Abstract
Discoidin I and Discoidin II (DiscI and DiscII), are N-acetylgalactosamine-binding proteins from Dictyostelium discoideum. They consist of two domains with an N-terminal discoidin domain and a C-terminal H-type lectin domain. They were cloned and expressed in high yield in recombinant form in Escherichia coli. Although both lectins bind galactose and N-acetylgalactosamine, glycan array experiments performed on the recombinant proteins displayed strong differences in their specificity for oligosaccharides. DiscI and DiscII bind preferentially to Gal/GalNAcβ1-3Gal/GalNAc- and Gal/GalNAcβ1-4GlcNAcβ1-6Gal/GalNAc- containing glycans respectively. The affinity of the interaction of DiscI with monosaccharides and disaccharides was evaluated using isothermal calorimetry experiments. The tree-dimensional structures of native DiscI and its complexes with GalNAc, GalNAcβ1-3Gal and Galβ1-3GalNAc were solved by X-ray crystallography. DiscI forms trimers with involvement of calcium at the monomer interface. The N-terminal discoidin domain presents structural similarity to F-type lectins such as the eel agglutinin where an amphiphilic binding pocket suggests a possible carbohydrate-binding activity. In the C-terminal H-type lectin domain, the GalNAc residue establishes specific hydrogen bonds that explain the observed affinity (Kd = 3 10-4 M). The different specificities of DiscI and DiscII for oligosaccharides were rationalized from the different structures obtained by either X-ray crystallography or molecular modelling.
Keywords: H type lectin, F type lectin, discoidin domain, adhesion, oligosaccharide
Introduction
Lectins are defined as ubiquitous proteins of non-immune origin that interact reversibly and specifically with carbohydrates without modifying them. A large panel of lectins has been identified in invertebrates and considerable attention is focused on their roles in biological recognition. Invertebrate lectins are often involved in specific binding to bacterial polysaccharides, playing a role in establishment of symbiosis or innate immunity 1,2,3. In other cases, they are involved in self-recognition and in cell-cell aggregation that are required for the building of multicellular organisms such as corals and sponges.4 They can be useful tools in cancer diagnosis/prognosis as histochemical markers or in cancer therapy thanks to antitumoral activities.5,6,7
The slime mold Dictyostelium discoideum grows as free-living amoebae in the soil, feeding on bacteria. Upon starvation, it undergoes a complex developmental cycle in which about 10.000-50.000 individual amoebae aggregate to form a multicellular fruiting body that is able to produce spores. The aggregation of individual amoeba occurs by chemotaxis to periodic cAMP signals (reviewed in8). During differentiation, numerous new proteins are synthesized, and some of the most abundant of these new products are the two N-acetylgalactosamine (GalNAc)-binding proteins Discoidin I and II (DiscI and II).9,10 They are virtually undetectable in cells growing on bacteria but constitute over 1% of the cellular proteins in aggregated cells.9 They present overlapping but distinct sugar specificities.10 The production of DiscI is prominent in aggregating cells, which accumulates intracellularly and upon externalisation in multilamellar bodies while DiscII is prominent during fruiting body formation and is localized in the prespore vesicules.11,12 The precise function of these lectins as well as their biological ligands remain unknown.
DiscI is polymorphic with at least three isoforms per strain coded by genes forming a small, coordinately regulated multigene family.13,14 The three genes dscA, dscC and dscD are found on chromosome 2 and are duplicated on the same chromosome in some laboratory strains (AX3 and AX4). They are often used as a marker of early development in D. discoideum.15,16 DiscI expression is developmentally-regulated at the transcriptional level by several signal transduction pathways involving cAMP, prestarvation factor (PSF) and conditioned medium factor (CMF).17,18,19 Several mutants and antisense transformants analysis showed that DiscI was apparently not necessary for aggregation since mutants plated at high density can aggregate and form fruiting bodies.17 The lack of gene product impairs the process of cell streaming in early development, where it seems important in the formation of head-to-tail streams by aggregating cells.20 DiscI was also shown to be involved in cell substratum attachment and ordered cell migration during aggregation by a fibronectin-like mechanism.21 It contains the RGD motif, which is the cell attachment site found in large collection of adhesive proteins.22,23 The proposed receptor for this RGD sequence is a developmentally regulated cell-surface glycoprotein of 67 KDa.24 DiscI also recognises glycoconjugates that contain GalNAc in the slime coat around aggregates and in multilamellar bodies since the interaction is completely blocked in the presence of GalNAc.25,26,27 The sugar binding site was shown to be different of the cell adhesion site and requires a divalent cation.24,28
DiscI is a two domain protein of 253 amino acids with higher affinity for GalNAc than for galactose (Gal).10 It displays 48% sequence identity with DiscII whose trimeric structure has been recently solved in unliganded state and in complex with Gal and GalNAc.29 The N-terminal domain, referred as the discoidin domain (DS domain), is a structural and functional motif found in various proteins from both eukaryotic and prokaryotic origins. This domain shows considerable functional diversity via interactions with a wide range of molecules and is mainly involved in cell surface mediated regulatory events and glycoconjugates binding.30,31,32 The C-terminal domain belongs to the H-type lectin family recently identified in invertebrates such as snails and corals.33
We present here the specificity and affinity of DiscI and DiscII for a variety of carbohydrate ligands as well as the crystal structures of DiscI in unliganded form and in complex with GalNAc, GalNAcβ1-3Gal and Galβ1-3GalNAc. The specificities, affinities, structures and binding sites of both discoidins are compared and discussed. DiscI and DiscII present interesting differences in term of oligosaccharide binding, which may be correlated to their differences in localisation and to their role in the slime mold.
Results
Specificity of D. discoideum lectins with Glycan Microarray Resource
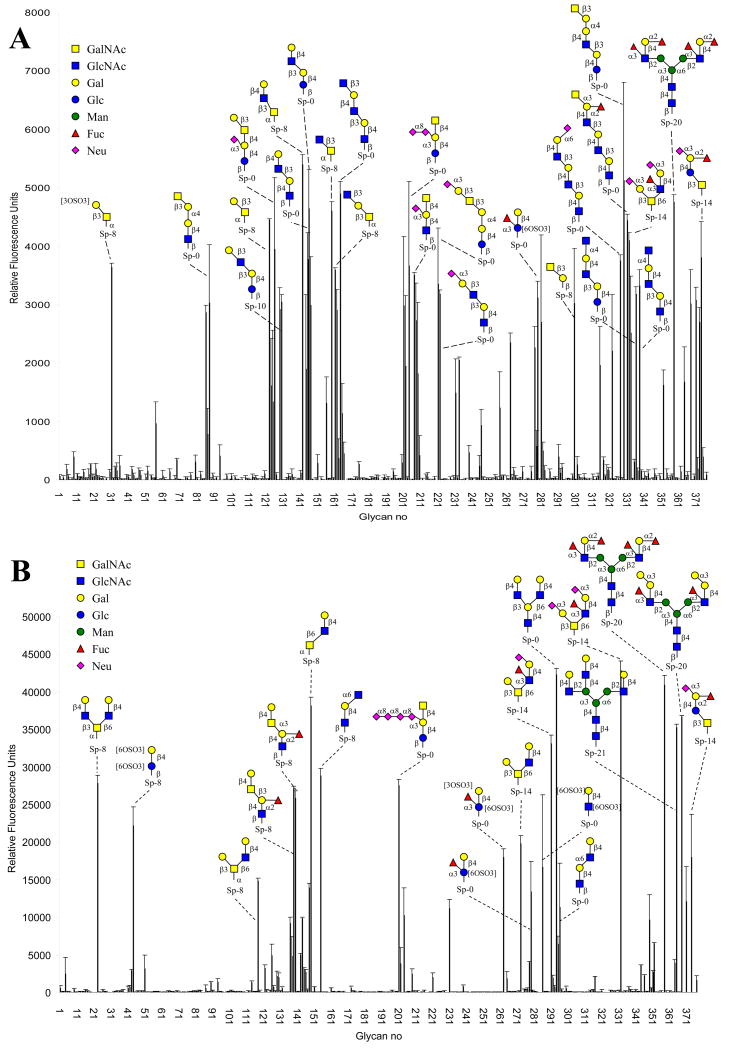
Recombinant DiscI was obtained with a yield of 20 mg l-1 of culture and its molecular weight, checked by mass spectrometry, corresponded to the expected value. Recombinant DiscII was produced as previously described.29 Both lectins were labelled with Alexa 488 and assayed on Glycan Microarray available at the Consortium for Functional Glycomics using concentrations varying from 0.01 to 200 μg ml-1. DiscI gave excellent signals at very low concentration and the analysis displayed in Figure 1A has been obtained for 0.04 μg ml-1. Strong binding to the glycochips was also observed for DiscII and Figure 1B has been obtained with 0.5 μg ml-1. These concentrations were selected to optimize the quality of the figure. Both proteins gave signal that increased in a parallel way with the concentration and reached a maximum at approx 0.5 to 1 μg ml-1.
Figure 1. Glycan array analysis.
