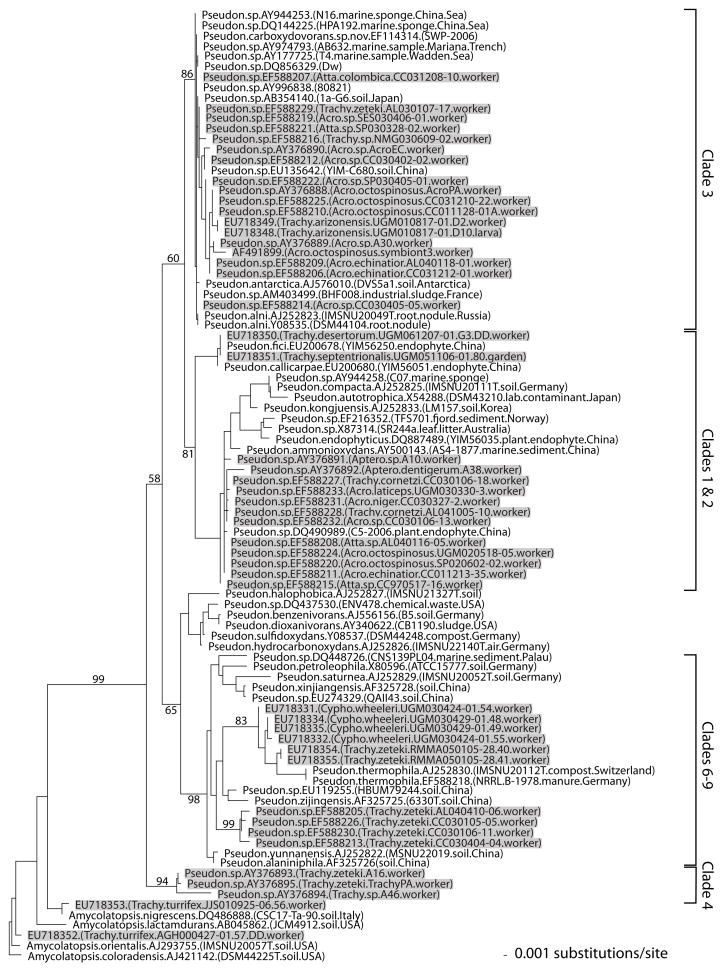
Using 16S rRNA gene sequence information from 45 ant-associated and 41 environmental Pseudonocardia, Mueller et al. (2008) recently began to reconstruct the phylogenetic relationships between these two types of Pseudonocardia within the genus. This previous study identified a number of ant associated Pseudonocardia that are sequence-identical at the 16S gene to environmental Pseudonocardia, and documented that well-supported clades include closely related environmental and ant-associated Pseudonocardia that differ in only a few base pairs (Fig. 1). However, phylogenetic analyses also revealed some Pseudonocardia clades that appeared derived and that consisted exclusively of Pseudonocardia isolated from ants (Fig. 1). These derived Pseudonocardia clades could be ant-specific either because (a) these Pseudonocardia were anciently acquired by the ants from environmental sources and since then had been exclusively associated with attine ants; or (b) the clades appear only ant-specific because free-living Pseudonocardia are undersampled. This later explanation predicts that more comprehensive sampling of free-living Pseudonocardia will eventually show that unknown environmental strains exist that are nested within the putative antspecific clades or that are very closely related to them. Because over 500 Pseudonocardia sequences have accumulated to date at GenBank (compared to 85 sequences in the analysis of Mueller et al. 2008), the above predictions of the Acquisition model can now be tested with a more comprehensive sampling of Pseudonocardia diversity. Additionally, we show here how 16S rRNA secondary structure modeling of Pseudonocardia can identify sequencing errors and help resolve ambiguities in alignments for Phylogenetic analyses.
Fig. 1.
Phylogeny of Pseudonocardia reconstructed by Mueller et al. (2008) from 16S rRNA gene sequence information. This reconstruction was based on a limited sample of 85 Pseudonocardia sequences available at GenBank in September 2007 and excluded sequence information in regions of unambiguous alignment (i.e., the reconstruction was not based on the 16S secondary-structure modeling employed in the present study). The clade labels on the far right correspond to the labels in Fig. 2a and b of the present study (Clade 1&2 was called “compacta-group” and Clade 3 was called “alni-group” in Mueller et al. 2008). This previous analysis revealed close affinities between many ant-associated (shaded) and environmental Pseudonocardia (unshaded), and predicted that future surveys will discover environmental Pseudonocardia nesting in groups appearing “ant-specific” at that time. This prediction is tested in the phylogenetic reconstruction shown in Fig. 2a and b, which is based on four times the sequence information that has become available for Pseudonocardia since the study by Mueller et al. (2008)
Methods
Utility of the 16S rRNA gene
Our phylogenetic analysis takes advantage of the exponentially increasing number of 16S rRNA gene sequences that have become available at the NCBI GenBank (http://www.ncbi.nlm.nih.gov/nuccore) for reconstruction of phylogenetic relationships. Some of these 16S sequences were generated for taxonomic descriptions of Pseudonocardia species (summarized in Huang and Goodfellow 2011), but the majority of the Pseudonocardia sequences derived from environmental surveys characterizing general bacterial diversity with either culture-dependent methods (e.g., specialized isolation media) or culture-independent methods (e.g., cloning and sequencing of 16S amplicons; direct pyrosequencing of 16S amplicons). Reliance on only the 16S gene in our phylogenetic analyses has disadvantages because this single gene does not fully resolve all relationships between closely related taxa, because bacterial genomes can contain several 16S genes that differ minimally in sequence (although this is currently unknown for Pseudonocardia), because of potential artifacts in multitemplate PCR on whole microbial communities in environmental surveys (Acinas et al. 2005), and because of the possibility of lateral gene transfer among bacteria (although lateral gene transfer is thought to be very unlikely for an isolated 16S gene because of the integrated, central role of this gene within a cell’s genomic network; Woese 2000). Despite these potential disadvantages, our 16S-based approach is justified for three reasons.
First, the amount of 16S information available from environmental surveys exceeds by several orders of magnitude that of any other gene, and the most comprehensive sample on Pseudonocardia diversity from diverse sources is critical to address close Phylogenetic affinities between environmental and ant-associated Pseudonocardia lineages. Second, the main subgroups of Pseudonocardia identified in our Phylogenetic reconstruction using only 16S information essentially matches the groupings inferred also by four protein-coding housekeeping genes also available at GenBank (UG Mueller, unpublished), showing that 16S and these four housekeeping genes have recorded parallel evolutionary histories that have not been eroded by lateral gene transfer. Third, our analysis does not aim to resolve relationship at all levels between all lineages of Pseudonocardia, but aims to identify closely-related lineages (i.e., we are less interested in resolving basal relationships between major subgroups within the genus) and, most importantly, aims to test for nesting of previously unknown environmental lineages within known clades of Pseudonocardia that currently are believed to be specialized to associate only with attine ants.
