Abstract
Histone deacetylase 6 (HDAC6), a member of the HDAC family whose major substrate is α-tubulin, has become a target for drug development to treat cancer due to its major contribution in oncogenic cell transformation. Overexpression of HDAC6 correlates with tumorigenesis and improved survival; therefore, HDAC6 may be used as a marker for prognosis. Previous work demonstrated that in multiple myeloma cells, inhibition of HDAC6 results in apoptosis. Furthermore, HDAC6 is required for the activation of heat-shock factor 1 (HSF1), an activator of heat-shock protein encoding genes (HSPs) and CYLD, a cylindromatosis tumor suppressor gene. HDAC6 contributes to cancer metastasis since its upregulation increases cell motility in breast cancer MCF-7 cells and its interaction with cortactin regulates motility. HDAC6 also affects transcription and translation by regulating the heat-shock protein 90 (Hsp90) and stress granules (SGs), respectively. This review will discuss the role of HDAC6 in the pathogenesis and treatment of cancer.
1. Introduction
Inhibition of histone deacetylation has become an accepted target for cancer therapy [1–4]. Cancer, the second leading cause of death in the United States [5], involves the uncontrolled growth of abnormal cells that can spread to other areas of the body [6]. It is generally treated with chemotherapy, such as vincristine, vinblastine, and colchicine bind tubulin and inhibit microtubule (MT) polymerization, which block mitosis. Taxol, on the other hand, stabilizes the MT and blocks cell division [7]. Despite chemotherapy, many patients relapse and die from cancer. The American Cancer Society estimated 565,650 deaths this year [5]; therefore, new targets for cancer therapy need to be investigated.
Histone deacetylases (HDACs) are primarily involved in the deacetylation of histones [8, 9] but some HDACs, such as HDAC6 can also affect the function cytoplasmic nonhistone proteins. This review will discuss the interaction of HDAC6 with these nonhistone proteins, and discuss how HDAC6 is a key regulator of many signaling pathways that are linked to cancer, thereby making HDAC6 an attractive target.
HDACs are grouped into three classes based on their primary homology to three Saccharomyces cerevisiae HDACs [10, 11]. The class I HDACs, which include HDAC1, HDAC2, HDAC3, and HDAC8, are most related to the yeast transcriptional regulator yRPD3. Class II HDACs, which include HDAC4, HDAC5, HDAC6, HDAC7, HDAC9, and HDAC10, share domains similar to yHDA1. HDAC11 is most closely related to class I HDACs; however, no classification has been given since its sequence similarity is too low. Class III HDACs are similar to the NAD+-dependent ySIR2 [10, 11].
The deacetylase-trichostatin A (TSA) and deacetylase-suberoylanilide hydroxamic acid (SAHA) crystal structures of a homologue from the bacterium Aquifex aeolicus revealed the histone acetylase catalytic core and established a general mechanism for HDAC inhibition [12]. TSA and SAHA are two types of HDACs inhibitors that have antitumor effects since they can inhibit cell growth and induce terminal differentiation [12]. The conserved catalytic domain of HDAC involves about 390 amino acids, forming a tube-like shape pocket with a wider bottom [12]. Removal of an acetyl group from substrate occurs through a “charge-relay system” consisting of two adjacent histidine residues, two aspartic residues, and one tyrosine residue at the bottom of the pocket [12]. A Zn2+ cation binds near the bottom of the pocket and it is coordinated by two additional aspartates and one histidine and also by a water molecule; in general, HDAC inhibitors function by chelating the zinc ion, making the charge-relay system dysfunctional [12].
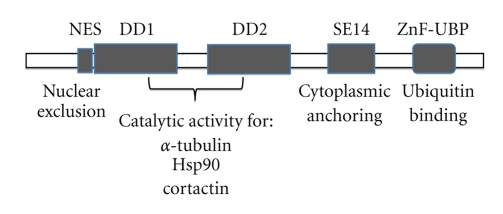
HDAC6 is a unique member of class II because it contains two homologous catalytic domains [8, 13]. Both catalytic domains of HDAC6 are fully functional HDACs and contribute independently to the overall activity of the HDAC6 protein, (see [8, 14–17] Figure 1). In one study, both HDAC6 catalytic domains were required for full tubulin deacetylase (TDAC) activity [18, 19]; however, in other studies the TDAC activity was attributed to the second domain [20–22]. HDAC6, like all other HDACs, is inhibited by TSA; however, HDAC6 is uniquely resistant to the potent HDAC inhibitors trapoxin-B and sodium butyrate. These drugs were used to show that HDAC6 is a deacetylase for α-tubulin in vivo, and in vitro [20, 23]. HDAC6 was found not to interact with histones, in vivo, however, in vitro, HDAC6 is able to deacetylate histones [8]. This result led to the investigation of an HDAC inhibitor that mediates acetylation of proteins other than histones. An HDAC6 inhibitor, known as tubacin (Tubulin acetylation inducer), was isolated through a multidimensional chemical genetic screen of 7,392 small molecules and cell-based assay targeting acetylation activity of proteins other than histones [21, 24]. Unlike other histone deacetylase inhibitors, tubacin was found to inhibit the deacetylation of α-tubulin in mammalian cells without affecting the level of histone acetylation, gene-expression, or cell cycle progression [21, 24].
The HDAC6 gene is localized in Xp11.23 (http://www.ncbi.nlm.nih.gov/) and was first cloned by two different groups Grozinger et al. [8] and Verdel and Khochbin [13]. It is the largest protein yet identified in humans with 1,216 amino acids [8]. Most HDACs are located in the nucleus; however, class II HDACs, are able to translocate to the cytoplasm [10]. HDAC6 predominates in the cytoplasm due to the NES and the SE14 motifs [10, 14], (Figure 1). HDAC6 is also found on the perinuclear and leading-edge subcellular regions, associated with p150glued-containing motor complex [23]. It contains a unique ubiquitin-binding zinc-finger domain (ZnF-UBP domain, also known as the PAZ, BUZ or DAUP domain) in its C-terminal region [25] and a dynein-binding domain (DBD) [7] (Figure 1). Other HDACs such as HDAC10 or a spliced variant of HDAC9 can be localized both in the nucleus and cytoplasm, and HDAC11, which localizes in the nucleus, has been reported to coprecipitate with HDAC6 in the cytoplasm [10]. The expression levels of HDAC6 were observed in the heart, liver, kidney, testis, brain, and, pancreas [8, 13].