A: DiscI at 0.04 μg ml-1 and B: DiscII at 0.5 μgml-1 as measured by fluorescence intensity. Complete results including comprehensive oligosaccharide list (Plate Array) are available from the Consortium of Functional Glycomics (http://www.functionalglycomics.org/).
Quite surprisingly, there is almost no superimposition in the binding specificity of the two discoidins. When looking at disaccharides, Discoidin I binds to several β1-3 linked disaccharides with a galacto residue at the reducing position such as GlcNAcβ1-3GalNAc (n° 159), Galβ1-3GalNAc (n° 123) and less strongly to GalNAcβ1-3Gal (n° 300). Elongation by additional residues or by sulphate groups are well tolerated on position 3 or 4 of the non-reducing sugar while the reducing sugar can be only elongated at position 1 and no branching is accepted. Polyllactosamine oligosaccharides containing repetitive Galβ1-4GlcNAcβ1-3-sequences are high affinity ligands. In few cases, such as in ganglioside of the GM2 to GT2 series (n° 201 to 209), the terminal GalNAc is also recognized.
On the opposite, DiscII does not recognize efficiently any disaccharide. GlcNAcβ1-6Gal/GalNAc is the preferred motif but is recognized only when part of larger oligosaccharides. The trisaccharide with the highest affinity is Galβ1-4GlcNAcβ1-6GalNAc (n° 149). In most of the high affinity oligosaccharides, this trisaccharide is branched with a Gal or GlcNAc residue on position 3 of the GalNAc. The glycan array data indicate therefore that DiscI has higher affinity for GalNAc-containing oligosaccharide elongated on position 3, while DiscII prefers Gal/GalNAc-containing motif substituted on position 6. It could be noted that both lectins bind to large fucosylated and/or sialylated glycans such as n° 358 and 371 but those could be ligands of the discoidin domain as discussed below.
Affinity studies by ITC microcalorimetry
The affinity of DiscII towards GalNAc and β–Methyl-galactoside (βMeGal) was previously determined using titration microcalorimetry, leading to dissociation constants of 1.15 and 0.95 mM, respectively.29 When DiscI is assayed with the same approach, higher affinity is observed towards GalNAc (Kd = 0.3 mM) while galactose and βMeGal have lower affinity (Kd in the range of 3 to 4 mM) (Table 1). DiscI affinity for disaccharides confirm the preference for β1-3Gal/GalNAc linked disaccharides. Among tested disaccharides, Galβ1-3GalNAc is the best ligand with a Kd of 27 μM, which is a rather high affinity for lectin/glycan interaction. GalNAcβ1-3Gal is also a strong ligand with a Kd of approx 50 μM, while Galβ1-4GlcNAc (LacNAc) is a poor ligand with a Kd in the millimolar range.
Table 1.
Microcalorimetry data. All values are averaged over two or three experiments. The reported standard deviations are calculated upon averaging of those experiments, unless otherwise specified.
| Protein | Ligand | KD (μM) |
n | -ΔG (kJ/mol) |
-ΔH (kJ/mol) |
-TΔS (kJ/mol) |
|---|---|---|---|---|---|---|
| DiscI | GalNAc | 302 ± 27 | 1a | 20.0 ± 0.2 | 33.4 ± 5.8 | 13.3 ± 6.0 |
| Galactose | 3770 ± 200 | 1 a | 13.8 ± 0.1 | 31 ± 7.4 | 17.1 ± 7.5 | |
| αMeGalactoside | 8390 ± 700 | 1 a | 11.8 ± 0.2 | 42.4 ± 5.9 | 30.5 ± 6.1 | |
| βMeGalactoside | 2960 ± 270 | 1 a | 14.4 ± 0.2 | 40.3 ± 2.4 | 25.9 ± 2.2 | |
| Gal1β1-4GlcNAc (LacNAc) | 2390 ±170 | 1 a | 15.0 ± 0.2 | 38.4 ± 2.2 | 23.3 ± 2.0 | |
| Gal1β-3GalNAc | 27.2 ± 4.6 | 1.03± 0.01 | 26.1 ± 0.4 | 45.6 ± 1.1 | 19.5 ± 1.5 | |
| GalNAcβ1-3Gal | 49.4 ± 1.5 | 0.81 ± 0.01 | 24.5± 0.3 | 53.2 ± 2.9 | 28.81 ± 2.8 | |
| DiscII | GalNAcb | 1115 | 1 a | 16.8 | 20.3 | 3.5 |
| βMeGalactosideb | 952 | 1 a | 17.4 | 24.3 | 6.7 | |
| Gal1β1-4GlcNAc (LacNAc) | 1221 ±23 | 1 a | 16.7 ± 0.5 | 37.0 ± 0,3 | 20.5 ± 0.4 | |
| Gal1β1-3GalNAcc | 1377 | 1 a | 16.3 | 42.3 | 26.0 | |
| GalNAcβ1-3Galc | 1113 | 1 a | 16.8 | 45.6 | 28.8 | |
fixed stoichiometry during fitting procedure for low affinity interaction,
taken from 29,
only one experiment due to lack of material.
The affinity of DiscII towards disaccharides was also tested and it was confirmed that none of the disaccharides cited above display high affinity since all observed dissociation constants where in the millimolar range. The trisaccharidic ligand best recognised by DiscII is not available and could therefore not be tested.
Titration curves of DiscI interacting with GalNAc and Galβ1-3GalNAc are shown in Figure 2. The thermograms display a large decrease in the exothermic heat of binding while saturation is achieved. For all ligands, the interaction observed upon binding to the lectin was found to be strongly driven by enthalpy as a result of the high number of hydrogen bonds with a strong unfavourable entropy contribution classically attributed either to solvent rearrangement or to the loss of ligand conformational flexibility.34
Figure 2. Isothermal titration microcalorimetry.
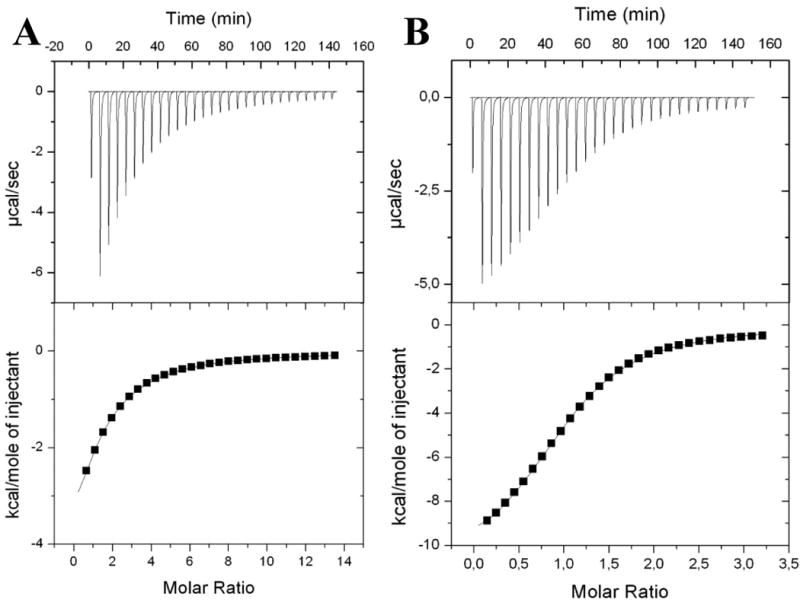
A: Results for GalNAc monosaccharide (10 mM) binding to DiscI. B: Results for Galβ1,3GalNAc disaccharide (2 mM) binding to DiscI. Experiments with 0.15 mM of protein in 20 mM Tris/HCl pH 7.5 and 150 mM NaCl buffer solution at 25°C. Top: data from 30 automatic injections of sugar of 10 μl each into the cell containing DiscI. Bottom: plot of the total heat released as a function of ligand concentration for the titration shown above (squares). The solid line represents the best least-square fit for the obtained data.
Crystal structure of DiscI
DiscI gives rise in four days to bipyramidal crystals, which diffracted to high resolution. They belong to the C2221 spacegroup with one trimer in the asymmetric unit. The final model of the native structure has a crystallographic R value of 16.7 %, with a corresponding Rfree of 20.6 % for all observed data between 28.6 and 1.8 Å resolution. In this model, all 253 amino acids could be observed in the electron density as well as some 2-methyl-2,4-pentanediol (MPD) molecules, sulphate, calcium and nickel ions (refinement statistics in Table 2). In chain A, an additional alanine resulting from the cleavage of the N-terminal His-tag is found. The overall fold of DiscI is comparable to the one observed for DiscII with the formation of a trimer through the intertwining of the two domains of the protein, Figure 3.29 The native structures of both recombinant discoidins present an rmsd of 1.2 Ǻ for the 245 aligned Cα calculated by Secondary Structure Matching.35
Table 2.