Selection of sequence information
To compile a comprehensive sample of 16S sequences for Pseudonocardia, we obtained all 456 sequences listed under Pseudonocardia by 1. November 2009 at the Ribosomal Database Project (RDP; Center for Microbial Ecology, Michigan State University; http://rdp.cme.msu.edu/treebuilder/treeing.spr). This list included all taxa classified under Pseudonocardia at the NCBI Taxonomy Browser (sequences obtained from isolated cultures; www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1847), as well as additional Pseudonocardia sequences from environmental surveys deposited at the NCBI Core Nucleotide Database (CND; www.ncbi.nlm.nih.gov/sites/entrez?db=nuccore). To find additional sequences not listed at the RDP (e.g., very recently deposited sequences at CND), we also searched the CND by BLASTing full sequence information ([1400 bp) of Pseudonocardia carboxydivorans, callicarpae, ammonioxydans, halophobica, spinosispora, petroleophila, thermophila, and zijingensis. These Pseudonocardia species were chosen because they span the phylogenetic diversity of the genus, and because they are closely related to Pseudonocardia lineages previously isolated from attine ants (Mueller et al. 2008). Because BLASTing of longer sequences can miss shorter sequences deposited at the CND, we also BLASTed, for each of these eight species, 16S sequence segments of about 300–500 bp length that spanned one or two of the V2, V3, V4, or V9 regions of the 16S gene. These regions were chosen because they represent the most variable regions of 16S within the genus Pseudonocardia. We added to our dataset two sequences (GU318369, GU318370) of Pseudonocardia isolated in an ongoing survey of actinomycete diversity in nests of the attine ant Trachymyrmex septentrionalis (Miller, Sen, Ishak, Mueller unpublished), and 16S sequences for P. compacta (NRRL B-16170; GU318372) and P. nitrificans (NRRL B-1664; GU318371; strains courtesy of David Labeda at the ARS Culture and Patent Culture Collections, United States Department of Agriculture, Peoria, Illinois). This search strategy accumulated a total of 519 sequences.
We excluded from this dataset those sequences actually belonging to other pseudonocardiaceous genera (e.g., some sequences currently listed at the RDP under Pseudonocardia actually belong into Crossiella, such as NCBI accessions FN297974, EU299989, FN297967, EF447067, EF447073, EF447078, EF188338, EF447079; sequence AJ871440 belongs into the genus Actinostreptospora; etc.), and we excluded poor-quality sequences flagged as such at the RDP. Second, to ensure sufficient overlap across phylogenetically informative regions of the 16S gene, we excluded all sequences shorter than 500 bp in length. This criterion of 500 bp minimum length also excluded the short sequences generated with next-generation sequencing platforms (e.g., 454 pyrosequencing) that are currently deposited at the NCBI Short Read Archive (www.ncbi.nlm.nih.gov/sites/entrez?db=sra). Third, we excluded all sequences that did not span both the V3 and the V4 regions. These two variable regions were targeted for three reasons: first, the great majority of partial 16S sequences available for Pseudonocardia included these two regions; second, these two regions are among the most variable 16S regions differentiating species in the genus Pseudonocardia; third, we believed it is important for tree stability in the phylogenetic analysis that all taxa overlapped in at least two contiguous variable regions (i.e., V3&V4). This criterion of V3&V4 sharing of all taxa means that these regions may have greater influence on the inferred phylogenetic relationships than other, similarly variable regions (e.g., V9). We excluded identical sequences that had been obtained in the same environmental survey, such as duplicate sequences from a large marine sediment survey from the Mariana Trench (AY974775-AY974796; retaining only AY974776, AY974783, and AY974793 in our analysis), and duplicate sequences from a large soil survey in Czechoslovakia (EF506948-EF507159; retaining from these only 13 non-identical Pseudonocardia sequences). Exclusion of these duplicate taxa does not affect any of our conclusions, but reduced computation time in our analyses. After exclusion of redundant taxa, our dataset included 325 Pseudonocardia taxa.
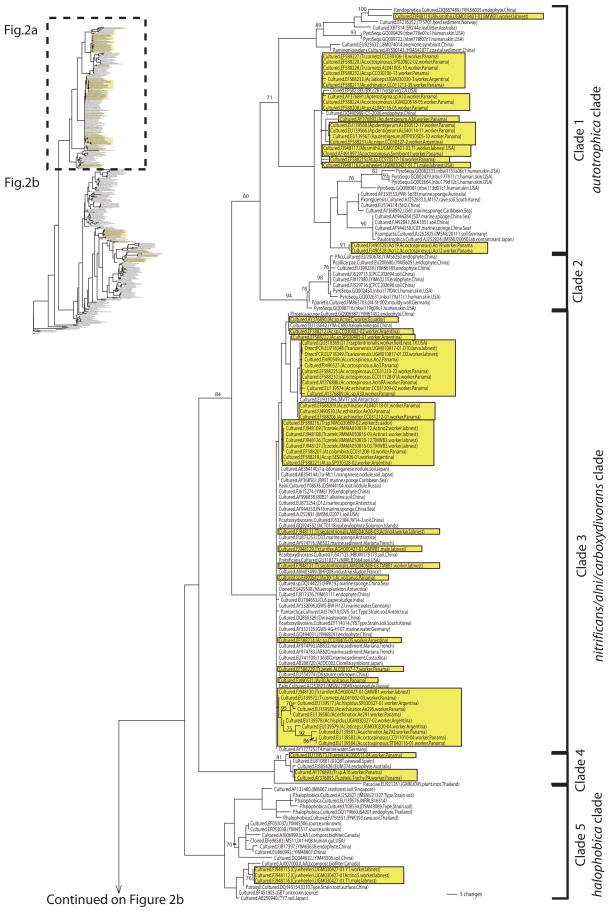
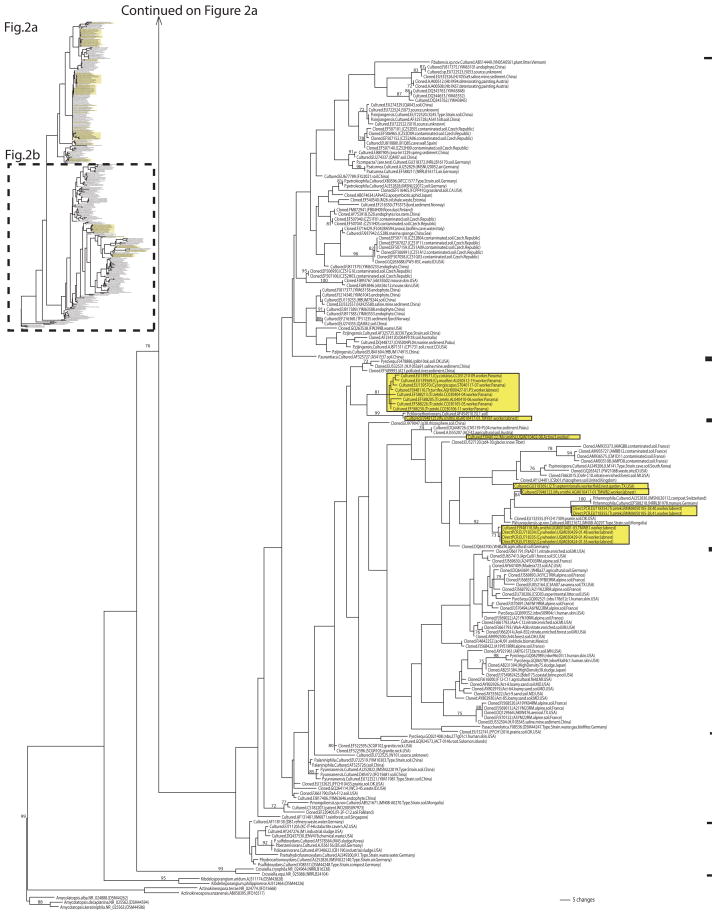
In the final dataset of 325 sequences, 67.4% of the Pseudonocardia sequences had been obtained by culture-dependent methods, 25.8% from cloning of 16S sequences amplified from microbial communities, 2.2% from direct PCR, and 4.6% from 454-pyrosequencing of 16S amplicons (Fig. 2a, b). Also from these 325 sequences, 26.8% of the sequences had been obtained from attine ants, 9.2% from plant material (e.g., endophytes, root bacteria), 3.4% from marine organisms (e.g., sponge symbionts), 5.2% from human or mouse skin, 31.7% from soil, 1.5% from rock surfaces, 5.2% from marine sediments, 5.5% from industrial sources (e.g., sludge, biofilters), and 11.4% from other sources (e.g., art paintings, house dust, air, etc.). Two Kibdelosporangium, two Crossiella, two Actinokineospora, and three Amycolatopsis were included as outgroups (Fig. 2b), covering pseudonocardiaceous genera most closely related to the genus Pseudonocardia (Huang and Goodfellow 2011; UGM unpublished).