Figure 1.
HDAC6 functional domains. The only HDAC with two tandem catalytic domains, deacetylase domains (DD1 and DD2). Tubulin, Hsp90, and cortactin have been found to be HDAC6 substrates. The nuclear export signal (NES) prevents accumulation of the protein in the nucleus and the Ser-Glu-containing tetrapeptide (SE14) region ensures a stable anchorage of the enzyme in the cytoplasm. The linker between both DDs is able to bind dynein and the high affinity ubiquitin-binding zinc finger domain (ZnF-UBP).
2. HDAC6 Expression and Cancer
Expression profiling analysis, with cDNA microarray in MCF-7 cells, showed that the HDAC6 gene is an estrogen-regulated gene [26]. Estrogens play an important role in the normal growth of mammary glands, as well as in the development of estrogen-dependent breast cancer [27]. The estrogen receptor α (ERα) status of breast cancer patients is widely used as an indicator for endocrine therapy responsiveness and for prognosis, but 30–40% of the ERα positive patients do not respond to the therapy, and 10% who are ERα negative do respond [3, 27].
The estrogen-mediated up-regulation of HDAC6 suggests that there might be a link between the levels of HDAC6 expression and metastasis of breast cancer that could be useful in the prognosis of patients. One study involving quantitative real-time reverse transcription-PCR, as well as immunohistochemistry in 135 female patients with invasive breast cancer showed that higher levels of expression of HDAC6 mRNA correlated with tumor size (less than 2 cm), low histologic grade, and ERα and progesterone receptor-positive tumors [27]. The high level of HDAC6 mRNA expression also correlated with better prognosis in terms of disease-free survival and better response to endocrine treatment. In a different study, the expression levels of HDAC6 were evaluated by immunohistochemical staining of 139 consecutively archived human breast cancer tissues [3]. In this study, a subset group of ER-positive patients with higher levels of HDAC6 expression who received adjuvant treatment with tamoxifen (TAM), a selective estrogen receptor modulator, resulted in improved survival. These results suggest that HDAC6 could potentially serve as an additional marker for prognosis in breast cancer patients.
The overexpression of HDAC6 has been identified in a variety of other cancer cell lines and mouse tumor models. The expression levels of HDAC6 in ovarian cancer cells and tissues were higher in low-grade and high-grade ovarian carcinomas compared with benign lesions and immortalized ovarian surface epithelium cell lines [28]. HDAC6 expression was also investigated in retrovirus expressing SV40 early region or/and RasG12V transformed and nontransformed human embryonic kidney cells (HEK), prostate epithelial cells (PrEC), and mouse embryonic fibroblast (MEF) cells and it was found that oncogenic Ras can lead to up-regulation of HDAC6 [29]. HDAC6 expression was also up-regulated in primary oral squamous cell lines and its level of expression correlated with primary tumor stage [30]. In a different study, HDAC6 was consistently overexpressed in primary acute myeloid leukemia (AML) blasts and in some myeloblastic cell lines [31]. The up-regulation of HDAC6 in diverse tumors and cell lines suggests an important role in cancer and it is therefore being widely investigated.
3. HDAC6 Is Required for Oncogenic Cell Transformation
A study performed by Lee et al. [29] found that HDAC6 is required for efficient oncogenic tumorigenesis by providing anchorage-independent proliferation to transduced cells. MEFs derived from wild type HDAC6-null embryos and transduced with retrovirus expressing SV40 early region and RasG12V resulted in >10-fold fewer colonies in the HDAC6-null oncogenic transduced MEFs samples than in the wild type oncogenic transduced MEFs. These results strongly indicated that HDAC6 is required for Ras-induced oncogenic transformation by providing anchorage-independent proliferation, a hallmark of malignant transformation. Anchorage-independent proliferation allows the cells to survive by escaping anoikis, a type of cell death due to lack of sufficient cell adhesion to the surrounding basement membrane and extracellular matrix (ECM) [29].
HDAC6 is required to maintain the transformed phenotypes of a number of established oncogenic cell lines and for tumor growth. Knockdown of HDAC6 in SKOV3 ovarian cancer, SKBR3 breast carcinoma, and MCF7 breast cancer cell lines inhibited the anchorage-independent growth of those cancer cells by 5-fold to 30-fold [29]. This suggests that HDAC6 plays a major role in the survival of tumor cells. To test whether HDAC6 was critical for tumorigenesis, immunocompromised severe combined immunodeficient (SCID)-Beige mice were subcutaneously injected with either HDAC-6 specific shRNA or scrambled control and it was found that the growth of HDAC6 knockdown cells was significantly retarded [29]. Furthermore, HDAC6 knockdown cells reconstituted with wild type HDAC6, but not with catalytically inactive mutant HDAC6, regained its phenotype, indicating that HDAC6 is specifically required for tumorigenic growth. Further studies of the role of HDAC6 and tumorigenesis were performed using 7,12-dimethylbenzanthracene (DMBA) and 12-O-tetradecanoylphorbor-13-acetate (TPA) carcinogen-induced spontaneous tumorigenesis [29]. After tumor induction, the tumors decreased by half and the formation of tumors was retarded by two weeks in the HDAC6 null mice. These results suggested that HDAC6 is required for optimal tumor growth. RAS/MAPK signaling pathway is known to be required for tumorigenesis, and this study also showed that HDAC6 plays a major role in this pathway. HDAC6 knockout mice demonstrated reduced phosphorylation of AKT and ERK1/2, signaling pathways involved in tumor growth, and lower levels of activated Ras than those derived from wild-type mice. Thus, HDAC6 is considered to be required for the efficient activation of the oncogenic Ras signaling pathway. These results indicate that HDAC6 is a major contributor of tumorigenesis and maintenance of the transformed phenotype.