Data collection and refinement statistics.
| rDiscI | rDiscI/GalNAc | rDiscI/GalNAcβ1-3Gal | rDiscI/Galβ1-3GalNAc | |
|---|---|---|---|---|
| Spacegroup | C2221 | C2221 | C2221 | C2221 |
| Cell dimensions | ||||
| a,b,c (Å) | 72.5, 274.3, 85.9 | 72.4, 274.0, 86.1 | 72.6, 274.0, 85.8 | 72.5, 273.7, 85.8 |
| α, β, γ (deg.) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (outer shell), Å | 28.6-1.8 (1.84-1.8) | 47.4-1.75 (1.79-1.75) | 43.7–1.6 (1.67-1.6) | 47.3 1.8 (1.92-1.82) |
| Measured/Unique reflections | 299983/75469 | 390844/86561 | 362907/114648 | 615779/76712 |
| Average multiplicity | 4.0 (3.8) | 4.5 (4.1) | 3.2 (3.0) | 8.0 (7.7) |
| Rmerge | 0.070 (0.234) | 0.069 (0.389) | 0.065 (0.283) | 0.094 (0.376) |
| Completeness (%) | 94.8 (94.8) | 99.9 (99.9) | 99.2 (99.1) | 99.7 (98.7) |
| Average I/σI | 5.9 (2.0) | 8.1 (1.8) | 9.8 (2.6) | 15.1 (4.7) |
| Refinement | ||||
| Rcryst/Rfree | 16.7/20.6 | 15.9/19.3 | 15.2/18.0 | 14.1/18.0 |
| Reflections/Free reflections | 71665/3802 | 82187/4351 | 108859/5754 | 72846/3849 |
| Rmsd bonds, Å | 0.017 | 0.016 | 0.017 | 0.017 |
| Rmsd angles,° | 1.5 | 1.55 | 1.6 | 1.6 |
| Protein atoms | 1995/1990/1990 | 1995/1990/1995 | 2019/2008/2002 | 2019/2008/2002 |
| Bfac, Å2 | 17.5/18/18.1 | 14.7/15.3/15.3 | 12.2/12.6/13.8 | 12.2/12.6/13.8 |
| Water molecules | 348/327/306 | 371/350/332 | 409/369/343 | 384/363/333 |
| Bfac, Å2 | 30.7/30.9/31.3 | 29.2/29.1/29.2 | 26.7/27.0/28.8 | 25.0/24.0/25.8 |
| Ligand atoms | 15/15/15 | 26/26/26 | 41/41/41 | |
| Bfac, Å2 | 24.9/23.3/29.2 | 23.9/28.2/28.5 | 12.3/13.5/14.7 | |
| Hetero atoms | 28/14/22 | 27/33/35 | 28/12/13 | 35/19/19 |
| Bfac, Å2 | 29/28/26.4 | 26.9/25.7/26.9 | 18.9/28.2/24.6 | 22.1/29.6/28.7 |
| Ramachandran Plot | ||||
| Most favour regions | 88 | 88 | 87.9 | 87.3 |
| Allowed regions | 12 | 12 | 12 | 12.7 |
| Generous regions | 0.1 | |||
| PDBcode | 2W94 | 2W95 | 2WN3 | 2WN2 |
Figure 3.
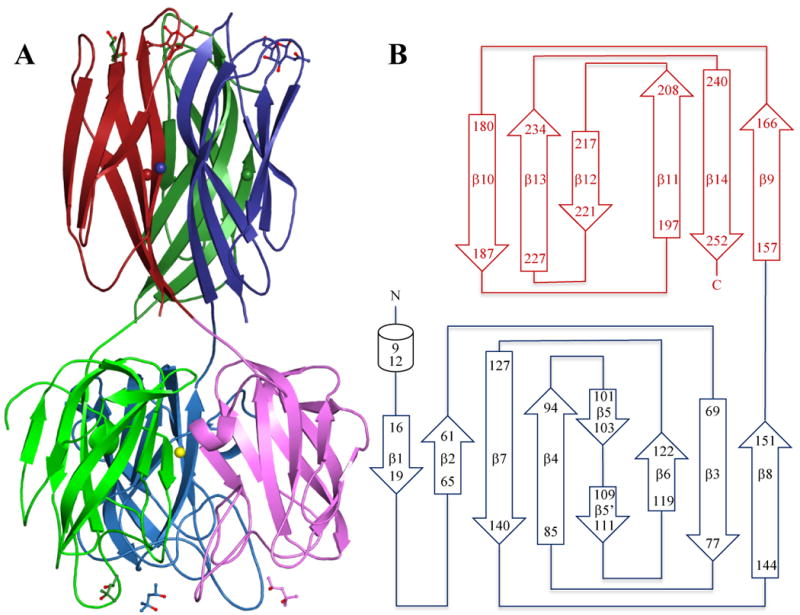
A: Crystal structure of the DiscI trimer coloured by chain with the N-terminal domain in light colours and C-terminal domain in dark colours. MPD and GalNAc ligands are represented in ball and sticks. Calcium ions are represented as spheres according to chain colours and nickel ions as a yellow sphere. B: Schematic representation of DiscI topology.
The N-terminal domain (1-155) presents the classical topology of the discoidin domain with a β-sandwich consisting of eight antiparallel strands arranged in two β-sheets of five and three β-strands packed against one another to form a distorted barrel, Figure 3.36,37 Prominent loops protrude like spikes on the opposite side of the termini creating a ligand binding site, where either a molecule of MPD or glycerol depending on the structure, is observed. At the level of the pseudo 3-fold axis of the trimer, a strong peak of positive density appears between the three His16 side chains. It was modelled as nickel and is probably a residual from the affinity chromatography. In the disaccharide complexes resulting from another batch of protein, it was modelled as water since there was no extra positive density and a slight movement of the His16 side chain yielded to coordination distances more relevant to water than nickel.
The C-terminal domain presents the immunoglobulin-like β-sandwich fold typical of H-type lectins first observed for Helix pomatia agglutinin (HPA) and then for DiscII.29,33 It consists of six antiparallel β-strands, which form two sheets of three β-strands stacked against one another.33 Inspection of the electron density map also reveals the presence of three ions identified as calcium according to the distance values and coordination numbers.38 Each calcium is found in the middle of the C-terminal domain at the interface between two monomers. It presents seven coordinations, five of them in a plane, one above the plane and one under.38 The contacts involve bidentate Asp246 (β14), monodentate Asp164 (β9) and Asp203 (β11), Gln219 (β12) from the neighbouring monomer and two water molecules coordinating the main chain oxygen atoms of Leu198 and Val200 (β11) from the neighbouring monomer. The interactions that involve four β-strands over six would help to maintain the trimeric arrangement of the lectin domain. That would explain the dependence for divalent cation for carbohydrate binding and the destabilising role of EDTA and EGTA observed for DiscI.28 In DiscII, which is not calcium-dependant, the position of the calcium ion is occupied by the NZ of Lys206 and other interactions stabilise the trimer.
Discoidin domain
The N-terminal discoidin domain of DiscI is found in a variety of extracellular and membrane proteins from about 100 eukaryotes and 300 prokaryotes such as coagulation factors, neuropilins, discoidin domain receptors (DDRs) or carbohydrate binding modules (CBMs) associated with enzymes.30,31 Such domains evolved from a common ancestor to bind very diverse ligands (collagen, lipids, carbohydrates or growth factors) via a binding site formed by surface loops (spikes) that protrude at the β-barrel opening. The nature and length of those spikes define therefore the protein specificity and are responsible for its biological function. Search of the protein structure database using DALI shows that besides DiscII, coagulation factors, neuropilin, lactadherin and several carbohydrate binding modules share the DS domain fold.39 The latter belong to families CBM32 and CBM47 in the CAZy database (http://www.cazy.org), which bind mainly galactose and fucose respectively.40 The structural homologues of discoidins that also forms trimers are F-type fucose binding lectins comprising the Anguilla anguilla agglutinin and the F-lectin from striped bass (PDB 3CQO).41
The overlay of the monomer of those different proteins shows that their carbohydrate binding site is located at the same position than the observed MPD/glycerol binding site of DiscI and DiscII. The ligand environment in AAA, CBMs and discoidins presents similarity, Figure 4. A hydrophobic residue (Tyr23 in DiscI, Leu23 in AAA, Tyr18 in the F-lectin and Trp/Tyr in CBMs), which makes stacking interactions with the ligand, is always found in the first spike of the first loop. Several charged residues (His27, His142, Gln84 in DiscI), which would be involved in the hydrogen bond network, are also observed but they are different from those of the F-type lectins. At position 82, an alanine is observed in isoform A contrary to a histidine in the other isoforms and in DiscII. This residue could also play a role in substrate binding and its possible regulation by phosphorylation has been proposed in DiscII.29 The third loop in discoidins contains the RGD motif, which promotes cell attachment to substrate in mammalian cells.23 The arginine of this motif is conserved in homologous F-type lectins and CBMs. In F-type lectins, this residue makes hydrogen bonds with the ring oxygen and an axial hydroxyl group at carbon 4 selecting the L-galacto configuration of fucose, Figure 4B. The binding site of the DS domain of discoidins present all required characteristics to bind carbohydrates.
Figure 4. Superposition of the discoidin domain binding site.