Fig. 2.
Phylogeny of Pseudonocardia reconstructed for 325 taxa under the maximum-likelihood criterion from 16S rRNA gene sequence information. Ant-associated Pseudonocardia are boxed (shaded background); environmental Pseudonocardia are unboxed. Attine-associated lineages are closely related to soil-dwelling or endophytic Pseudonocardia lineages, indicating frequent recruitment of free-living Pseudonocardia into association with ants. Support values are inferred from 100 ML bootstrap pseudoreplicates. Each taxon label gives the method of characterization (sequencing of 16S amplicons from cultured isolates; sequencing of cloned 16S amplicons from environmental surveys; pyrosequencing of 16S amplicons; direct PCR from ants) and the GenBank accession, followed in parentheses by the ant species name from which a bacterium was sampled, the ant collection ID (e.g., UGM010817-01), the source from which the sequence was obtained (worker, garden, soil, plant, etc.), and the collection location. The attine ant genera are abbreviated as: Ac. = Acromyrmex; Ap. = Apterostigma; At. = Atta; Cy. = Cyphomyrmex; My. = Mycocepurus; Tr. = Trachymyrmex
On 30. November 2009, 16S sequences from an additional 24 ant-associated Pseudonocardia were released at GenBank (accessions EU283917- EU283940; Poulsen Cafaro, Erhardt, Little, Gerardo, Currie, unpublished, Host-pathogen dynamics in an ancient antibiotic system). While it was too late to incorporate these sequences in our Phylogenetic analyses, comparison of these new sequences with those in our phylogenetic reconstruction permitted precise placement of the new sequences into our phylogenetic framework (UG Mueller unpublished). Placement of these new sequences into the Phylogenetic context (Fig. 2) does not change any of the conclusions emphasized below.
Alignment and character selection
Sequences were aligned in MacClade version 4.06 (Maddison and Maddison 2000). The initial alignment of 1521 characters was adjusted manually to correct obvious alignment errors. To validate and adjust our initial alignment, we used the sequence alignment editor AE2 (Olsen et al. 1992) to manually align characters according to similar positioning in the RNA secondary structure. Regions of the 16S rRNA sequence that have significant sequence similarity can be aligned solely based on this identity. However, regions of the alignment that do not contain sufficient sequence similarity require additional constraints to accurately place nucleotides in columns that represent a similar part of the overall rRNA higher-order structure. The secondary structure diagrams were drawn with the interactive graphics program XRNA (B Weiser, H Noller unpublished). Inspection of our alignment revealed that sequences from ant-associated Pseudonocardia and P. halophobica (accessions EU139566.1-EU139584.1; these sequences were later replaced after completion of our analyses and once we had pointed out sequencing errors; see Supplemental Information) submitted by Poulsen et al. (2007) contained highly unusual character scorings, some of them at positions within the 16S gene that are thought to be invariable or highly conserved across all Eubacteria (see Supplemental Information). The secondary structure analysis of these suspect 16S sequences revealed that these unusual characters are likely sequencing artifacts. These unusual characters were furthermore rendered suspect because they also appeared in a sequence deposited by Poulsen et al. (2007) for P. halophobica (EU139576.1, from strain NRRL B-16514), whereas all other sequences deposited at GenBank for P. halophobica (AJ252827 = strain IMSNU21327; Z14111 = strain DSM43089T; Y08534 = strain DSM43089T) do not show the unusual character scorings appearing in Poulsen et al. 2007 (see Figures S3 and S7 in the Supplemental Information for secondary-structure models of the P. halophobica 16S rRNA sequences with and without the sequencing errors). We therefore excluded from our phylogenetic analyses 11 suspect characters introduced by the Poulsen et al. (2007) sequences (these characters did not vary among all the other taxa, and exclusion therefore did not affect phylogenetic inferences about the remaining taxa). However, we were unsure about the error of four additional suspect characters in the Poulsen et al. (2007) sequences, which therefore were retained in our phylogenetic analyses and were expected to render the corresponding taxa as phylogenetically derived. The derived phylogenetic positions of some of the sequences from Poulsen et al. (2007) (EU139566 - EU139584) in Fig. 2 therefore may be an artifact of uncorrected sequence-scoring errors.
16S RNA secondary structure modeling
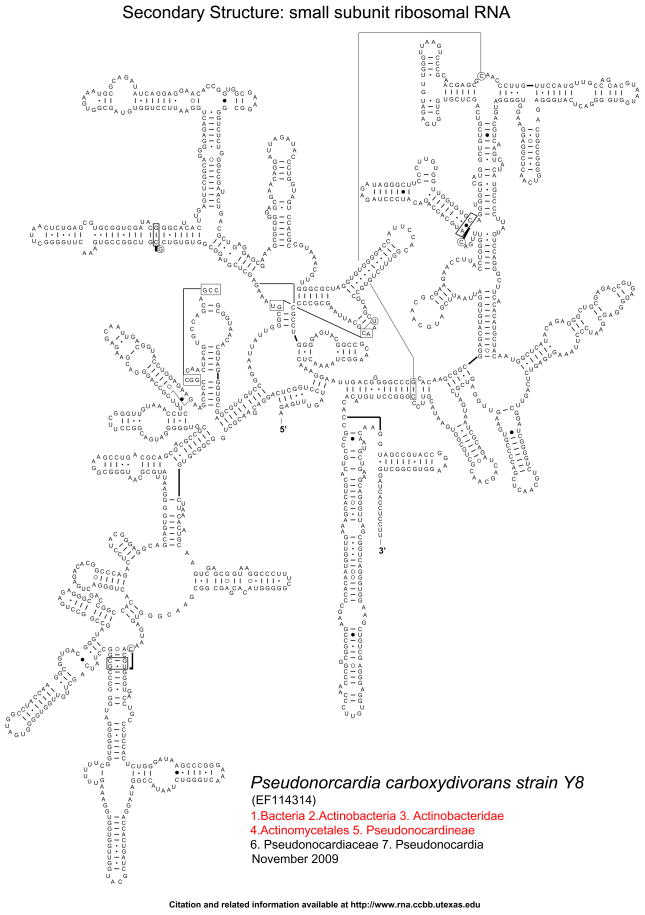
To facilitate future 16S RNA alignments for Phylogenetic reconstruction, we generated secondary structure models of 16S rRNA for five Pseudonocardia species spanning the diversity of the genus: P. carboxydivorans (GenBank accession EF114314; type strain Y8; Fig. 3), P. kongjuensis (AJ252833; type strain LM 157), P. halophobica (AJ252827; type strain IMSNU 21327), P. spinosispora (AJ249206; type strain LM 141), and P. yunnanensis (AJ252822; type strain IMSNU 22019) (see Supplemental Information). The 16S rRNA secondary structures were predicted with comparative methods for a secondary structure that is common to all of the known 16S rRNA sequences, using Covariation analysis (Woese et al. 1980, 1983; Gutell et al. 1985, 1992, 1994; Cannone et al. 2002). We are confident that the 16S rRNA secondary structure predictions are accurate, because, in a previous analysis, approximately 98%of the predicted model base pairs were present in the high-resolution crystal structure of the ribosomal subunit (Gutell et al. 2002). Bacterial 16S rRNA secondary structure diagrams for the modeled Pseudonocardia species are available at the Comparative RNA Web Site http://www.rna.ccbb.utexas.edu/DAT/3C/Structure/index.php, http://www.rna.ccbb.utexas.edu/SIM/4D/TOL/.