4. HDAC6 Inhibition and CYLD Activation Lead to a Delay in the Cell Cycle
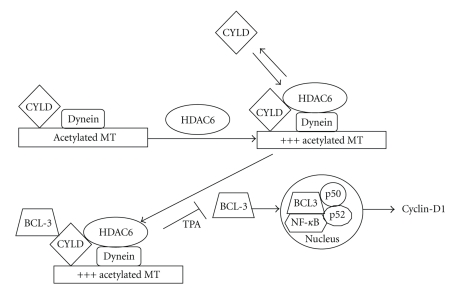
In a different study of HDAC6 and its role in tumorigenesis, it was found that CYLD-mediated inhibition of HDAC6 was essential for CYLD activation and delay in the cell cycle. CYLD is a tumor-suppressor gene present in patients suffering from cylindromatosis, a benign skin tumor [32]. In CYLD-transduced melanoma cells, as well as other tumor cell lines, CYLD constitutively colocalizes with acetylated MTs in the perinuclear region, where as in primary mouse keratinocytes melanoma cells or primary human melanocytes, CYLD perinuclear localization occurs after TPA treatment or exposure to UV light [30]. In this study, it was found that keratinocytes and melanoma cells that were treated with TSA or tubacin yielded increased levels of acetylated tubulin in enhanced green fluorescent protein (EGFP)-tagged expressing melanoma cells, while treatment of EGFP-CYLD-expressing melanoma cells with TSA or tubacin failed to increase acetylated ∝-tubulin. These results indicated that CYLD endogenously induces acetylation of ∝-tubulin by inhibiting HDAC6. Furthermore, it was found that inhibition of HDAC6 by the N-terminal cytoskeleton-associated protein-glycine-conserved (CAP-GLY) domains of CYLD mediated inhibition of HDAC6 [32].
CYLD-mediated HDAC6 inhibition was observed to enhance its association with the MTs, reduce the rate of cytokinesis, and delay cell cycle (Figure 2). Inhibition of BCL-3, a proto-oncogene, by the deubiquitinase activity of CYLD is mediated by an additional signal induced by TPA and requires intact acetylated MTs [32]. CYLD-mediated inhibition of HDAC6 results in perinuclear translocation of CYLD, and this in turn mediates BCL-3 inhibition by blocking its nuclear translocation. The lack of BCL-3 in the nucleus inhibits the transcriptional activity of NF-κB p50/p52, reduces the expression of cyclin-D1, and delays G1/S transition of the cell cycle [32] (Figure 2). These studies demonstrated how inhibition of HDAC6 can prevent tumor growth, indicating that HDAC6 should be considered as a major target in transformed cells.
Figure 2.
CYLD-mediated inhibition of HDAC6 leads to a delay in the cell cycle. CYLD binds acetylated microtubules and inhibits HDAC6. Inhibition of HDAC6 leads to increased levels of acetylated ∝-tubulin and this induces more binding of CYLD to MTs. CYLD binds BCL-3, and in the presence of TPA blocks its nuclear translocation, thereby inhibiting cyclin-D1.
5. The Role of HDAC6 during Cellular Stress Response and Tumorigenesis
A defense mechanism known to counteract the toxic effects of misfolded protein accumulation is carried out by the proteasome and the aggresome pathway; this in turn gives a survival advantage to transformed cells. A purified HDAC6-containing complex from mouse testis cytosolic extracts revealed the presence of vasolin-containing protein/protein 97 (VCP/p97) and phospholipase A2-activating protein/ubiquitin fusion degradation protein 3 (PLAP/UFD3), two proteins that are known to control protein ubiquitination [25]. HDAC6 contains a ZnF-UBP domain and a dynein-binding domain (Figure 1). These findings led to the hypothesis that HDAC6 participates in cargo sorting and transport by regulating the acetylation of MTs and by binding ubiquitinated proteins; therefore, the effect of HDAC6 on MTs and its role in protein degradation in response to cell stress are widely being investigated.
6. Microtubule Stabilization Leads to Increased Levels of HDAC6-Mediated Acetylated α-Tubulin
MTs play a role in cell cycle division (mitotic spindle), intracellular protein trafficking, shape maintenance, cell motility and attachment, and in signal transduction [44], hence, they are validated targets for anticancer therapy [45]. MTs are components of the cytoskeleton and they are composed of the tubulin molecule, αβ tubulin dimer (Mr ~ 100,000), which polymerizes to form 13 linear protofilaments with a rigid hollow rod of approximately 25 nm in diameter and several microns in length [7, 46]. The MTs also bind proteins that are collectively known as the MT associated proteins (MAPs). In animal cells, the major MT organizing center (MTOC) is the centrosome [7, 46]. The MTs undergo rapid cycles of assembly and disassembly and it has been established that their instability arises from the hydrolysis of GTP bound to β-tubulin during or shortly after polymerization [7]. Cytoplasmic MTs, which polymerize and depolymerize at high rates, in the presence of taxol, an antimitotic drug, bind the drug and are stabilized. This MT assembly allows for acetylation of α-tubulin to occur [20, 23, 47]. Therefore, the reversible acetylation on the ε-amino group of Lys40 near the N-terminus of α-tubulin is coupled to MT turnover [47].
HDAC6 plays a role in regulating the stability of dynamic MTs [18, 20, 23]. It was observed that in vivo, depolymerized tubulin is rapidly deacetylated while tubulin acetylation generally occurs on stable or polymerized MTs [18, 20, 23]. Furthermore, a group of researchers reported that combination of farnesyl transferase inhibitor lonafarnib and taxol enhance HDAC6-dependent tubulin deacetylation in breast and cell lung carcinoma cells [2].