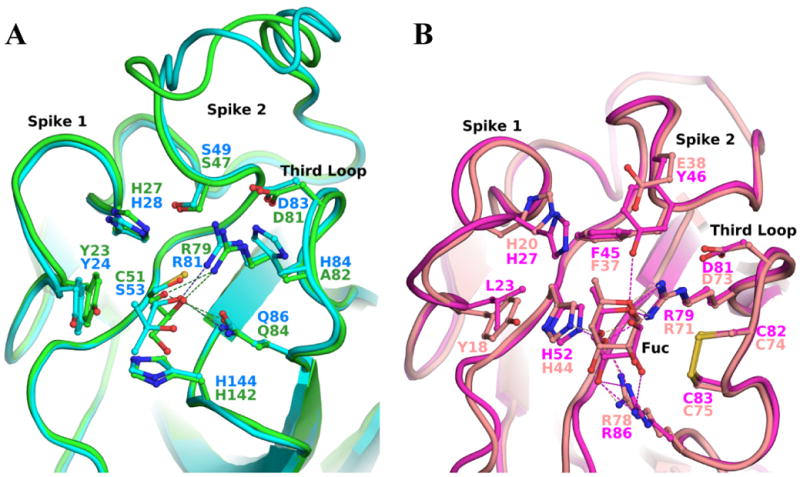
A: Blow up of the glycerol/MPD binding site of DiscI (green) and DiscII (cyan) respectively. B: Blow up of fucose binding site of AAA (magenta) and the F-Lectin from striped Bass (salmon) in the same orientation than discoidins.
H-Lectin binding site in complex with carbohydrates
Crystallization studies of complexes with mono and disaccharides were performed in order to understand the tenfold increase in DiscI affinity for disaccharides Galβ1-3GalNAc and GalNAcβ1-3Gal, when compared to GalNAc monosaccharide. The complex structure of DiscI with GalNAc at 1.75 Å resolution was obtained by soaking while those with the disaccharides Galβ1-3GalNAc and GalNAcβ1-3Gal at 1.8 and 1.6 Å respectively were obtained by cocrystallisation. All crystals belong to the C2221 space group and data and refinement statistics are summarised in Table 2. Inspection of the initial electron-density map revealed for each monomer unambiguous density for the sugars in a shallow groove at the interface between two monomers. In the DiscI/GalNAcβ1-3Gal complex, the density for the Gal residue was of good enough quality only after positioning and refinement of the GalNAc moiety.
In the DiscI/GalNAc complex, the monosaccharide is found in the same position as the one observed previously in the other H-type lectins.29,33,42 It is mainly involved in direct hydrogen bonds with the protein, Figure 5A. The O3 hydroxyl interacts with Trp235 NE1 and Asp206 OD2 (neighbouring monomer), the O4 hydroxyl with Asp206 OD1 (neighbouring monomer) and Arg215 NE and NH2, the ring oxygen with Arg215 NH2, the O7 and O1 hydroxyls with Asn236 ND2 (Details of distances are listed in Table 3). A single water-mediated interaction is usually observed between the O6 hydroxyl and Asn174 ND2. Tyr241 side chain (neighbouring monomer) is involved in hydrophobic interaction with part of the sugar ring which includes C3, C4, C5 and C6. Its orientation is quite different from the one observed in HPA where its equivalent Tyr89 has rotated of about 145° around the Cα-Cβ bond and only stacks against the O6 hydroxymethyl group of GalNAc.
Figure 5. Analysis of the GalNAc binding site of H-type lectins.

A: Representation of the maximum-likelihood weighted 2mFo-DFc electron density around the GalNAc complexed to DiscI contoured at 1σ (0.41 eA3). Hydrogen bonds are represented as dotted lines and waters as magenta spheres. B: Overlay of the GalNAc binding site of DiscI (cyan/marine), DiscII (green/yellow) and HPA (salmon/magenta). The residues are labelled according to their polypeptide chain and number.
Table 3.
Contacts between DiscI and its ligand in the complex crystal structures with standard deviation calculated for the three binding sites of the trimer between brackets.
| Complex | GalNAc | GalNAcβ1-3Gal | Galβ1-3GalNAc | |
|---|---|---|---|---|
| Sugar atom | Protein atom | Distances (Å) | ||
| GalNAc site | ||||
| O1 | Asn236 ND2 | 3.15 (0.10) | 3.03 (0.01) | |
| O7 | Asn236 ND2 | 3.01 (0.10) | 2.90 (0.03) | 3.04 (0.02) |
| Wat - Asn236 N | 2.73 (0.10) - 2.89 (0.07) | |||
| O3 | Trp235 NE1 | 2.94 (0.09) | 2.86 (0.05) | 3.06 (0.06) |
| Asp206a OD2 | 2.55 (0.10) | 2.54 (0.03) | ||
| O4 | Asp206a OD1 | 2.55 (0.00) | 2.58 (0.04) | |
| Arg215 NE | 2.78 (0.08) | 2.96 (0.10) | 2.86 (0.04) | |
| Arg215 NH2 | 3.15 (0.10) | 3.17 (0.01) | ||
| Trp235 NE1 | 3.09 (0.04) | |||
| Asp206a OD2 | 2.60 (0.06) | |||
| O5 | Arg215 NH2 | 3.17 (0.03) | 2.98 (0.08) | 2.91 (0.02) |
| O6 | Wat - Asn174 ND2a | 2.4b - 3.05b | 2.54 (0.06) - 2.95 (0.06) | 2.75 (0.13) - 3.15 (0.10) |
| Asp206a OD1 | 2.50b | |||
| Gal site | ||||
| O3 | Asn236 ND2 | 3.00 (0.06) | ||
| O4 | Asn236 ND2 | 2.97 (0.07) | ||
| Wat - Gln204a OE1 | 2.90 (0.27) 2.77 (0.13) | |||
| O5 | Trp235 NE1 | 3.24 (0.04) | ||
| O6 | Ser242a OG | 2.70 (0.02) | ||
| Asp206a OD2 | 2.72 (0.03) | |||
residue from the neighbouring molecule
hydrogen bond not present in each monomer
The specificity of DiscI and other H-type lectins for monosaccharides with D-galacto configuration is determined by the fact that most observed interactions involve the O3 and O4 hydroxyls. Those two have to adopt equatorial and axial orientation respectively for optimal binding since any other stereochemistry at those positions would lead to steric conflict with Tyr241 and to the lost of crucial hydrogen bonds with the protein. The amino acids involved are strictly conserved among H-type lectins (Asp206, Arg215 and Trp235 in DiscI).
Comparison of the structures of H-type lectins in complex with GalNAc indicates that the anomeric preference is influenced by the residue following the conserved Trp in the last surface loop, Figure 5B. In HPA, His84 side chain would hinder the accommodation of the β-anomer justifying HPA specificity for αGalNAc.33 In DiscII, Gly239 allows the binding of both anomers as observed in its GalNAc complex structure.29 In DiscI, both anomers can bind as observed in the Galβ1-3GalNAc complex structure. Asn236 favours however the binding of the β-anomer as demonstrated in the ITC experiments where DiscI has higher affinity for βMeGal than for αMeGal (Table 1). Asn236 also interacts with the N-acetyl group of GalNAc, which explains the better affinity of DiscI for GalNAc compare to Gal.
In the DiscI/GalNAcβ1-3Gal complex structure, the GalNAc moiety is found in the previously described position and presents the same hydrogen bond network. The galactose moiety is oriented outwards at the protein surface and only makes two hydrogen bonds between the O3 and O4 hydroxyls and the Asn236 ND2, Figure 6A. In the DiscI/Galβ1- 3GalNAc complex, the GalNAc moiety is also in the primary binding site but in a different orientation since it is rotated by about -30° around the anomeric carbon. At the protein level, no changes in amino acids conformation are detected. This mostly modifies the interactions involving the O3 and O4 hydroxyls. The hydrogen bond between the O3 hydroxyl and Trp235 is weaker whilst the one with Asp206 is lost. At the level of the O4 hydroxyl, the loss of the hydrogen bond with Arg215 NH2 is compensated by a new one with Trp235 NE1. It interacts now with the Asp206 OD2 (neighbouring monomer) instead of the OD1, Figure 6B. The O6 hydroxyl can present a second conformation, which interacts with the Asp206 OD1 (neighbouring monomer). The GalNAc shift allows the good positioning of the galactose moiety in the binding site. The hydrogen bond network created involves the O5 hydroxyl with Trp235 NE1, the O6 hydroxyl with Ser242 OG (neighbouring monomer) and Asp206 OD2 (neighbouring monomer) and the O4 hydroxyl with Gln204 OE1 (neighbouring monomer) via a water molecule. No interaction is observed for the O2 and O3 hydroxyls. The additional interactions with the galactose moiety explain the better affinity of DiscI for the disaccharides than for the monosaccharide. The better recognition of Galβ1-3GalNAc compare to GalNacβ1-3Gal by DiscI is related to the more enclosed environment of its galactose moiety that makes twice as many and stronger hydrogen bonds with the protein, Figure 6C.
Figure 6. Disaccharides binding to DiscI H-type lectin domain.