Fig. 3.
16S rRNA secondary structure model for P. carboxydivorans (GenBank accession EF114314; type strain Y8). The Supplemental Information presents additional secondary structure models for four additional species within the genus Pseudonocardia (P. kongjuensis, P. halophobica, P. spinosispora, P. yunnanensis). Canonical base pairs (G:C, A:U) are shown with tick marks, wobble (G:U) base pairs with small closed circles, A:G base pairs with large open circles, and all other non-canonical base pairs with large closed circles 2002). Bacterial 16S rRNA secondary structure diagrams for the modeled Pseudonocardia species are available at the Comparative RNA Web Site http://www.rna.ccbb.utexas.edu/DAT/3C/Structure/index.php, http://www.rna.ccbb.utexas.edu/SIM/4D/TOL/.
Phylogenetic analyses
Our final alignment utilized all characters of the 16S gene, but excluded the 11 suspect characters from Poulsen et al. (2007; see above) and excluded 72 characters from the conserved region at the beginning and end of the 16S gene because the great majority of the sequences were missing character information in these end regions. Our final alignment included 1436 characters (289 informative, 77 autapomorphic, and 1070 invariable characters). Phylogenetic relationships were inferred under the maximum likelihood (ML) criterion using the General Time Reversible (GTR ? I ? G) model of nucleotide substitution (Garli version 0.951; Zwickl 2006; www.zo.utexas.edu/faculty/antisense/garli/Garli.html). The appropriate model of nucleotide substitution was selected using the Akaike Information Criterion (AIC) implemented in Modeltest 3.7 (Posada and Crandall 1998). We performed twenty separate likelihood searches to explore likelihood space more thoroughly. Branch support was calculated by performing bootstrap analyses with 100 repetitions. Figure 2 reports the best phylogenetic topology under the ML criterion (lowest negative log-likelihood score) among the twenty ML searches.
Results and discussion
Sub-groupings within the genus Pseudonocardia
The maximum-likelihood (ML) reconstruction shown in Fig. 2a and b places 87 known ant-associated Pseudonocardia within the larger phylogenetic context of 238 environmental Pseudonocardia obtained from diverse sources (e.g., soil, marine sediment, plants, marine invertebrates, mammal skin, industrial material, etc.). We recover the main sub-groupings of Pseudonocardia that already emerged in the 16S analyses of Lee et al. (2000) and Mueller et al. (2008), but our increased taxon sampling tentatively identifies at least 10 subgroups within the genus (labeled as Clades 1–10 in Fig. 2a, b). The genus Pseudonocardia appears to be split basally into two main groups, one clade comprised of five subgroups that contain the great majority of the known ant associated Pseudonocardia (Fig. 2a); and a second, less satisfactorily resolved clade comprised of at least 5 subgroups (Fig. 2b). However, this basal split into these two main groups is only weakly supported in our ML analyses, as Clades 4 and 5 (shown in Fig. 2a) exhibited also affinities with the other main group (shown in Fig. 2b) in some of the less-likely ML reconstructions. Only few subgroups within the genus received adequate support in the ML bootstrap analyses, but two subgroups with acceptable support values (Clade 1 and a larger clade comprised of Clades 1–3; Fig. 2a) contain the great majority of the ant-associated Pseudonocardia. Clearly, information from additional genes will be necessary to resolve all subgroupings and affiliations with confidence. Fortunately, Wen-Jun Li’s Lab at Yunnan University, China, is currently compiling sequence information of four protein-coding genes (rpoB, fusA, gyrB, and lepA) for a multilocus phylogenetic analysis of key species in the genus Pseudonocardia. This framework should be useful in future studies to place the ant-associated Pseudonocardia into the subgroups of the genus. Preliminary analyses (UG Mueller unpublished) of the sequence information available at GenBank for these four protein-coding genes also revealed the monophyly of Clade 1 and the monophyly of the larger clade comprised of Clades 1–3, as shown in Fig. 2a.
Nesting of environmental Pseudonocardia lineages within clades previously thought to be ant-specific
Our analysis confirms the prediction of the Acquisition model that there exist previously unknown environmental Pseudonocardia that insert themselves into clades that originally were thought to be “ant-specific”. For example, Cafaro and Currie (2005) had identified a three-taxon clade of Pseudonocardia (isolated from Trachymyrmex ants from Panama) that had emerged also as phylogenetically unique and “antspecific” in the phylogenetic analysis by Mueller et al. (2008; Fig. 1); however, we found two environmental Pseudonocardia nested within that group of allegedly Trachymyrmex-specific Pseudonocardia (Clade 4 in Fig. 2a). One of these is an endophytic Pseudonocardia that was isolated from the surface-sterilized root of an Eucalyptus tree in Australia (GenBank accession FJ805426; 99.8% sequence-identical to AY376893 isolated from a Trachymyrmex worker from Panama), the second Pseudonocardia was isolated in a survey of microbial growth that degraded prehistoric cave paintings in Spain (Stomeo et al. 2008, 2009; EU810881; 99.3% sequence-identical to AY376893 isolated from a Trachymyrmex worker). The nesting of these two environmental Pseudonocardia inside a clade of ant-associated Pseudonocardia is less compatible with the phylogenetically derived position of ant-associated Pseudonocardia predicted by the Coevolution-Codivergence Hypothesis. Moreover, representatives of Clade 4 have so far been isolated only six times (four times from Trachymyrmex ants, twice from other sources; details in Supplemental Information) and they have so far not been detected in any culture-independent screen. This suggests that it may be sometimes inherently difficult to find environmental Pseudonocardia that are nested within antspecific clades, either because these environmental Pseudonocardia are difficult to isolate, or they are rare, or a combination of both factors. Because of this inherent difficulty of detection, the absence of evidence for environmental Pseudonocardia that are nested within ant-associated Pseudonocardia clades should not be interpreted as evidence for absence, and therefore also not as evidence in support for the Coevolution-Codivergence hypothesis (e.g., Cafaro and Currie 2005; Poulsen et al. 2009).