7. HDAC6 and its Role in the Aggresome Pathway in Response to Cellular Stress
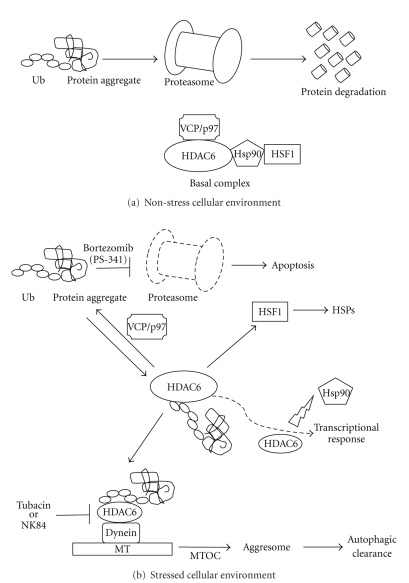
Targeting the proteasome or the aggresome to induce apoptosis or programmed cell death is one of the methods to target cancer cells. Transformed cells accumulate more misfolded proteins, which are disposed of by the proteasome and the aggresome [33, 48, 49]; therefore, cancer cells are more dependent on the proteasome and aggresome activity than nonmalignant cells for cell survival [33]. Upon inhibition of the proteasome, the high binding affinity of HDAC6 to ubiquitinated proteins activates the aggresome pathway [14] (Figure 3). Misfolded proteins form particulate (~200 nm) miniaggregates [36]. HDAC6 binds the ubiquitinated proteins and delivers them to the motor protein dynein, which it turn, transports the cargo along the MTs to the MTOC, where intermediate filament cytoskeleton surrounds the particle to form an aggresome [33, 36]. The aggresome is dynamic and it recruits chaperones and proteasomes [36]. Once this occurs, it activates the autophagic clearance and the autophagosomes fuse to lysosomes resulting in the degradation of the misfolded proteins [50]. The HDAC6 partner VCP/p97 is a chaperone with segregase activity. It disassembles the ubiquitinated aggregate/HDAC6 complex by extracting or sequestering HDAC6, so that the ubiquitinated proteins can be degraded through the proteasome instead of the aggresome [14, 36].
Figure 3.
Cell stress response: activation of the aggresome pathway, HSPs, and transcriptional response. (a) Under nonstress conditions, protein degradation occurs through the proteasome. HDAC6 is sequestered by VCP/p97 and forms a basal complex with HSF1 and HSP90. This keeps HSF1 in its inactive form. (b) Inhibition of the proteasome leads to an increase of misfolded proteins. HDAC6 binds these ubiquitinated protein aggregates and the basal complex disassembles. This complex dissociation activates HSF1, which eventually activates HSPs. The protein aggregates are transported along the MTs to MTOC to form an aggresome and autophagic clearance is activated. HDAC6 can also modulate Hsp90 acetylation levels and affect the maturation of client proteins, such as the glucocorticoid and androgen receptors, impairing gene transcription activation.
To study the effects of HDAC6 inhibition on the stress response, the HDAC6 inhibitor, tubacin, was used to test its effect on multiple myeloma cells. Tubacin-mediated α-tubulin acetylation disrupted HDAC6-dynein complex and synergistically increased the bortezomib-induced apoptotic effect in myeloma cells by c-Jun NH2-terminal kinase/caspase activation [37]. Multiple myeloma is cancer of plasma cells and can become resistant to conventional chemotherapy. The combination of bortezomib and tubacin was able to inhibit the proliferation of myeloma cells in vitro. [37]. More importantly, it was found that tubacin did not have an effect on normal peripheral blood mononuclear cells [37], hence the inhibition of HDAC6 by tubacin may be used for combined chemotherapy to treat myeloma or other types of cancer.
In ovarian cancer cells, their elevated metabolic activity results in increased ubiquitin proteasome-system (UPS) stress and this in turn, leads to an up-regulation of HDAC6 expression [28]. In one study, ovarian cancer cells were treated with HDAC6-specific inhibitors, tubacin and its derivative NK84. The treatment significantly inhibited the growth/survival of the ovarian cancer cell lines relative to the immortalized ovarian surface epithelial (IOSE) cell lines. Combined inhibition of both the proteasome and HDAC6-mediated aggresome degradation pathways is targeted with PS-341 and NK84, respectively, and the results showed synergistic cell death induced by both drugs. Furthermore, immunofluorescence analysis of the polyubiquitinated proteins in ovarian cancer cells, treated with both PS-341 and NK84 drugs, showed that HDAC6 inhibition prevented aggresome formation [28].
8. HDAC6 Is Required for Activation of HSF1 and Heat-Shock Protein Encoding Genes Response
An elegant study performed by Boyault et al. [34] using primary cells and established cell lines isolated from HDAC6-deficient mice, or human HDAC-6 deficient cells re-expressing wild type or mutated HDAC6, found that VCP/p97 is recruited by HDAC6 to a heat-shock protein 90/heat-shock factor 1 (Hsp90/HSF1) complex to form a “basal complex” in unstressed cells (Figure 3). At baseline, Hsp90 maintains HSF1 in its inactive form. HSF1 is a transcription factor that is essential in the activation of HSPs in response to the accumulation of misfolded proteins caused by heat shock or proteasome inhibition [29, 34] (Figure 3). The HSP response helps to reduce protein aggregate toxicity by increasing the expression of major cellular chaperones that either disaggregate proteins and help in their normal refolding or deliver them to the proteasome [35, 51]. Upon proteasome inhibition, HDAC6 binds the ubiquitinated aggregates and dissociation of HDAC6 and Hsp90 from HSF1 occurs. In the absence of VCP/p97, the Hsp90/HSF1 complex remains; therefore, the VCP/p97 ATPase activity seems to play a role in the dissociation of the Hsp90/HSF1 complex [34].
The essential role of HDAC6 in the activation of HSF1 is in accordance with the findings that similar to HDAC6-deficient cells and HDAC6-null mice, HSF-1-null MEFs were found to suppress skin tumor formation induced by DMBA-TPA treatment and oncogenic Ras-mediated anchorage-independent proliferation [29, 35]. Both HSF1 and HDAC6 regulate the oncogenic RAS/MAPK signaling pathway, and their presence is required for maintenance of transformed phenotypes in established tumor cells [29, 35].