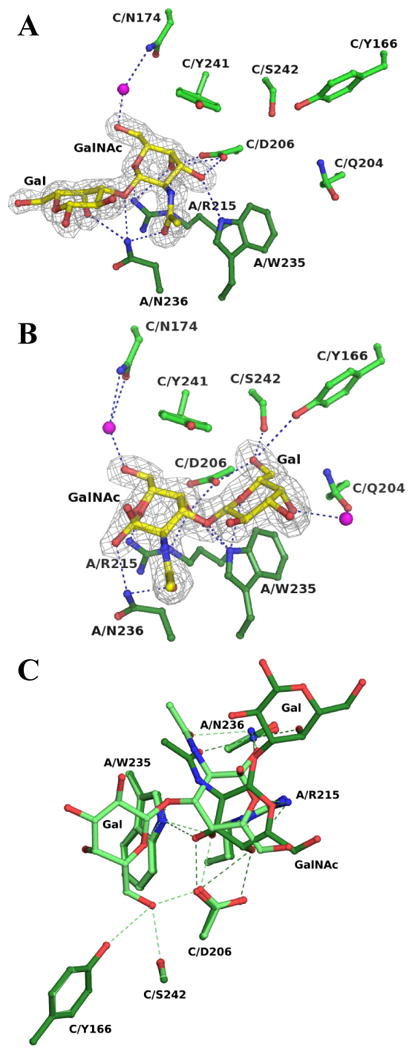
Representation of the maximum-likelihood weighted 2mFo-DFc electron density around the GalNAcβ1-3Gal (A) and Galβ1-3GalNAc (B) complexed to DiscI contoured at 1σ (0.44 and 0.45 eA3 respectively). Hydrogen bonds are represented as dotted lines and waters as magenta spheres. C: Overlay of the interactions of the disaccharides with DiscI. The Galβ1-3GalNAc and GalNAcβ1-3Gal disaccharides are coloured in light and dark green respectively. The residues are labelled according to their polypeptide chain and number.
Molecular modelling of oligosaccharides in DiscII
DiscII has no strong affinity for disaccharide (excepted sulphated ones) but binds efficiently to Galβ1-4GlcNacβ1-6βGalNAc trisaccharide present on core 2 and core 4 mucins. Since the trisaccharide was not available for ITC and cocrystallization experiments, it was modelled into DiscII binding site, with the reducing GalNAc sitting in the position observed in the crystal structures. The trisaccharide is accommodated in a shallow groove at the top of the β-sandwich and extends towards the surface of the lectin trimer. This area is rather hydrophilic and two hydrogen bonds from the O6 hydroxyl of GlcNAc to Asn177 side chain and Ala242 carbonyl group are predicted. Additional stabilising interactions are established by van der Waals interaction between the non-reducing galactose of the trisaccharide and the aromatic group of His211. The absence of this residue in DiscI (Thr208) may explain its lack of affinity for 1-6 branched GalNAc.
Specificity loops
The main difference in the overall structure and binding site architecture of H-type lectins is observed in the regions of the first and third surface loops that play a role in tuning the specificity and affinity towards oligosaccharides. The first loop varies in sequence, length and structure from one member to the other while the third one varies in sequence and conformation, Figure 7A. Their N-terminal part in one monomer creates with their C-terminal part of the neighbouring monomer a shallow groove that goes along the side of the β-sandwich and continues across its top. Along the side, it is formed on one side by the N-terminal part of the first loop (167-174 in DiscI, 169-177 in DiscII and 9-13 in HPA) and on the other side by its neighbouring C-terminal one (177-179 in DiscI, 180-182 in DiscII and 17-28 in HPA). At the top of the binding surface, one side entails both ends of the third loop and the other the part of the first loop not listed above.
Figure 7. Binding site of H-type lectins.
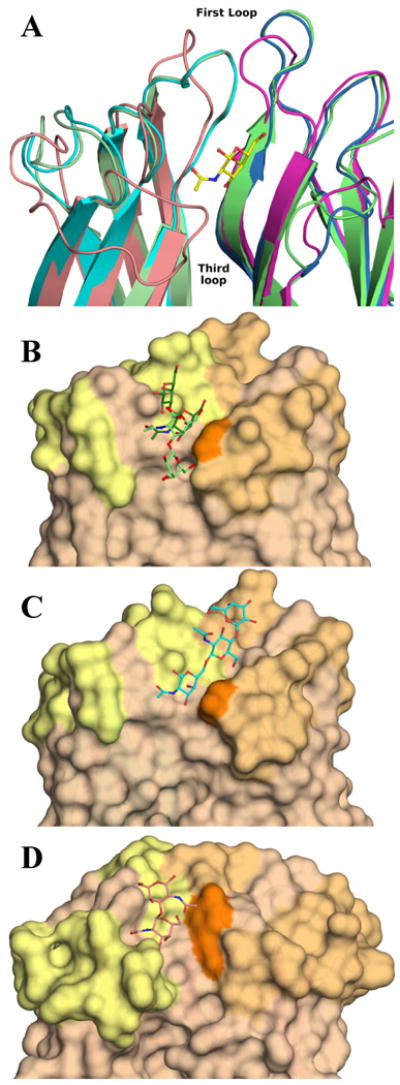
A: Cartoon representation of the superimposition of DiscI (light green/green), DiscII (cyan/blue) and HPA (salmon/magenta) crystal structures. Zoom on one binding site form by two monomers coloured by chain and representation of the GalNAc moiety bound to DiscI in ball and sticks. Surface representation of the binding site of DiscI (B), DiscII (C) and HPA (D). The disaccharides, the modelled trisaccharide and Forsman antigen bound to DiscI, DiscII and HPA (PDB 2CGY) respectively are represented in ball and stick. The Galβ1-3GalNAc and GalNAcβ1-3Gal disaccharides are coloured in light and dark green respectively.
The binding side has almost an inverted L shape in discoidins and is deeper, narrower and more extended in DiscI than in DiscII along the side of β-sandwich, Figure 7B/C. This allows DiscI to recognise type 3/4 oligosaccharides such as polylactosamine contrary to DiscII which lacks affinity for those since its binding side is far too open on that area to discriminate any sugar. Across the top of the binding surface, the differences in sequence in the third loop are determinant for the recognition of 1,6 oligosaccharides by DiscII with in particularly the presence of His211. The model of Galβ1-4GlcNacβ1-6βGalNAc in the binding site of DiscII also illustrates that the O3 of GalNAc is available for branching residue, which could extend the interaction area.
Following the differences in conformation of third loop and of Tyr89, the binding site of HPA extends towards the top of the trimer. The orientation of Tyr89 side chain prevents the binding of 1,6 branched oligosaccharides in a similar way to discoidins. The C-terminal end of the first loop is longer by at least seven amino acids in HPA compared to discoidins that leads to a narrower, deeper and more extended groove with a twist on the side than in discoidins, Figure 7D. This would result in a more restricted specificity for 1,3 branched GalNAc as observed on the glycan arrays.33 This part of the loop is also involved in HPA specificity for monosaccharide since it surrounds and stabilises the N-acetyl group and the O3 hydroxyl of GalNAc via several hydrogen bonds that would favour the binding of GalNAc to galactose. In discoidins, it is short and does not interact with the monosaccharide.
Discussion
Discoidins I and II are two domain proteins assembled as trimers. We reported previously the trimeric structure of DiscII, while the present work focuses on isoform A of DiscI. The structure of isoforms C and D of DiscI should be almost identical to this one since they only differ by 13 and 9 amino acids respectively. These mutations are mainly located at the protein surface and only one is not found in the DS domain. It is very likely that heterotrimers combining the different isoforms are formed in the slime mold but no experimental details are available.
The binding site of the DS domain is very similar for both discoidins and the nature of its ligand is unknown. The presence of MPD and glycerol molecules, often observed in the carbohydrate binding sites, together with the overall similarity to many carbohydrate binding protein, suggests that it could bind carbohydrates. According to the glycan array results, fucosylated and/or sialylated N and O-glycans could be candidates since they are the only ones recognised by both discoidins and do not have the required characteristics to be ligands for the H-type lectin domain. Such ligand would be also in agreement with the proposed glycoprotein receptor for DiscI DS domain.24 The strong resemblance in fold, quaternary structure and binding site properties with the fucose-binding F-type lectins indicates that the DS domain of discoidins could belong to this lectin family. Specificity studies on the whole proteins and their isolated DS domain are under progress to resolve this issue.