Figure 2a and 2b reveal additional cases where environmental Pseudonocardia nest within clades that were previously thought to be largely ant-specific. For example, for the main group of ant-associated Pseudonocardia (Clade 3 in Fig. 2a), only one environmental counterpart was known previously (Fig. 1), whereas now a number of environmental affiliates are known (sequence identical or near sequence identical at [99% similarity), such as the endophytic P. tropica sp. nov. from China, EU931094 from Antarctic soil, P. carboxydivorans from soils in China and Korea, P. nitrificans from soil in the USA, etc. (Fig. 2a). For the second main group of ant-associated Pseudonocardia (Clade 1 in Fig. 2a), one environmental affiliate was known previously (Fig. 1) whereas now three environmental affiliates are known (Fig. 2a), including the endophytic P. endophytica from China, DQ490989 from an endophyte in China, and EF682939 from a hotspring in the USA. Another interesting case is a conglomerate of ant-associated Pseudonocardia from three species of lab-reared attine ants, whose Pseudonocardia have close affinities with P. khuvsgulensis sp. nov. from soil in Mongolia, P. thermophila from compost in Germany, and the cloned EU132555 from prairie soil in the USA (Fig. 2b). Overall, the Phylogenetic patterns conform with the prediction of the Acquisition model that, because the attine integument is frequently colonized by bacteria from the environment, ant-associated Pseudonocardia have close phylogenetic links to diverse environmental Pseudonocardia. As we predicted before (Mueller et al. 2008), future studies are likely to uncover many additional close links between environmental counterparts and ant-associated Pseudonocardia, especially if more environmental Pseudonocardia are surveyed across the Neotropics (the realm of attine ants) and if more of the neglected basal attine ant lineages are surveyed for their integumental microbes.
Single Pseudonocardia lineages colonize the integument of a wide diversity of attine ants
In previous analyses (Cafaro and Currie 2005; Zhang et al. 2007; Mueller et al. 2008), Clade 3 appeared to contain Pseudonocardia that had been isolated mostly from Atta and Acromyrmex leafcutter ants. This near exclusive association between leafcutter ants and Clade 3 Pseudonocardia could be interpreted as evidence for co-diversification between the leafcutter ant clade and Clade 3 Pseudonocardia (sensu Poulsen et al. 2009). However, the accumulated evidence now documents also the presence of Clade 3 Pseudonocardia in the microbial communities of the integument of many Trachymyrmex ants (Fig. 2a), and it is possible that a more thorough survey of basal attine lineages could reveal associations of Clade 3 Pseudonocardia with ant lineages across the entire diversity of attine ants. As a parallel case, Sen et al. (2009) recently isolated from Clade 1 Pseudonocardia from laboratory nests of the lowerattine species Mycocepurus smithii (Fig. 2a), showing that more comprehensive sampling uncovered the first exceptions to the original “specificity” of Clade 1 Pseudonocardia on leafcutter and Apterostigma ants. Likewise, a group of closely related Clade 7 Pseudonocardia isolated from three species of Cyphomyrmex (costatus, muelleri, longiscapus) was interpreted by Poulsen et al. (2007, 2009) as possibly Cyphomyrmex-specific, but our phylogenetic analyses now reveal Pseudonocardia from Clade 7 that had been isolated from diverse Trachymyrmex ant species (Zhang et al. 2007; Sen et al. 2009; Fig. 2b). The isolation of Clade 7 Pseudonocardia from Trachymyrmex laboratory colonies by Sen et al. (2009) could potentially mean that the screened Trachymyrmex workers were colonized by Pseudonocardia derived from Cyphomyrmex species that had been kept in the same lab room for several years. If so, this then would document how readily Pseudonocardia can colonize the integument of Trachymyrmex species from other sources. We conclude, therefore, that tests for ant-Pseudonocardia specificities and codiversification should avoid conclusions based on small sample sizes (which too readily can underestimate the true diversity of Pseudonocardia associates). Rather, tests for specificities should first carefully document the full diversity of Pseudonocardia that colonize the integument of a single ant and different workers from the same colony, evaluate potential isolation biases (e.g., preferential isolation of autotrophic Pseudonocardia with minimum-nutrient medium; see below), and rule out cross-contamination from environmental sources.
Culturable versus unculturable Pseudonocardia
Perhaps the most striking difference between the two main Pseudonocardia clades in Fig. 2a and b is that 99% of the sequences in Fig. 2a (Clades 1–5) derived from Pseudonocardia that had been cultured, whereas the sequences in Fig. 2b derived from a mix of cultured and uncultured Pseudonocardia (cloned, pyrosequenced). In fact, the sizable Clade 9 in Fig. 2b consists entirely of uncultured Pseudonocardia, with the exception of the cultured P. asaccharolytica. Absence or underrepresentation of cultured Pseudonocardia in a clade suggests that these types of bacteria may be inherently difficult to isolate and were therefore not captured with the culture-dependent methods used so far. In contrast, the near absence of cloned Pseudonocardia in Clades 1–5 (Fig. 2a) suggests potential cloning biases, for example due to disruption of E. coli growth by the 16S sequences inserted into the E. coli genome during cloning (e.g., Hamady and Knight 2009, and references therein). Interestingly, the great majority of ant-associated Pseudonocardia characterized to date belong to two clades (Clade 1 and Clade 3) that are heavily biased towards culturable Pseudonocardia (this bias is also true for all the environmental Pseudonocardia in these clades; Fig. 2a), suggesting that these kind of Pseudonocardia are readily captured with the isolation methods used in the surveys so far, whereas the less culturable Pseudonocardia (i.e., from Clades 6–8) may have been missed by the same surveys.
The only study that compared culture-dependent and culture-independent methods for surveying antassociated Pseudonocardia was Sen et al. (2009), who documented that, whereas the standard culture-dependent method of Cafaro and Currie (2005) yielded only a single Pseudonocardia type from workers of the ant Trachymyrmex zeteki, a culture-independent pyrosequencing survey of T. zeteki workers revealed the presence of several species from different Pseudonocardia subgroups. Taken together, the Phylogenetic distribution of the cultured and uncultured Pseudonocardia (Fig. 2a versus 2b) and the preliminary findings of Sen et al. (2009) suggest that the standard isolation method of attine-associated Pseudonocardia with minimum-carbon, chitin medium (Cafaro and Currie 2005) may underestimate the true Pseudonocardia diversity present on attine ants. If so, the isolation of only one kind of Pseudonocardia from the integument of a particular ant (e.g., a Pseudonocardia from Clades 1 and 3) should therefore not be taken as evidence that this kind of Pseudonocardia is the only Pseudonocardia species in the microbial community on the ant integument. We therefore caution that previous studies that relied on minimum-nutrient isolation methods might have underestimated the true diversity of Pseudonocardia in the microbial communities growing on the integument of attine ants.