9. HDAC6-Mediated Transportation of Stress Granules Leads to Reversible Translational Suppression in Response to Cellular Stress
HDAC6 also plays an essential role in a different cellular response induced by environmental stress. Kwon et al. [43] found that either of the two HDAC6 domains is sufficient to interact with cytoplasmic stress granule (SG) components, which are involved in reversible translational suppression in response to cellular environmental stress. The deacetylase activity and the ubiquitin-binding function of HDAC6 were found to be essential in the formation of SGs [43]. HDAC6 recruits the SGs to motor protein via binding of another SG protein, Ras-GTPase-activating protein SH3 domain-binding protein 1 (G3BP1) to allow SGs movement along the MTs. SGs are also ubiquitin positive and this allows direct binding to HDAC6 and MTs. In this study, disruption of MTs or motor proteins was found to inhibit individual SG components along the MTs. This type of HDAC6-mediated stress response strongly suggested a role of HDAC6 in the control of RNA metabolism and translation. It is worth mentioning that in a different study using cultured HSF-1 deficient MEFs under growth factor-depleted conditions or serum starvation, reduced levels of three ribosomal subunits, L26RP, L28RP, and S6RP, as well as those of phosphorylated ribosomal protein S6 kinase (p70 S6K), were observed [35]. p70 S6K is known to be a regulator of translational activity [35]. These results suggest that HSF1, which is activated by HDAC6, plays a role in ribosome biogenesis and protein translation, pathways that contribute to cancer [35].
10. HDAC6 Modulates Cell Motility
Cell motility plays a key role in tumor metastasis; hence, fibroblast motility has been used as a model to study the mechanisms of tumor cell migration in vivo [52]. Motile fibroblast contains two different types of MTs: The highly dynamic MTs predominate randomly throughout the cell periphery, and the less dynamic ones are found between the fibroblast nucleus and its leading edge [53]. In fibroblast crawling, actin is a key player, however, the MTs also play important roles, since MTs are required for the establishment and maintenance of cell polarity and directional movement [52]. Fibroblasts were observed to require dynamic MTs for the remodeling and turnover of their focal adhesions [54]. The detachment from the extracellular matrix can slow down invasion migration, so adhesion turnover is rate-limiting for rapid migration of fibroblasts [55]. Nevertheless, dependence for cell polarity on MT dynamism is seen only in some cell types; for example, fibroblasts and epithelial cells, which require an intact MT cytoskeleton to migrate during wound healing [56]. Faster migrating cells, such as leukocytes, do not require an intact MT cell cytoskeleton [56].
Studies of cell motility revealed that overexpression of HDAC6 increases chemotactic cell motility, suggesting that deacetylation of at least one cytoplasmic HDAC protein enhances motility [23]. HDAC inhibitors, such as TSA and SAHA block invasive cell motility by altering gene expression as a result of hyperacetylated HDAC nuclear substrates, such as histones or transcription factors [38]. Studies have shown that, as expected, tubacin leads to a reduction in cell motility [21, 38]. The inhibition of HDAC6 by tubacin, which leads to the hyperacetylation of tubulin, blocks fibroblast invasion motility similarly to the inhibition of all HDACs by TSA [38]. It was observed that in HDAC6-inhibited cells, MT dynamics is decreased, leading to an increase of focal adhesion accumulation, thus a decrease in fibroblast motility [38]. Another group performed a scratch assay and trans-Matrigel migration assays and showed that NK84-mediated inhibition of HDAC6, in ovarian cancer cell lines retarded spreading and inhibited migration, respectively [28].
In a different study, treatment of ERα positive breast cancer MCF-7 cells with estradiol or stably transfected with HDAC6 revealed that HDAC6 is a novel estrogen up-regulated gene and its expression led to increased cell motility [3]. Saji et al. [3] also found that either TAM, the pure antiestrogen ICI 182,780, or tubacin, prevented the estradiol-induced HDAC6 accumulation and deacetylation of α-tubulin, leading to reduced motility.
HDAC6 can also interact with a different substrate, cortactin, in vivo and in vitro, and both HDAC6 catalytic domains are necessary for interaction [17], (Figure 1). Cortactin is an acetylated protein found at areas of dynamic actin assembly, such as the leading edge of migrating cells [17]. This protein is originally known as a substrate of Src tyrosine kinase and it plays a role in regulating cell motility [57, 58]. Cortactin has been found to be overexpressed in several carcinomas [58]. It interacts with F-actin to promote polymerization and branching by modulating a “charge patch” in the cortactin repeat region [15]. Zhang et al. [15] found that inhibition of HDAC6 leads to hyperacetylation of cortactin and prevents its translocation to the cell periphery, blocks association with F-actin, and impairs cell motility.
In a different study, HDAC6 was found to play an important role in the chemotaxis of T-lymphocytes, independent of its deacetylase activity. The knockdown of HDAC6 expression in T-cells decreased their chemotactic capability; however, HDAC6 inhibitors, such as TSA and tubacin, showed that the deacetylase activity of HDAC6 was not involved in the modulatory effect of HDAC6 on cell migration [59]. Cabrero et al. [59] suggested hat HDAC6 might act as an on/off switch, regulating the rapid formation (10 to 20 folds faster than fibroblasts) of molecular complexes required for lymphocyte migration. In favor of that hypothesis the results of two studies were mentioned. First, HDAC6 reversibly complexes with phosphatases such as protein phosphatase 1 (PP1) and upon HDAC inhibition, release of PP1 from its complex with HDAC6 leads to the dephosphorylation of Akt, which in turn affects cell migration. The second finding was that tubulin acetylation decreases with inhibition of RhoA activity and since Rho GTPases coordinate both poles of migrating T-lymphocytes and control the dynamics of actin and tubulin cytoskeleton, HDAC6 may act as a mediator between actin- and tubulin-associated proteins to regulate motility [59].
A recent study involving the effect of HDAC6 on MT found that the impaired catalytic domain of HDAC6 is responsible for MT dynamics. In two different studies using NIH 3T3 fibroblast or A549 cells, it was found that MT stabilization is not promoted by tubulin acetylation [21, 60]. Palazzo et al. [60] reported that reduced motility is a consequence of alterations in the degree of tubulin acetylation and not of MT stabilization. Zilberman et al. [39] showed that the impaired catalytic domain of HDAC6, induced by HDAC6 inhibitors, TSA or tubacin, or point mutations in either of the two catalytic domains, is responsible for the regulation of MT dynamics, rather than tubulin acetylation alone. In this study, HDAC6 knockdown increased tubulin acetylation, but had no effect on the MT growth velocity, in the presence or absence of tubacin.
