Abstract
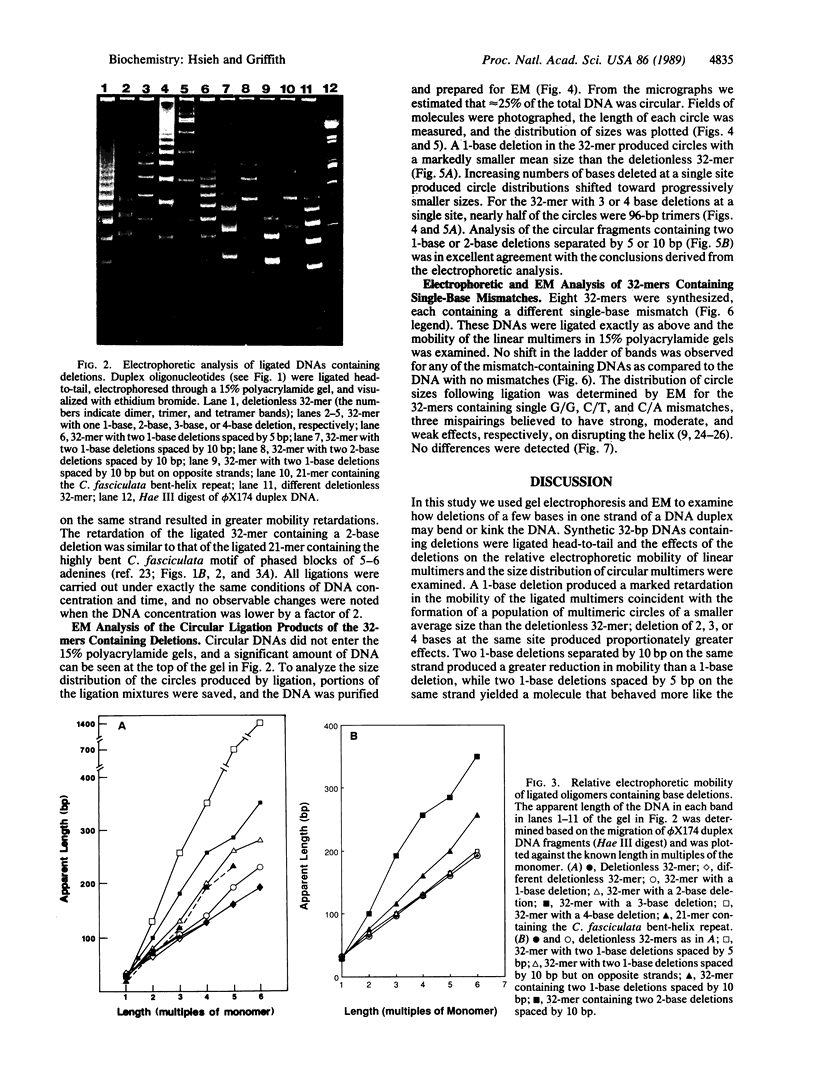
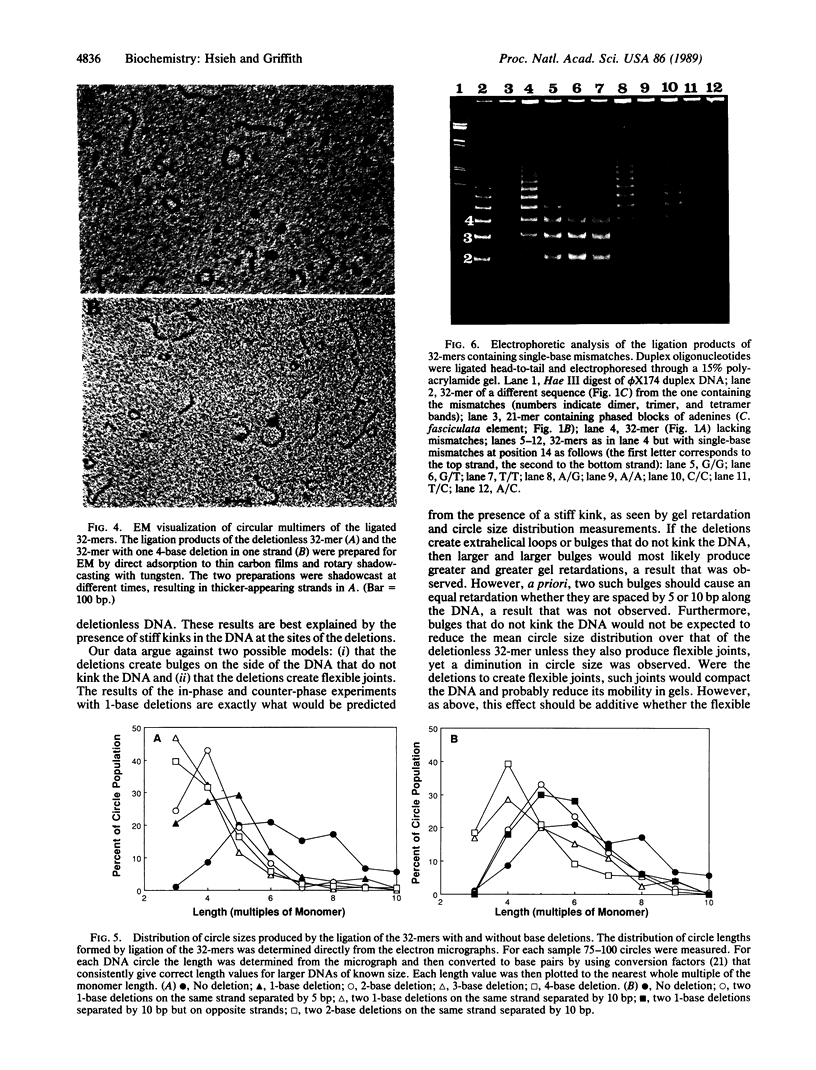
A 32-base-pair (bp) DNA duplex with deletions in one strand, and thus extra bases in the opposing strand, was ligated head-to-tail to produce linear and circular multimers. The electrophoretic mobility of the linear multimers was analyzed in polyacrylamide gels and the size of the circular DNA was determined by electron microscopy. A 1-base deletion produced a marked retardation in the mobility of the linear multimers coincident with the formation of a population of multimeric circles of a smaller average size than the deletionless 32-mer; 2-, 3-, or 4-base deletions at the same site produced proportionately greater effects. Two 1-base deletions separated by 10 bp on the same strand produced a greater reduction in mobility than a 1-base deletion, whereas two 1-base deletions spaced by 5 bp on the same strand yielded a molecule that behaved more like the deletionless DNA. We conclude that deletions of 1-4 bases at a single site on duplex DNA produce molecules that behave as if they contain sharp bends or kinks. In contrast, single mismatches in the 32-bp duplex produced no abnormality in behavior relative to normally base-paired DNA in the gel mobility and electron microscopic assays. The possible role of such structures in organizing the three-dimensional folding of single-stranded nucleic acids is considered.