Colonization of the ant integument by Pseudonocardia from plant and soil sources
As already noted by Mueller et al. (2008), many antassociated Pseudonocardia are closely related to Pseudonocardia from soil and plant material (e.g., endophytic Pseudonocardia), but several additional close affinities now emerge from our larger phylogenetic analysis. For example, ant-associated Pseudonocardia that have sequences identical to P. carboxydivorans, P. tropica sp. nov., and P. endophytica are nowknown (Fig. 2a), all ofwhich have been cultured as endophytes from plant material. Likewise, environmental Pseudonocardia from soil can be sequence-identical in 16S to attine-associated Pseudonocardia (Fig. 2a). Both soil-dwelling and endophytic Pseudonocardia therefore remain possible sources of Pseudonocardia that colonize the attine integument. This calls for experimental evaluation of the colonization rate of the attine integument bymicrobes fromsuch environmental sources, which may cause fast or slow microbial turnover on the integument depending on the nature of the resident microbial community, the nature of the colonizing microbes, and the general disturbance in the nest. Such studies will require in-depth screening of the entire microbial community on the integument, as well as understanding of any biases of culturedependent and culture-independent Pseudonocardia screens (see above Culturable versus unculturable Pseudonocardia).
Pseudonocardia symbiosis beyond fungusgrowing ants
Pseudonocardia has been found not only in association with fungus-growing ants, but also in association with a wide diversity of other organisms, including plants, marine sponges, an anemone, a Clorella alga, an aphid, and mammals (human gut; human and mouse skin) (Fig. 2). Excluding the 87 ant-associated Pseudonocardia in Fig. 2, 12.6% of the surveyed Pseudonocardia derived from plants (endophytes), 4.6% from marine invertebrates, 0.4% from terrestrial arthropods (27% when including ants), and 7% frommammal skin (human, mouse). The absence or rarity of Pseudonocardia in other well-characterized microbial communities (e.g., human gut, termite gut, ungulate rumen) indicates that Pseudonocardia are not omnipresent and capable of life in all symbiotic niches. However, the frequent isolation from marine sponges and plants is intriguing and suggests that Pseudonocardia evolution may have been shaped by symbiosis with a wide array of organisms.
Autotrophic versus heterotrophic Pseudonocardia on the ant integument
Many Pseudonocardia isolated from attine ants are closely related to autotrophic Pseudonocardia, such as P. carboxydivorans, P. autotrophica, P. nitrificans (Fig. 2a), suggesting the possibility that the integumental Pseudonocardia of attine ants may also include autotrophs that are not limited in growth (or are less limited) by nutrients supplied by the ants, as assumed by the Coevolution-Codivergence hypothesis. The traditional formulation of the Coevolution- Codivergence hypothesis assumes that the ants have precise control over growth (upregulation of Pseudonocardia growth to combat an acute Escovopsis infection of the garden; Currie et al. 2003); such control may be less possible if Pseudonocardia are autotrophs that can thrive in nutrient-poor niches. The possibility of autotrophic Pseudonocardia on the ant integument could explain why it is relatively easy to isolate slow-growing Pseudonocardia with minimum- nutrient chitin medium (e.g., preferential selection of autotrophs), perhaps with similar biochemical properties as the closely related P. carboxydivorans that can fix carbon from carbon-monoxide (Park et al. 2008). While one could speculate that removal of toxic carbon-monoxide could be a potential function of Pseudonocardia in the interior of nests of fungus growing ants, it is equally possible that the anoxic conditions in the bottom of fungus gardens promotes non-adaptive proliferation of autotrophic Pseudonocardia (e.g., on the immobilized Acromyrmex workers at the bottom of fungus gardens, as documented by Currie et al. 2003).
Phylogenetic position of Pseudonocardia compacta and P. nitrificans
Our new 16S sequence of P. nitrificans (strain NRRL B-1664) places this species into a clade with P. alni, P. carboxydivorans, P. antarctica, and P. tropica sp. nov. (Clade 3, Fig. 2a). Our P. nitrificans sequence (GU318371) is identical to AJ252831 (= strain IMSNU 22071 = P. nitrificans; Lee et al. 2000), which currently is reported at GenBank as an undescribed Pseudonocardia.We believe that these two sequences are of much better quality than sequence X55609 (=strain IFAM 379 = P. nitrificans), which has been used in previous phylogenetic analyses (Warwick et al. 1994; McVeigh et al. 1994; Huang and Goodfellow 2011) but whose phylogenetic placement in these analyses is now dubious because of the poor sequence quality of X55609. More surprising was the placement of our 16S sequence of P. compacta (NRRL B-16170; GU318372) close to P. saturnea (Fig. 2b), that is, in a different subgroup of Pseudonocardia than accession AJ252825 (=IMSNU 20111; Fig. 2a) of P. compacta that is generally used in phylogenetic analyses of the genus. Interestingly, the placement of P. compacta in the vicinity of P. saturnea conforms approximately with the placement of P. compacta (DSM 43592) when using information from four protein-coding genes available at GenBank (EU722624, EU722594, EU722569, EU722535; UG Mueller unpublished observation), suggesting the possibility of strain mixup between culture collections. [Note inserted at proof stage: We recently obtained a strain labeled P. saturnea (NRRL B-16172) from the ARS Culture and Patent Culture Collections (USDA, Peoria, Illinois) and the 16S sequence of this strain was identical to the sequence of P. compacta (AJ252825; IMSNU 20111T) (H Ishak and UG Mueller unpublished). Because we had received the strain labeled NRRL B-16170 (believed to be P. compacta, but sequencing to P. saturnea) and the strain labeled NRRL B-16172 (believed to be P. saturnea, but sequencing to P. compacta) in two separate shipments from the ARS-USDA culture collection, and because we completed sequencing of NRRL B-16170 before we received NRRL B-16172, it seems that these two strains were mixed up prior to shipment from the ARSUSDA culture collection to our lab.] To resolve these issues, it will be necessary to re-sequence the type strains of P. compacta, P. nitrificans, and P. saturnea for 16S and other informative genes.
Conclusion
The two models juxtaposed in our analyses – the Coevolution-Codivergence model and the Acquisition model—represent two extreme viewpoints. The traditional Coevolution-Codivergence model stresses mutualism and long-term ant-Pseudonocardia-Escovopsis co-evolution and co-divergence, the more recent Acquisition model stresses diversity in ecological association and close links between antassociated and environmental Pseudonocardia. These two models make distinct predictions regarding the phylogenetic proximity of ant-associated and environmental Pseudonocardia. Our expanded Phylogenetic analyses conform more with the predictions of the Acquisition model, adding to recent antibiotic assays and phylogenetic analyses (Kost et al. 2007; Mueller et al. 2008; Haeder et al. 2009; Sen et al. 2009) that had started to question the plausibility of ant-Pseudonocardia-Escovopsis co-evolution and codivergence. Specifically, our phylogenetic analyses reveal previously unknown cases of 16S-sequenceidentical ant-associated and environmental Pseudonocardia (from soil and plants), and environmental Pseudonocardia that are nested within clades previously thought to contain only specialized, ant-associated Pseudonocardia. These phylogenetic patterns indicate frequent and continuous colonization of the attine integument by Pseudonocardia from environmental sources, microbial turnover on the ant integument over evolutionary time, and consequently disruption of long-term ant-Pseudonocardia associations. Such processes greatly reduce the potential for ant-Pseudonocardia co-evolution and for adaptive modification of Pseudonocardia to serve the fitness interests of the ants. Inadequate inclusion of environmental Pseudonocardia in previous analyses appears to have led to the premature identification of “ant-specific” Pseudonocardia clades. More sampling of environmental Pseudonocardia is needed to fully address the ecological links between ant-associated and environmental Pseudonocardia. Likewise needed are genomic and biochemical studies that evaluate genetic and phenotypic differences (or absence of differences) between ant-associated and environmental Pseudonocardia, particularly in the Neotropics where only few environmental bacterial surveys have been carried out to date. Because previous culture-dependent methods characterizing Pseudonocardia diversity on attine ants may have been biased (see Culturable and unculturable Pseudonocardia), future studies should also attempt to develop unbiased methods to characterize the true microbial diversity on attine ants. We encourage studies that test for both non-adaptive and adaptive roles of integumental microbes in carefully designed experiments, and we concur with the recent assessment by Boomsma and Aanen (2009) that the traditional Coevolution-Codivergence model greatly oversimplifies the nature and ecological diversity of ant-Pseudonocardia associations.