The natural ligands of the H-type lectin domain of DiscI and DiscII are still unknown but the high specificity observed for some oligosaccharide present on the glycan arrays opens the route for hypothesis. There are not many data on the composition and structures of glycoconjugates present in D. discoideum. In addition to classical N- and O-glycans, αGlcNAc-1-PO4 phosphoglycosylation has been observed on serine residues of cysteine proteinases and a complex glycan containing a core tetrasaccharide Galα1-3Fucα1-2βGalα1-3GlcNAc-, has been found attached to hydroxyproline on Skp1, a subunit of ubiquitin ligase protein complex.43,44 A polysaccharide rich in Gal and GalNAc extracted from D. Discoideum was proposed to be DiscII endogenous ligand since it resembled the spore coat polysaccharide and is produced in late development when DiscII is very prominent and accumulates in pre-spore vesicles.26 The spores formed upon D. discoideum starvation are indeed enclosed in a specialized cell wall, the spore coat, which contains crystalline cellulose together with a Gal/GalNAc-rich polysaccharide (GPS).45 GPS is stored in the pre-spore vesicles prior secretion and incorporation in the spore coat and the matrix around. It is essential for spore integrity and plays a role in stress-resistance.46 GPS of the related species D. mucoides has been demonstrated to be a branched polysaccharide with repeating unit of [-3Galβ1-3(Galβ1-6)GalNAcα1-].47 Antisera cross-reactivity indicates that this GPS is related to the GPS of D. discoideum. Interestingly this branched trisaccharide corresponds closely to the glycan with highest affinity for DiscII in its glycan array. This strengthens the suggestion that GPS would be DiscII natural ligand.
DiscII appears to be externalised along with glycoconjugates of the spore coat as the spores mature.11 Similarly, in early development, DiscI was found extracellularly around aggregates and slugs in the so-called slime coat where it binds to an uncharacterized glycoconjugate.25 The externalisation of discoidins in association with their complementary glycoconjugates during differentiation lets suppose a role in the formation of Dictyostelium extracellular matrices. Alternatively, discoidins could play a role in the recognition of polysaccharides on bacteria, which are abundant in soil. They could be involved in defence mechanisms by aggregating bacteria, as it has been proposed for the related Helix pomatia lectin that protects fertilized eggs from infection.48 They could also play a role in phagocytosis, the process by which wild-type strains of D. discoideum feed upon bacteria. A proteomics analysis of the phagosome indicated the presence of DiscII and of the three isoforms of DiscI.49 More recently, DNA microarray comparison between the wild type strain V12M2, which is strictly dependent on phagocytosis, and the axenic strain AX2, which is not, demonstrated that both DiscI and DiscII are strongly up-regulated in V12M2, indicating a possible role of these proteins in phagocytosis.50 Discoidins could have different role and different ligands depending on their localisation since they are broadly distributed in the cytoplasm, within vesicles, and at the cell surface.
The present study highlighted the differences in specificity for both discoidins from Dictyostelium discoideum, proteins, which are spatially and temporally regulated during the life cycle of the slime mold. The structural work rationalised the observed differences in affinity for the lectin domain. The possibility of a carbohydrate binding site in the DS domain is now also opened. In such case, discoidins would be a mix of F and H-type lectins with distinct specificities. They present six binding sites as HPA and the striped bass F-lectin. Those trimeric proteins present different domain organisation. HPA creates intermolecular disulfide bridges between two trimers of H-type lectin, the F-type lectin domain is repeated in tandem in the striped bass F-lectin trimer (PDB 3CQO) while in discoidins, one domain of each is stabilised by intertwining during the trimer formation, Figure 8.33 This will greatly enhance the multivalency of discoidins as they will present two binding surfaces on the opposite sides of the protein. They would be able to bind simultaneously and non-simultaneously to their different ligands. The determination of the endogenous ligands of both discoidin domains will require additional knowledge on D. discoideum glycoconjugates. Such study will be necessary to shed some light on the biological function of discoidins, which remains unknown.
Figure 8.
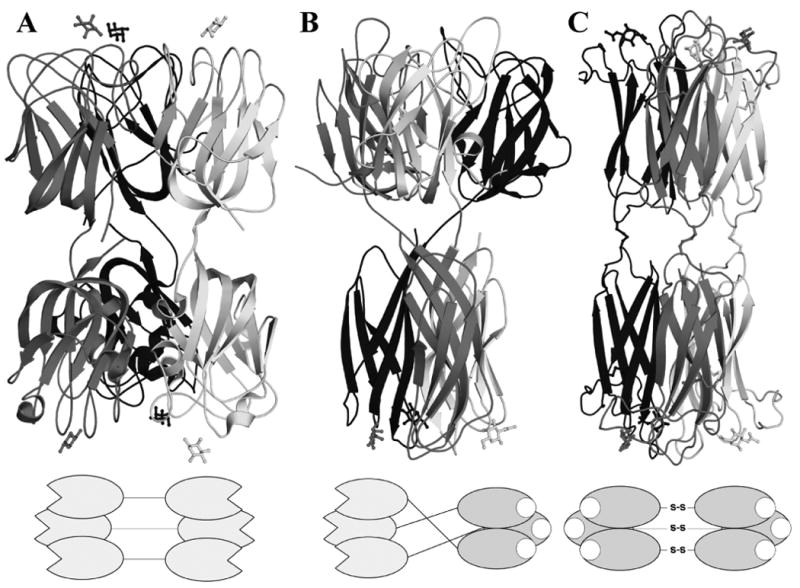
Cartoon representation of the quaternary structure of the F-Lectin from striped Bass (PDB 3CQO, A), Discoidin I (B) and HPA (PDB 2CCV, C). A diagram of the domain layout is drawn beneath each protein.
Materials and Methods
Construction of plasmid for expression and purification of recombinant DiscI
The discoidin I gene of isoform A (P02886) was cloned in-frame with an N-terminal hexahistidine tag (Histag) and the tobacco-etch virus (TEV) protease cleavage site of the prokaryotic expression vector pProEXHTb (Invitrogen) using polymerase chain reaction and standard molecular biology techniques. The primers used were the following: 5′-GTG CCA TGG CTA CCC AAG GTT TAG TTC-3′ and 5′-GAC TCG AGT TAT TCC AAA GCG GTA GC-3′. Sequence analysis was in accordance with the one deposited.
The protein was overexpressed in Rosetta2(DE3)plysS bacterial cells (Novagen) and in Luria broth medium (LB). Cells were cultured at 37°C up to an optical density of 1.2–1.3 at 600 nm before induction with 0.2 mM IPTG overnight at 16°C and harvested by centrifugation. Each gram of cells was resuspended in 3 ml of buffer A: 20 mM Tris/HCl pH 8.5, 500 mM NaCl and 10 mM imidazol, before using a cell disrupter (Constant system Ltd). After centrifugation 30 min at 40,000g, the supernatant was loaded on a 1 ml Histrap™ column (GE healthcare). After a wash step of five volumes with buffer A, DiscI was eluted with a 10-500 mM imidazol gradient. The Histag was cut by addition of the TEV protease at the ratio 1/100 overnight at 20°C in the presence of 2 mM DTT. Imidazol was removed using a PD10 column (GE healthcare) equilibrated with 20 mM Tris-HCl pH 7.5 and 150 mM NaCl before reloading on the affinity column. The pure protein without tag eluted in the flow-through was concentrated around 5.5 mg ml-1 using Microsep™ 10K (Pall). Each purification step was analysed on 12% SDS-PAGE.
Isothermal Titration Microcalorimetry analysis
ITC experiments were performed using a Microcal VP-ITC microcalorimeter. All titrations were carried out in 20mM Tris-HCl pH 7.5 and 150 mM NaCl, at 25 °C. Sugars and proteins were dissolved in the same buffer. A total of 30 injections of 10 μL of sugar solution at concentrations varying from 2 to 40 mM were added at intervals of 5 min to the lectin solution present in the calorimeter cell (1.4478 ml) with stirring at 310 rpm. The protein concentration was 0.15 mM.
Control experiments performed by injection of sugar solution into the microcalorimeter cell in the absence of protein yielded insignificant heats of dilution. Integrated heat effects were analysed by non-linear regression using a single-site binding model (Origin 7.0). The experimental data were fitted and the association constant (Ka) and the enthalpy of binding (ΔH) were then obtained. The stoichiometry n was fixed to 1 for low affinity ligands and fitted to the curve for high affinity ligands. Other thermodynamic parameters, i.e. changes in free energy (ΔG), entropy (ΔS), and number of binding sites per monomer (n) were calculated from the equation:
where T is the absolute temperature, R=8.32 J mol-1K-1 and K is the association constant. The experiment was at least duplicated for each ligand.
The glycan microarray analysis
Recombinant Discoidin I and II were labelled with Alexa Fluor® 488–TFP (carboxylic acid, 2,3,5,6 tetrafluorophenyl, Invitrogen) according to the manufacturer's instructions and purified on a D-Salt™ polyacrylamide 6000 desalting column (Pierce). The specificity of the two proteins was then analysed using the printed glycan arrays version 3.1 of the Consortium for Functional Glycomics (http://www.functionalglycomics.org/) following the standard procedure.