11. HDAC6 Modulates the Acetylation Status of Hsp90 and Represses Transcription
HDAC6 was also shown to have a third substrate, heat shock protein 90 (Hsp90), and affect transcriptional activation (Figure 3). Hsp90 functions by facilitating the structural maturation and complex formation of client proteins, including steroid hormone receptors and selected kinases [40, 41]. HDAC6 was found to regulate the activity of Hsp90. Inactivation or knockdown of HDAC6 led to Hsp90 hyperacetylation, its dissociation from cochaperone p23, and loss of chaperone activity. The loss of Hsp90 activity prevents the maturation of the glucocorticoid receptor (GR), affecting ligand binding, nuclear translocation, and transcriptional activation [40, 42]. In a different study, knockdown of HDAC6 in prostate cancer (PCa) C4-2 cells led to the hyperacetylation of Hsp90, impaired ligand-independent nuclear localization of endogenous androgen receptor (AR), inhibited prostate-specific antigen (PSA) expression, and cell growth in the absence or presence of dihydrotestosterone (DHT) [41]. These findings suggest that HDAC6 potentially affects the activity of other client proteins by regulating Hsp90. Nevertheless, other studies have shown that HDAC6 is related to cell migration rather than to transcriptional regulation [21, 23].
12. Conclusions
HDAC6 should be considered a target for cancer therapy due to its essential role in many signaling pathways that provide an advantage to malignant cells to survive and maintain its phenotype (Table 1). HDAC6 as well as its substrate HSF1 are essential for efficient oncogenic transformation by supporting anchorage-independent proliferation to prevent anoikis. HDAC6 and HSF1 can also regulate oncogenic Ras/MAPK signaling pathways, which are required for efficient tumor growth. High levels of HDAC6 expression were found to correlate with tumorigenesis, and ERα positive breast cancer patients who received adjuvant treatment with TAM showed better survival prognosis. Thus, HDAC6 could potentially be used as an additional marker for prognosis. HDAC6 is required for the activation of HSF1, a master activator of HSPs, leading to massive expression of chaperones; thereby, contributing to oncogenic transformed cell survival. HDAC6 is involved in the activation of SGs leading to reversible translational suppression, thus, helping transformed cells survive cellular environmental stress. HDAC6 can also regulate other proteins such as Hsp90, leading to the inhibition of steroid receptors activity and transcriptional activation.
Table 1.
Overview of HDAC and its role in cancer.
| Tumorigenesis | HDAC6 expression and its mediated HSF1 activation are essential for tumor growth and maintenance of oncogenic phenotype by promoting anchorage-independent proliferation to transformed cells [3, 26–35]. |
| CYLD-mediated HDAC6 inhibition leads to delay in the cell cycle and reduced rate of cytokinesis [30, 32]. | |
| Cell survival | HDAC6 binds ubiquitinated protein aggregates and this leads to the dissociation of the basal complex, and eventual activation of the aggresome pathway [28, 33, 36]. |
| HDAC6 inhibition leads to apoptosis [37]. | |
| Cell motility or metastasis | HDAC6 overexpression leads to increased cell motility [3, 21, 28, 38]. |
| HDAC6 inhibition leads to hyperacetylated cortactin and impaired cell motility [15, 17]. | |
| HDAC6 inhibition also leads to its impaired catalytic domain affecting MT dynamism [39]. | |
| Transcriptional response | HDAC6 deacetylates Hsp90 and prevents the maturation of glucocorticoid and androgen receptors [40–42]. |
| Translational response | HDAC6 forms a complex with SGs and G3BP1 and eventually induces a reversible translational suppression [43]. |
Studies of HDAC6 and its role in cell motility suggest its participation in cancer metastases and migration. The impaired catalytic domain of HDAC6 was found to be responsible for the regulation of MT dynamics. Recent studies have also shown that the inhibition of HDAC6 by tubacin leads to the accumulation of misfolded ubiquitinated proteins and apoptosis in multiple myeloma cells. The effects of HDAC6 on protein degradation and apoptosis revealed that this protein participates in cargo sorting and transport by regulating the acetylation of MTs and binding to ubiquitinated proteins. It is possible that the same HDAC6-mediated protective effect in response to cellular stress is observed in other cells; therefore, more studies using different cancer cell lines should be performed. Furthermore, since inhibition of HDAC6 stabilizes the MTs, other potential apoptotic pathways that inhibit oncogenic tumorigenesis need to be investigated.
Acknowledgments
K. M. Sakamoto is supported by the Leukemia and Lymphoma Society of America, National Institutes of Health (NIH) HL75826 and HL83077, William Lawrence and Blanche Hughes Foundation, and the St. Baldrick's Foundation. G. I. Aldana-Masangkay is supported by the Eugene Cota-Robles Fellowship (ECR) grant. The authors would also like to thank Bryan Mitton for reading the manuscript.