Supplementary Material
Acknowledgments
We thank Linquan Bai and Amanda Jones for the invitation to contribute to this special issue honoring Mike Goodfellow and his lifetime contribution to actinomycete ecology, systematics, and evolution. We thank Linda Kinkel, Liz Wellington, and Linquan Bai for the invitation to give a plenary talk at the 2009 ISBA Congress in Shanghai, for which we developed many ideas presented here. We thank two anonymous reviewers for constructive comments on the manuscript. The research was supported by NSF awards DEB-0919519, DEB-0639879, and DEB-0110073 to UGM; and NIH award R01GM067317 to RRG.
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s10482-010-9427-3) contains supplementary material, which is available to authorized users.
Contributor Information
Ulrich G. Mueller, Email: umueller@mail.utexas.edu, Section of Integrative Biology, University of Texas at Austin, Austin, TX 78712, USA. Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, TX 78712, USA
Heather Ishak, Section of Integrative Biology, University of Texas at Austin, Austin, TX 78712, USA.
Jung C. Lee, Section of Integrative Biology, University of Texas at Austin, Austin, TX 78712, USA. Center for Computational Biology and Bioinformatics, University of Texas at Austin, Austin, TX 78712, USA. Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, TX 78712, USA
Ruchira Sen, Section of Integrative Biology, University of Texas at Austin, Austin, TX 78712, USA.
Robin R. Gutell, Section of Integrative Biology, University of Texas at Austin, Austin, TX 78712, USA. Center for Computational Biology and Bioinformatics, University of Texas at Austin, Austin, TX 78712, USA. Institute for Cellular and Molecular Biology, University of Texas at Austin, Austin, TX 78712, USA
References
- Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF. PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl Environ Microbiol. 2005;71:8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacci M, Ribeiro SB, Casarotto MEF, Pagnocca FC. Biopolymer-degrading bacteria from nests of the leafcutting ant Atta sexdens rubropilosa. Braz J Med Biol Res. 1995;28:79–82. [Google Scholar]
- Boomsma JJ, Aanen DK. Rethinking crop-disease management in fungus-growing ants. Proc Natl Acad Sci USA. 2009;106:17611–17612. doi: 10.1073/pnas.0910004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cafaro MJ, Currie CR. Phylogenetic analysis of mutualistic filamentous bacteria associated with fungus-growing ants. Can J Microbiol. 2005;51:441–446. doi: 10.1139/w05-023. [DOI] [PubMed] [Google Scholar]
- Caldera EJ, Poulsen M, Suen G, Currie CR. Insect symbioses: a case study of past, present, and future fungus- growing ant research. Environ Entomol. 2009;38:78–92. doi: 10.1603/022.038.0110. [DOI] [PubMed] [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, Pande N, Shang Z, Yu N, Gutell RR. The comparative RNA Web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BioMed Cen Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreiro SC, Pagnocca FC, Bueno OC, Bacci M, Hebling MJA, da Silva OA. Yeasts associated with the nests of the leaf-cutting ant Atta sexdens rubropilosa Forel, 1908. Antonie van Leeuwenhoek. 1997;71:243–248. doi: 10.1023/a:1000182108648. [DOI] [PubMed] [Google Scholar]
- Currie CR. A community of ants, fungi, and bacteria: a multilateral approach to studying symbiosis. Annu Rev Microbiol. 2001;55:357–380. doi: 10.1146/annurev.micro.55.1.357. [DOI] [PubMed] [Google Scholar]
- Currie CR, Mueller UG, Malloch D. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci USA. 1999a;96:7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie CR, Scott JA, Summerbell RC, Malloch D. Fungus-growing ants use antibiotic-producing bacteria to control garden parasites. Nature. 1999b;398:701–704. [Google Scholar]
- Currie CR, Bot ANM, Boomsma JJ. Experimental evidence of a tripartite mutualism: bacteria protect ant fungus gardens from specialized parasites. Oikos. 2003;101:91–102. [Google Scholar]
- Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311:81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- Dattagupta S, Schaperdoth I, Montanari A, Mariani S, Kita N, Valley JW, Macalady JL. A novel symbiosis between chemoautotrophic bacteria and a freshwater cave amphipod. Int Soc Microb Ecol. 2009;3:935–943. doi: 10.1038/ismej.2009.34. [DOI] [PubMed] [Google Scholar]
- Diamond J. Virus and the whale: exploring evolution in creatures small and large. National Science Teachers Association Press; Arlington, VA: 2006. [Google Scholar]
- Fernández-Marín H, Zimmerman JK, Nash DR, Boomsma JJ, Wcislo WT. Reduced biological control and enhanced chemical pest management in the evolution of fungus farming in ants. Proc R Soc Lond B. 2009;276:2236–2269. doi: 10.1098/rspb.2009.0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow M, Williams ST. Ecology of actinomycetes. Annu Rev Microbiol. 1983;37:189–216. doi: 10.1146/annurev.mi.37.100183.001201. [DOI] [PubMed] [Google Scholar]
- Gould SJ, Lewontin RC. The spandrels of San Marco and the Panglossian Paradigm: a critique of the Adaptationist Programme. Proc R Soc Lond B. 1979;205:581–598. doi: 10.1098/rspb.1979.0086. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16S-like ribosomal RNA. Prog Nucl Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Gutell RR, Power A, Hertz G, Putz E, Stormo G. Identifying constraints on the higher-order structure of RNA: continued development and application of comparative sequence analysis methods. Nucleic Acids Res. 1992;20:5785–5795. doi: 10.1093/nar/20.21.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Larsen N, Woese CR. Lessons from an evolving ribosomal RNA: 16S and 23S rRNA structure from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell RR, Lee JC, Cannone JJ. The accuracy of ribosomal RNA comparative structure models. Curr Opin Struct Biol. 2002;12:301–310. doi: 10.1016/s0959-440x(02)00339-1. [DOI] [PubMed] [Google Scholar]