Crystallisation and structure determination
All crystals were obtained by the handing-drop vapour diffusion method using 2 μl drops containing a 50:50 (v/v) mix of the protein at 5.5 mg ml-1 and the reservoir solution at 20 °C. Rod crystals for unliganded experiments were obtained from the solution 16 of the Hampton crystallization screen I (Hampton research) containing 1.5 M lithium sulphate and 100 mM NaHepes pH 7.5 after almost one month. GalNAc was introduced by soaking the crystals for a short time in the mother liquor supplemented with 10 mM GalNAc. Crystals of the disaccharides complexes were obtained by co-crystallization after one week from a solution containing 100 mM NaHepes pH 7.5, 1.2 M lithium sulphate. The sugars (13 mM) were incubated for an hour at room temperature with the protein prior crystallisation. Before data collection, the crystals were transferred in a cryoprotecting solution (either 25% MPD for unliganded and GalNAc crystals or 20% glycerol for disaccharides crystals were added to the mother liquor), mounted in a rayon fiber loop and frozen at 100K. Native and GalNAc complex data were collected at 1.8 and 1.75 Å resolution respectively on ID14-2 beamline at ESRF (Grenoble, France) using an ADSC Q4 CCD detector (Quantum Corp.). GalNAcβ1-3Gal and Galβ1-3GalNAc complex data were collected at 1.8 and 1.6 Å resolution respectively on ID23-2 beamline at ESRF using a MarCCD detector. All data belong to the C2221 spacegroup, were processed and reduced using MOSFLM and SCALA 51 All further computing was performed using the CCP4 suite, unless otherwise stated.52
Structures determination
All DiscI structures were solved by molecular replacement using PHASER53. The coordinates of the DiscII monomer (PDB code 2VM9) and of the native DiscI trimer were used as search models for the native and complex structures respectively.29 For the refinement of each structure, 5% of the observations were immediately set aside for cross validation analysis and were used to monitor various refinement strategies such as the weighting of geometrical and temperature factor restraint. Manual corrections of the model were performed using Coot54 and were interspersed with cycles of maximum-likelihood refinement in REFMAC.55 Water molecules were added in Coot and checked manually. The quality of the model was assessed in PROCHECK before its deposition in the Brookhaven Protein Data Bank.56,57 Details of the data and model quality are given in the Table 2. All figures were drawn with PyMOL Molecular Graphics System (2002) DeLano Scientific, Palo Alto, CA, unless otherwise stated.
Molecular modelling
The trisaccharide Galβ1-4βGlcNAcβ1-6GalNAc was modelled in the binding site of DiscII using the crystal structure of the complex with GalNAc (PDB 2VMC) as starting point. The protein moiety was edited with the use of the Sybyl software (St Louis, US). Two adjacent lectin domains were considered. All the hydrogen atoms were added, the partial charges were included with the Pullman library and the position of hydrogen atoms were optimized with the use of the Tripos force-field.58 The trisaccharide was built using the monosaccharide library available at Glyco3D (http://www.cermav.cnrs.fr/glyco3d/) using the PIM parameters and partial charges for carbohydrates.59 The reducing GalNAc moiety was placed in the binding site by fitting on the GalNAc moiety observed in the crystal structure. Possible orientations of the remaining residues were calculated by systematic search around the glycosidic linkages. The lowest energy conformations were further optimized by energy minimization of the full oligosaccharide and the protein hydrogen atoms with the Tripos force field extended with the PIM parameters. Energy minimizations were carried out using the Powell procedure with a distance-dependent dielectric constant until a gradient deviation of 0.05 kcal/mol/A° was attained. The validity of the final conformations was checked by comparison with available energy maps (Glyco3D and 60).
Protein Data Bank accession number
Coordinates and structure-factor amplitudes have been deposited with the Protein Data Bank with accession codes 2W94 (native crystal), 2W95 (GalNAc complex), 2WN2 (Galβ1-3GalNAc complex) and 2WN3 (GalNAcβ1-3Gal complex).
Acknowledgments
The glycan microarray analysis was provided by the Consortium for Functional Glycomics funded by the National Institute of General Medical Sciences grant GM62116. We acknowledge the European Synchrotron Radiation Facility for provision of synchrotron radiation facilities. We thank Michel Satre and Christopher West for helpful discussions about Dictyostelium discoideum. This work was supported by CNRS and French Ministry of Research. KSA was the recipient of a CNPq grant from Brazilian government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Iwanaga S, Lee BL. Recent advances in the innate immunity of invertebrate animals. J Biochem Mol Biol. 2005;38:128–50. doi: 10.5483/bmbrep.2005.38.2.128. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez L, Alpuche J, Maldonado G, Agundis C, Pereyra-Morales A, Zenteno E. Immunity mechanisms in crustaceans. Innate Immun. 2009;15:179–88. doi: 10.1177/1753425909102876. [DOI] [PubMed] [Google Scholar]
- 3.Chaston J, Goodrich-Blair H. Common trends in mutualism revealed by model associations between invertebrates and bacteria. FEMS Microbiol Rev. 2010;34:41–58. doi: 10.1111/j.1574-6976.2009.00193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller WE, Dorn A, Uhlenbruck G. The molecular mechanisms of the distinct calcium-dependent aggregation systems in marine sponges and corals. Acta Histochem Suppl. 1985;31:37–46. [PubMed] [Google Scholar]
- 5.Mody R, Joshi S, Chaney W. Use of lectins as diagnostic and therapeutic tools for cancer. J Pharmacol Toxicol Methods. 1995;33:1–10. doi: 10.1016/1056-8719(94)00052-6. [DOI] [PubMed] [Google Scholar]
- 6.Brooks SA, Wilkinson D. Validation of a simple avidin-biotin detection method for Helix pomatia lectin (HPA) binding as a prognostic marker in cancer. Acta Histochem. 2003;105:205–12. doi: 10.1078/0065-1281-00705. [DOI] [PubMed] [Google Scholar]
- 7.Queiroz AF, Silva RA, Moura RM, Dreyfuss JL, Paredes-Gamero EJ, Souza AC, Tersariol IL, Santos EA, Nader HB, Justo GZ, de Sales MP. Growth inhibitory activity of a novel lectin from Cliona varians against K562 human erythroleukemia cells. Cancer Chemother Pharmacol. 2009;63:1023–33. doi: 10.1007/s00280-008-0825-4. [DOI] [PubMed] [Google Scholar]
- 8.Kessin RH. Dictyostelium - Evolution, Cell Biology, and the Development of Multicellularity. Cambridge Univiversity Press; Cambridge, UK: 2001. [Google Scholar]
- 9.Rosen SD, Kafka JA, Simpson DL, Barondes SH. Developmentally regulated, carbohydrate-binding protein in Dictyostelium discoideum. Proc Natl Acad Sci USA. 1973;70:2554–7. doi: 10.1073/pnas.70.9.2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frazier WA, Rosen SD, Reitherman RW, Barondes SH. Purification and comparison of two developmentally regulated lectins from Dictyostelium discoideum. Discoidin I and II J Biol Chem. 1975;250:7714–21. [PubMed] [Google Scholar]
- 11.Barondes SH, Cooper DN, Haywood-Reid PL. Discoidin I and discoidin II are localized differently in developing Dictyostelium discoideum. J Cell Biol. 1983;96:291–6. doi: 10.1083/jcb.96.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barondes SH, Haywood-Reid PL, Cooper DN. Discoidin I, an endogenous lectin, is externalized from Dictyostelium discoideum in multilamellar bodies. J Cell Biol. 1985;100:1825–33. doi: 10.1083/jcb.100.6.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Poole S, Firtel RA, Lamar E, Rowekamp W. Sequence and expression of the discoidin I gene family in Dictyostelium discoideum. J Mol Biol. 1981;153:273–89. doi: 10.1016/0022-2836(81)90278-3. [DOI] [PubMed] [Google Scholar]
- 14.Tsang AS, Devine JM, Williams JG. The multiple subunits of discoidin I are encoded by different genes. Dev Biol. 1981;84:212–7. doi: 10.1016/0012-1606(81)90385-7. [DOI] [PubMed] [Google Scholar]
- 15.Kessin RH. Genetics of early Dictyostelium discoideum development. Microbiol Rev. 1988;52:29–49. doi: 10.1128/mr.52.1.29-49.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strmecki L, Bloomfield G, Araki T, Dalton E, Skelton J, Schilde C, Harwood A, Williams JG, Ivens A, Pears C. Proteomic and Microarray Analyses of the Dictyostelium Zak1-GSK-3 Signaling Pathway Reveal a Role in Early Development. Eukaryot Cell. 2007;6:245–252. doi: 10.1128/EC.00204-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alexander S, Shinnick TM, Lerner RA. Mutants of Dictyostelium discoideum blocked in expression of all members of the developmentally regulated discoidin multigene family. Cell. 1983;34:467–75. doi: 10.1016/0092-8674(83)90380-x. [DOI] [PubMed] [Google Scholar]