References
- 1.Marks PA, Miller T, Richon VM. Histone deacetylases. Current Opinion in Pharmacology. 2003;3:344–351. doi: 10.1016/s1471-4892(03)00084-5. [DOI] [PubMed] [Google Scholar]
- 2.Marcus AI, Zhou J, O'Brate A, et al. The synergistic combination of the farnesyl transferase inhibitor lonafarnib and paclitaxel enhances tubulin acetylation and requires a functional tubulin deacetylase. Cancer Research. 2005;65(9):3883–3893. doi: 10.1158/0008-5472.CAN-04-3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Saji S, Kawakami M, Hayashi S-I, et al. Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene. 2005;24(28):4531–4539. doi: 10.1038/sj.onc.1208646. [DOI] [PubMed] [Google Scholar]
- 4.Carew JS, Giles FJ, Nawrocki ST. Histone deacetylase inhibitors: mechanisms of cell death and promise in combination cancer therapy. Cancer Letters. 2008;269(1):7–17. doi: 10.1016/j.canlet.2008.03.037. [DOI] [PubMed] [Google Scholar]
- 5.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer Journal for Clinicians. 2008;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 6.Weinberg RA. The Biology of Cancer. New York, NY, USA: Taylor & Francis Group LLC; 2007. (Garland Science). [Google Scholar]
- 7.Cooper GM. The Cell—A Molecular Approach. 2nd edition. USA: Sinauer Associates Publishers; 2000. [Google Scholar]
- 8.Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proceedings of the National Academy of Sciences of the United States of America. 1999;96(9):4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marks PA, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Histone deacetylases and cancer: causes and therapies. Nature Reviews Cancer. 2001;1(3):194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 10.De Ruijter AJM, Van Gennip AH, Caron HN, Kemp S, Van Kuilenburg ABP. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochemical Journal. 2003;370(3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Verdin E, Dequiedt F, Kasler HG. Class II histone deacetylases: versatile regulators. Trends in Genetics. 2003;19(5):286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 12.Finnin MS, Donigian JR, Cohen A, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401(6749):188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 13.Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases: coordinate expression of differentiation-dependent chromatin modifiers. Journal of Biological Chemistry. 1999;274(4):2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- 14.Boyault C, Sadoul K, Pabion M, Khochbin S. HDAC6, at the crossroads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene. 2007;26(37):5468–5476. doi: 10.1038/sj.onc.1210614. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Yuan Z, Zhang Y, et al. HDAC6 Modulates cell motility by altering the acetylation level of cortactin. Molecular Cell. 2007;27(2):197–213. doi: 10.1016/j.molcel.2007.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawaguchi Y, Kovacs JJ, McLaurin A, Vance JM, Ito A, Yao T-P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115(6):727–738. doi: 10.1016/s0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 17.Wu H, Parsons JT. Cortactin, an 80/85-kilodalton pp60(src) substrate, is a filamentous actin-binding protein enriched in the cell cortex. Journal of Cell Biology. 1993;120(6):1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Li N, Caron C, et al. HDAC-6 interacts with and deacetylates tubulin and microtubules in vivo. EMBO Journal. 2003;22(5):1168–1179. doi: 10.1093/emboj/cdg115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Y, Gilquin B, Khochbin S, Matthias P. Two catalytic domains are required for protein deacetylation. Journal of Biological Chemistry. 2006;281(5):2401–2404. doi: 10.1074/jbc.C500241200. [DOI] [PubMed] [Google Scholar]
- 20.Matsuyama A, Shimazu T, Sumida Y, et al. In vivo destabilization of dynamic microtubules by HDAC6-mediated deacetylation. EMBO Journal. 2002;21(24):6820–6831. doi: 10.1093/emboj/cdf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective small-molecule inhibitor of histone deacetylase 6 (HDAC6)-mediated tubulin deacetylation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100(8):4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou H, Wu Y, Navre M, Sang B-C. Characterization of the two catalytic domains in histone deacetylase 6. Biochemical and Biophysical Research Communications. 2006;341(1):45–50. doi: 10.1016/j.bbrc.2005.12.144. [DOI] [PubMed] [Google Scholar]
- 23.Hubbert C, Guardiola A, Shao R, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417(6887):455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 24.Haggarty SJ, Koeller KM, Wong JC, Butcher RA, Schreiber SL. Multidimensional chemical genetic analysis of diversity-oriented synthesis-derived deacetylase inhibitors using cell-based assays. Chemistry and Biology. 2003;10(5):383–396. doi: 10.1016/s1074-5521(03)00095-4. [DOI] [PubMed] [Google Scholar]
- 25.Seigneurin-Berny D, Verdel A, Curtet S, et al. Identification of components of the murine histone deacetylase 6 complex: link between acetylation and ubiquitination signaling pathways. Molecular and Cellular Biology. 2001;21(23):8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inoue A, Yoshida N, Omoto Y, et al. Development of cDNA microarray for expression profiling of estrogen-responsive genes. Journal of Molecular Endocrinology. 2002;29(2):175–192. doi: 10.1677/jme.0.0290175. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Yamashita H, Toyama T, et al. HDAC6 expression is correlated with better survival in breast cancer. Clinical Cancer Research. 2004;10(20):6962–6968. doi: 10.1158/1078-0432.CCR-04-0455. [DOI] [PubMed] [Google Scholar]
- 28.Bazzaro M, Lin Z, Santillan A, et al. Ubiquitin proteasome system stress underlies synergistic killing of ovarian cancer cells by bortezomib and a novel HDAC6 inhibitor. Clinical Cancer Research. 2008;14(22):7340–7347. doi: 10.1158/1078-0432.CCR-08-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee Y-S, Lim K-H, Guo X, et al. The cytoplasmic deacetylase HDAC6 is required for efficient oncogenic tumorigenesis. Cancer Research. 2008;68(18):7561–7569. doi: 10.1158/0008-5472.CAN-08-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakuma T, Uzawa K, Onda T, et al. Aberrant expression of histone deacetylase 6 in oral squamous cell carcinoma. International Journal of Oncology. 2006;29(1):117–124. [PubMed] [Google Scholar]
- 31.Bradbury CA, Khanim FL, Hayden R, et al. Histone deacetylases in acute myeloid leukaemia show a distinctive pattern of expression that changes selectively in response to deacetylase inhibitors. Leukemia. 2005;19(10):1751–1759. doi: 10.1038/sj.leu.2403910. [DOI] [PubMed] [Google Scholar]