- Haeder S, Wirth R, Herz H, Spiteller D. Candicidinproducing Streptomyces support leaf-cutting ants to protect their fungus garden against the pathogenic fungus Escovopsis. Proc Natl Acad Sci USA. 2009;106:4742–4746. doi: 10.1073/pnas.0812082106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Goodfellow M. Pseudonocardia (Henssen 1957, 408AL) emend; Park, Park: 2011. Genus I. [Google Scholar]
- Kim 2008. 2477VP. In: Goodfellow M, Kämpfer P, Busse H-J, Trujillo M, Suzuki K, Ludwig W, Whitman WB, editors. Bergey’s manual of systematic bacteriology. 2. Vol. 5. Springer; New York: the actinobacteria. (in press) [Google Scholar]
- Kost C, Lakatos T, Böttcher I, Arendholz W-R, Redenbach M, Wirth R. Non-specific association between filamentous bacteria and fungus-growing ants. Naturwissenschaften. 2007;94:821–828. doi: 10.1007/s00114-007-0262-y. [DOI] [PubMed] [Google Scholar]
- Lee SD, Kim ES, Hah YC. Phylogenetic analysis of the genera Pseudonocardia and Actinobispora based on 16S ribosomal DNA sequences. FEMS Microbiol Lett. 2000;182:125–129. doi: 10.1111/j.1574-6968.2000.tb08885.x. [DOI] [PubMed] [Google Scholar]
- Maddison DR, Maddison WP. MacClade 4: analysis of phylogeny and character evolution. Sinauer Associates; Sunderland, MA: 2000. [DOI] [PubMed] [Google Scholar]
- McVeigh HP, Munro J, Embley TM. The phylogenetic position of Pseudoamycolata halophobica (Akimov et al. 1989) and a proposal to reclassify it as Pseudonocardia halophobica. Int J Syst Bacteriol. 1994;44:300–302. doi: 10.1099/00207713-44-2-300. [DOI] [PubMed] [Google Scholar]
- Mueller UG, Gerardo NM, Aanen DK, Six DL, Schultz TR. The evolution of agriculture in insects. Ann Rev Ecol Evol Sys. 2005;36:563–595. [Google Scholar]
- Mueller UG, Dash D, Rabeling C, Rodrigues A. Coevolution between attine ants and actinomycete bacteria: a reevaluation. Evolution. 2008;62:2894–2912. doi: 10.1111/j.1558-5646.2008.00501.x. [DOI] [PubMed] [Google Scholar]
- Oh D-C, Poulsen M, Currie CR, Clardy J. Dentigerumycin: a bacterial mediator of an ant-fungus symbiosis. Nat Chem Biol. 2009;5:391–393. doi: 10.1038/nchembio.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen GJ, Overbeek R, Larsen N, Marsh TL, McCaughey MJ, Maciukenas MA, Kuan WM, Macke TJ, Xing Y, Woese CR. The ribosomal database project. Nucleic Acids Res. 1992;20(suppl):2199–2200. doi: 10.1093/nar/20.suppl.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SW, Park ST, Lee JE, Kim YM. Pseudonocardia carboxydivorans sp. nov., a carbon monoxide-oxidizing actinomycete, and an emended description of the genus Pseudonocardia. Int J Syst Evol Microbiol. 2008;58:2475–2478. doi: 10.1099/ijs.0.65765-0. [DOI] [PubMed] [Google Scholar]
- Posada DK, Crandall A. Model selection and model averaging in phylogenetics: advantages of the AIC and Bayesian approaches over likelihood ratio tests. Bioinformatics. 1998;14:817–818. doi: 10.1080/10635150490522304. [DOI] [PubMed] [Google Scholar]
- Poulsen M, Erhardt DP, Molinaro DJ, Ting-Li L, Currie CR. Antagonistic bacterial interactions help shape hostsymbiont dynamics within the fungus-growing antmicrobe mutualism. PLoS ONE. 2007;2:e960. doi: 10.1371/journal.pone.0000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen M, Little AEF, Currie CR. Fungus-growing antmicrobe symbiosis: using microbes to defend beneficial associations with symbiotic communities. In: White JF, Torres MS, editors. Defensive mutualism in microbial symbiosis. CRC Press; Boca Raton: 2009. pp. 149–162. [Google Scholar]
- Rodrigues A, Bacci M, Mueller UG, Ortiz A, Pagnocca FC. Microfungal “weeds” in the leafcutter ant symbiosis. Microb Ecol. 2008;56:604–614. doi: 10.1007/s00248-008-9380-0. [DOI] [PubMed] [Google Scholar]
- Rodrigues A, Cable RN, Mueller UG, Bacci M, Pagnocca FC. Antagonistic interactions between garden yeasts and microfungal garden pathogens of leaf-cutting ants. Antonie Leeuwenhook. 2009;96:331–342. doi: 10.1007/s10482-009-9350-7. [DOI] [PubMed] [Google Scholar]
- Santos AV, Dillon RJ, Dillon VM, Reynolds SE, Samuels RI. Occurrence of the antibiotic producing bacterium Burkholderia sp. in colonies of the leaf-cutting ant Atta sexdens rubropilosa. FEMS Microbiol Lett. 2004;239:319–323. doi: 10.1016/j.femsle.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Sen R, Ishak HD, Estrada D, Dowd SE, Hong E, Mueller UG. Generalized antifungal activity and 454-screening of Pseudonocardia and Amycolatopsis bacteria in nests of fungus-growing ants. Proc Natl Acad Sci USA. 2009;106:17805–17810. doi: 10.1073/pnas.0904827106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomeo F, Portillo MC, Gonzalez JM, Laiz L, Saiz-Jimenez C. Pseudonocardia in white colonizations in two caves with Paleolithic paintings. Int Biodeterior Biodegradation. 2008;62:483–486. [Google Scholar]
- Suen G, Currie CR. Ancient fungal farmers of the insect world. Microbiol Today. 2008;35:172–175. [Google Scholar]
- Warwick S, Bowen T, McVeigh H, Embley TM. A phylogenetic analysis of the family Pseudonocardiaceae and the genera Actinokineospora and Saccharothrix with 16S rRNA sequences and a proposal to combine the genera Amycolata and Pseudonocardia in an emended genus Pseudonocardia. Int J Syst Bacteriol. 1994;44:293–299. doi: 10.1099/00207713-44-2-293. [DOI] [PubMed] [Google Scholar]
- Woese CR. Interpreting the universal phylogenetic tree. Proc Natl Acad Sci USA. 2000;97:8392–8396. doi: 10.1073/pnas.97.15.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Magrum LJ, Gupta R, Siegel RB, Stahl DA, Kop J, Crawford N, Brosius R, Gutell R, Hogan JJ, Noller HF. Secondary structure model for bacterial 16S ribosomal RNA: phylogenetic, enzymatic and chemical evidence. Nucleic Acids Res. 1980;8:2275–2294. doi: 10.1093/nar/8.10.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR, Gutell R, Gupta R, Noller HF. Detailed analysis of the higher-order structure of 16S-like ribosomal ribonucleic acids. Microbiol Rev. 1983;47:621–669. doi: 10.1128/mr.47.4.621-669.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Poulsen M, Currie CR. Symbiont recognition of mutualistic bacteria in Acromyrmex leaf-cutting ants. Int Soc Microb Ecol J. 2007;1:313–320. doi: 10.1038/ismej.2007.41. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. PhD dissertation. The University of Texas; Austin: 2006. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.