- 18.Burdine V, Clarke M. Genetic and physiologic modulation of the prestarvation response in Dictyostelium discoideum. Mol Biol Cell. 1995;6:311–25. doi: 10.1091/mbc.6.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blusch J, Alexander S, Nellen W. Multiple signal transduction pathways regulate discoidin I gene expression in Dictyostelium discoideum. Differentiation. 1995;58:253–60. doi: 10.1046/j.1432-0436.1995.5840253.x. [DOI] [PubMed] [Google Scholar]
- 20.Crowley TE, Nellen W, Gomer RH, Firtel RA. Phenocopy of discoidin I-minus mutants by antisense transformation in Dictyostelium. Cell. 1985;43:633–41. doi: 10.1016/0092-8674(85)90235-1. [DOI] [PubMed] [Google Scholar]
- 21.Springer WR, Cooper DN, Barondes SH. Discoidin I is implicated in cell-substratum attachment and ordered cell migration of Dictyostelium discoideum and resembles fibronectin. Cell. 1984;39:557–64. doi: 10.1016/0092-8674(84)90462-8. [DOI] [PubMed] [Google Scholar]
- 22.Ruoslahti E, Pierschbacher MD. New perspectives in cell adhesion: RGD and integrins. Science. 1987;238:491–7. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 23.Ruoslahti E. The RGD story: a personal account. Matrix Biol. 2003;22:459–65. doi: 10.1016/s0945-053x(03)00083-0. [DOI] [PubMed] [Google Scholar]
- 24.Gabius HJ, Springer WR, Barondes SH. Receptor for the cell binding site of discoidin I. Cell. 1985;42:449–56. doi: 10.1016/0092-8674(85)90102-3. [DOI] [PubMed] [Google Scholar]
- 25.Cooper DN, Barondes SH. Colocalization of discoidin-binding ligands with discoidin in developing Dictyostelium discoideum. Dev Biol. 1984;105:59–70. doi: 10.1016/0012-1606(84)90261-6. [DOI] [PubMed] [Google Scholar]
- 26.Cooper DN, Lee SC, Barondes SH. Discoidin-binding polysaccharide from Dictyostelium discoideum. J Biol Chem. 1983;258:8745–50. [PubMed] [Google Scholar]
- 27.Cooper DNW, Haywood-Reid PL, Springer WR, Barondes SH. Bacterial glycoconjugates are natural ligands for the carbohydrate binding site of discoidin I and influence its cellular compartmentalization. Dev Biol. 1986;114:416–425. [Google Scholar]
- 28.Alexander S, Cibulsky AM, Lerner RA. Ion dependence of the discoidin I lectin from Dictyostelium discoideum. Differentiation. 1983;24:209–12. doi: 10.1111/j.1432-0436.1983.tb01321.x. [DOI] [PubMed] [Google Scholar]
- 29.Saboia Aragão K, Satre M, Imberty A, Varrot A. Structure determination of Discoidin II from Dictyostelium discoideum and carbohydrate binding properties of the lectin domain. Proteins. 2008;73:43–52. doi: 10.1002/prot.22038. [DOI] [PubMed] [Google Scholar]
- 30.Baumgartner S, Hofmann K, Chiquet-Ehrismann R, Bucher P. The discoidin domain family revisited: new members from prokaryotes and a homology-based fold prediction. Protein Sci. 1998;7:1626–31. doi: 10.1002/pro.5560070717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiedzierska A, Smietana K, Czepczynska H, Otlewski J. Structural similarities and functional diversity of eukaryotic discoidin-like domains. Biochim Biophys Acta. 2007;1774:1069–78. doi: 10.1016/j.bbapap.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 32.Vogel WF, Abdulhussein R, Ford CE. Sensing extracellular matrix: an update on discoidin domain receptor function. Cell Signal. 2006;18:1108–16. doi: 10.1016/j.cellsig.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 33.Sanchez JF, Lescar J, Chazalet V, Audfray A, Gagnon J, Alvarez R, Breton C, Imberty A, Mitchell EP. Biochemical and structural analysis of Helix pomatia agglutinin (HPA): a hexameric lectin with a novel fold. J Biol Chem. 2006;281:20171–20180. doi: 10.1074/jbc.M603452200. [DOI] [PubMed] [Google Scholar]
- 34.Dam TK, Brewer CF. Thermodynamic studies of lectin-carbohydrate interactions by isothermal titration calorimetry. Chem Rev. 2002;102:387–429. doi: 10.1021/cr000401x. [DOI] [PubMed] [Google Scholar]
- 35.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–68. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 36.Macedo-Ribeiro S, Bode W, Huber R, Quinn-Allen MA, Kim SW, Ortel TL, Bourenkov GP, Bartunik HD, Stubbs MT, Kane WH, Fuentes-Prior P. Crystal structures of the membrane-binding C2 domain of human coagulation factor V. Nature. 1999;402:434–9. doi: 10.1038/46594. [DOI] [PubMed] [Google Scholar]
- 37.Pratt KP, Shen BW, Takeshima K, Davie EW, Fujikawa K, Stoddard BL. Structure of the C2 domain of human factor VIII at 1.5 A resolution. Nature. 1999;402:439–42. doi: 10.1038/46601. [DOI] [PubMed] [Google Scholar]
- 38.Harding MM. The geometry of metal-ligand interactions relevant to proteins. Acta Crystallogr D Biol Crystallogr. 1999;55:1432–43. doi: 10.1107/s0907444999007374. [DOI] [PubMed] [Google Scholar]
- 39.Holm L, Kaariainen S, Rosenstrom P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–8. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bianchet MA, Odom EW, Vasta GR, Amzel LM. A novel fucose recognition fold involved in innate immunity. Nature Struct Biol. 2002;9:628–34. doi: 10.1038/nsb817. [DOI] [PubMed] [Google Scholar]
- 42.Lescar J, Sanchez JF, Audfray A, Coll JL, Breton C, Mitchell EP, Imberty A. Structural basis for recognition of breast and colon cancer epitopes Tn antigen and Forssman disaccharide by Helix pomatia lectin. Glycobiology. 2007;17:1077–1083. doi: 10.1093/glycob/cwm077. [DOI] [PubMed] [Google Scholar]
- 43.Haynes PA. Phosphoglycosylation: a new structural class of glycosylation? Glycobiology. 1998;8:1–5. doi: 10.1093/glycob/8.1.1. [DOI] [PubMed] [Google Scholar]
- 44.West CM. Evolutionary and functional implications of the complex glycosylation of Skp1, a cytoplasmic/nuclear glycoprotein associated with polyubiquitination. Cell Mol Life Sci. 2003;60:229–40. doi: 10.1007/s000180300018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Erdos GW, West CM. Formation and organization of the spore coat ofDictyostelium discoideum. Experimental Mycology. 1989;13:169–182. [Google Scholar]
- 46.West CM, Nguyen P, van der Wel H, Metcalf T, Sweeney KR, Blader IJ, Erdos GW. Dependence of stress resistance on a spore coat heteropolysaccharide in Dictyostelium. Eukaryot Cell. 2009;8:27–36. doi: 10.1128/EC.00398-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sakurai MH, Kiyohara H, Nakahara Y, Okamoto K, Yamada H. Galactose-containing polysaccharides from Dictyostelium mucoroides as possible acceptor molecules for cell-type specific galactosyl transferase. Comp Biochem Physiol B. 2002;132:541–9. doi: 10.1016/s1096-4959(02)00067-2. [DOI] [PubMed] [Google Scholar]
- 48.Kohler W, Prokop O, Kuhnemund O. Routine identification of group-C streptococci by means of an agglutinin (protectin) from the albumen gland of the edible snail, Helix pomatia. J Med Microbiol. 1973;6:127–130. doi: 10.1099/00222615-6-1-127. [DOI] [PubMed] [Google Scholar]
- 49.Gotthardt D, Blancheteau V, Bosserhoff A, Ruppert T, Delorenzi M, Soldati T. Proteomics fingerprinting of phagosome maturation and evidence for the role of a Galpha during uptake. Mol Cell Proteomics. 2006;5:2228–43. doi: 10.1074/mcp.M600113-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Sillo A, Bloomfield G, Balest A, Balbo A, Pergolizzi B, Peracino B, Skelton J, Ivens A, Bozzaro S. Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium. BMC Genomics. 2008;9:291. doi: 10.1186/1471-2164-9-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leslie AGW. Recent changes to the MOSFLM package for processing film and image plate data. Jnt CCP4/ESF-EACMB Newsletter on Protein Crystallography. 1992;26 [Google Scholar]
- 52.Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;D52:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 53.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr D Biol Crystallogr. 2005;61:458–64. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 54.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 55.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 56.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 57.Bernstein FC, Koetzle TF, Williams GJB, Meyer ET, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The protein data bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 58.Clark M, Cramer RDI, van den Opdenbosch N. Validation of the general purpose Tripos 5.2 force field. J Comput Chem. 1989;10:982–1012. [Google Scholar]
- 59.Imberty A, Monier C, Bettler E, Morera S, Freemont P, Sippl M, Flockner H, Ruger W, Breton C. Fold recognition study of alpha3-galactosyltransferase and molecular modeling of the nucleotide sugar-binding domain. Glycobiology. 1999;9:713–22. doi: 10.1093/glycob/9.7.713. [DOI] [PubMed] [Google Scholar]
- 60.Imberty A, Delage MM, Bourne Y, Cambillau C, Perez S. Data bank of three-dimensional structures of disaccharides: Part II, N-acetyllactosaminic type N-glycans. Comparison with the crystal structure of a biantennary octasaccharide. Glycoconj J. 1991;8:456–83. doi: 10.1007/BF00769847. [DOI] [PubMed] [Google Scholar]