- 32.Wickström SA, Masoumi KC, Khochbin S, Fässler R, Massoumi R. CYLD negatively regulates cell-cycle progression by inactivating HDAC6 and increasing the levels of acetylated tubulin. The EMBO Journal. 2010;29(1):131–144. doi: 10.1038/emboj.2009.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodriguez-Gonzalez A, Lin T, Ikeda AK, Simms-Waldrip T, Fu C, Sakamoto KM. Role of the aggresome pathway in cancer: targeting histone deacetylase 6-dependent protein degradation. Cancer Research. 2008;68(8):2557–2560. doi: 10.1158/0008-5472.CAN-07-5989. [DOI] [PubMed] [Google Scholar]
- 34.Boyault C, Zhang Y, Fritah S, et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes and Development. 2007;21(17):2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai C, Whitesell L, Rogers AB, Lindquist S. Heat shock factor 1 is a powerful multifaceted modifier of carcinogenesis. Cell. 2007;130(6):1005–1018. doi: 10.1016/j.cell.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garcia-Mata R, Gao Y-S, Sztul E. Hassles with taking out the garbage: aggravating aggresomes. Traffic. 2002;3(6):388–396. doi: 10.1034/j.1600-0854.2002.30602.x. [DOI] [PubMed] [Google Scholar]
- 37.Hideshima T, Bradner JE, Wong J, et al. Small-molecule inhibition of proteasome and aggresome function induces synergistic antitumor activity in multiple myeloma. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(24):8567–8572. doi: 10.1073/pnas.0503221102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran AD-A, Marmo TP, Salam AA, et al. HDAC6 deacetylation of tubulin modulates dynamics of cellular adhesions. Journal of Cell Science. 2007;120(8):1469–1479. doi: 10.1242/jcs.03431. [DOI] [PubMed] [Google Scholar]
- 39.Zilberman Y, Ballestrem C, Carramusa L, Mazitschek R, Khochbin S, Bershadsky A. Regulation of microtubule dynamics by inhibition of the tubulin deacetylase HDAC6. Journal of Cell Science. 2009;122(19):3531–3541. doi: 10.1242/jcs.046813. [DOI] [PubMed] [Google Scholar]
- 40.Kovacs JJ, Murphy PJM, Gaillard S, et al. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Molecular Cell. 2005;18(5):601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 41.Ai J, Wang Y, Dar JA, et al. HDAC6 regulates androgen receptor hypersensitivity and nuclear localization via modulating Hsp90 acetylation in castration-resistant prostate cancer. Molecular Endocrinology. 2009;23(12):1963–1972. doi: 10.1210/me.2009-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Murphy PJM, Morishima Y, Kovacs JJ, Yao T-P, Pratt WB. Regulation of the dynamics of hsp90 action on the glucocorticoid receptor by acetylation/deacetylation of the chaperone. Journal of Biological Chemistry. 2005;280(40):33792–33799. doi: 10.1074/jbc.M506997200. [DOI] [PubMed] [Google Scholar]
- 43.Kwon S, Zhang Y, Matthias P. The deacetylase HDAC6 is a novel critical component of stress granules involved in the stress response. Genes and Development. 2007;21(24):3381–3394. doi: 10.1101/gad.461107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nogales E. Structural insights into microtubule function. Annual Review of Biochemistry. 2000;69:277–302. doi: 10.1146/annurev.biochem.69.1.277. [DOI] [PubMed] [Google Scholar]
- 45.Jordan MA, Wilson L. Microtubules as a target for anticancer drugs. Nature Reviews Cancer. 2004;4(4):253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]
- 46.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 5th edition. New York, NY, USA: Garland Science; 2008. [Google Scholar]
- 47.Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. Journal of Cell Biology. 1987;104(2):289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Miller CP, Ban K, Dujka ME, et al. NPI-0052, a novel proteasome inhibitor, induces caspase-8 and ROS-dependent apoptosis alone and in combination with HDAC inhibitors in leukemia cells. Blood. 2007;110(1):267–277. doi: 10.1182/blood-2006-03-013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hershko A, Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 50.Iwata A, Riley BE, Johnston JA, Kopito RR. HDAC6 and microtubules are required for autophagic degradation of aggregated Huntingtin. Journal of Biological Chemistry. 2005;280(48):40282–40292. doi: 10.1074/jbc.M508786200. [DOI] [PubMed] [Google Scholar]
- 51.Höhfeld J, Cyr DM, Patterson C. From the cradle to the grave: molecular chaperones that may choose between folding and degradation. EMBO Reports. 2001;2(10):885–890. doi: 10.1093/embo-reports/kve206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Glasgow JE, Daniele RP. Role of microtubules in random cell migration: stabilization of cell polarity. Cell Motility and the Cytoskeleton. 1994;27(1):88–96. doi: 10.1002/cm.970270110. [DOI] [PubMed] [Google Scholar]
- 53.Gundersen GG, Bulinski JC. Selective stabilization of microtubules oriented toward the direction of cell migration. Proceedings of the National Academy of Sciences of the United States of America. 1988;85(16):5946–5950. doi: 10.1073/pnas.85.16.5946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaverina I, Krylyshkina O, Small JV. Regulation of substrate adhesion dynamics during cell motility. International Journal of Biochemistry and Cell Biology. 2002;34(7):746–761. doi: 10.1016/s1357-2725(01)00171-6. [DOI] [PubMed] [Google Scholar]
- 55.Huttenlocher A, Palecek SP, Lu Q, et al. Regulation of cell migration by the calcium-dependent protease calpain. Journal of Biological Chemistry. 1997;272(52):32719–32722. doi: 10.1074/jbc.272.52.32719. [DOI] [PubMed] [Google Scholar]
- 56.Valenzuela-Fernández A, Cabrero JR, Serrador JM, Sánchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends in Cell Biology. 2008;18(6):291–297. doi: 10.1016/j.tcb.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 57.Bryce NS, Clark ES, Leysath JL, Currie JD, Webb DJ, Weaver AM. Cortactin promotes cell motility by enhancing lamellipodial persistence. Current Biology. 2005;15(14):1276–1285. doi: 10.1016/j.cub.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 58.Luxton GWG, Gundersen GG. HDAC6-pack: cortactin acetylation joins the brew. Developmental Cell. 2007;13(2):161–162. doi: 10.1016/j.devcel.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 59.Cabrero JR, Serrador JM, Barreiro O, et al. Lymphocyte chemotaxis is regulated by histone deacetylase 6, independently of its deacetylase activity. Molecular Biology of the Cell. 2006;17(8):3435–3445. doi: 10.1091/mbc.E06-01-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Palazzo A, Ackerman B, Gundersen GG. Tubulin acetylation and cell motility. Nature. 2003;421(6920):p. 230. doi: 10.1038/421230a. [DOI] [PubMed] [Google Scholar]