Full text
PDF

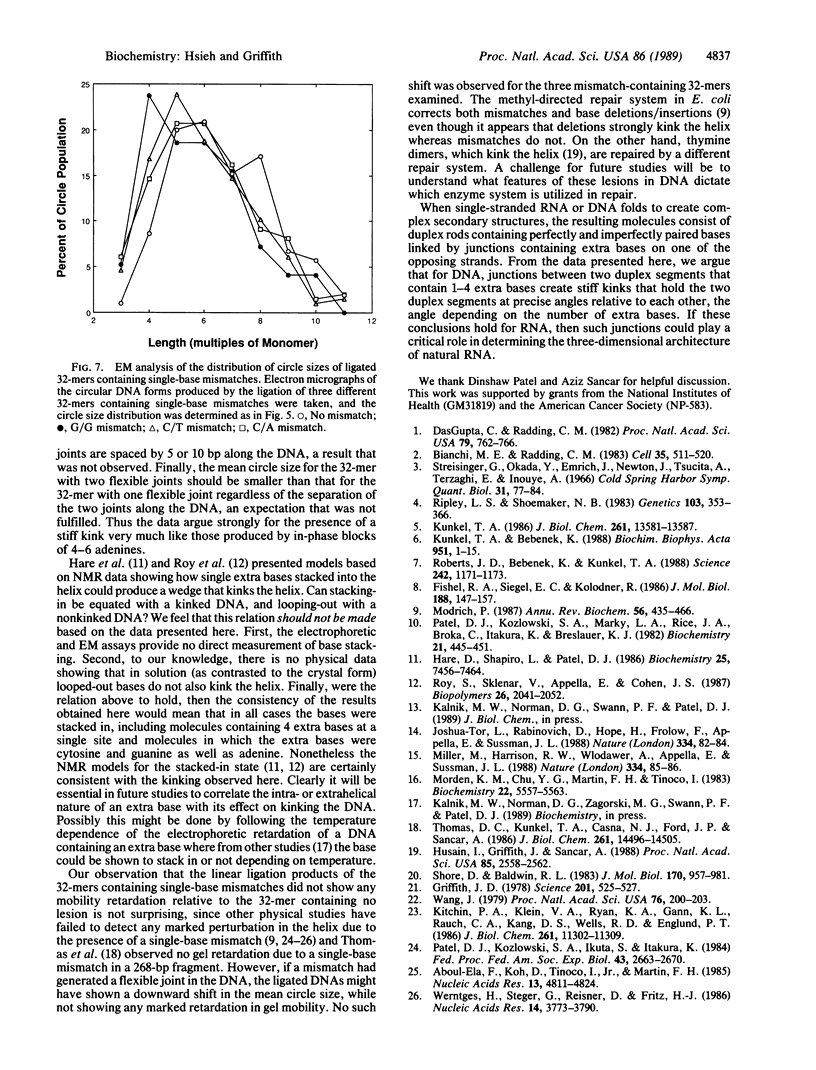
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aboul-ela F., Koh D., Tinoco I., Jr, Martin F. H. Base-base mismatches. Thermodynamics of double helix formation for dCA3XA3G + dCT3YT3G (X, Y = A,C,G,T). Nucleic Acids Res. 1985 Jul 11;13(13):4811–4824. doi: 10.1093/nar/13.13.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi M. E., Radding C. M. Insertions, deletions and mismatches in heteroduplex DNA made by recA protein. Cell. 1983 Dec;35(2 Pt 1):511–520. doi: 10.1016/0092-8674(83)90185-x. [DOI] [PubMed] [Google Scholar]
- DasGupta C., Radding C. M. Polar branch migration promoted by recA protein: effect of mismatched base pairs. Proc Natl Acad Sci U S A. 1982 Feb;79(3):762–766. doi: 10.1073/pnas.79.3.762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishel R. A., Siegel E. C., Kolodner R. Gene conversion in Escherichia coli. Resolution of heteroallelic mismatched nucleotides by co-repair. J Mol Biol. 1986 Mar 20;188(2):147–157. doi: 10.1016/0022-2836(86)90300-1. [DOI] [PubMed] [Google Scholar]
- Griffith J. D. DNA structure: evidence from electron microscopy. Science. 1978 Aug 11;201(4355):525–527. doi: 10.1126/science.663672. [DOI] [PubMed] [Google Scholar]
- Hare D., Shapiro L., Patel D. J. Extrahelical adenosine stacks into right-handed DNA: solution conformation of the d(C-G-C-A-G-A-G-C-T-C-G-C-G) duplex deduced from distance geometry analysis of nuclear Overhauser effect spectra. Biochemistry. 1986 Nov 18;25(23):7456–7464. doi: 10.1021/bi00371a030. [DOI] [PubMed] [Google Scholar]
- Husain I., Griffith J., Sancar A. Thymine dimers bend DNA. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2558–2562. doi: 10.1073/pnas.85.8.2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshua-Tor L., Rabinovich D., Hope H., Frolow F., Appella E., Sussman J. L. The three-dimensional structure of a DNA duplex containing looped-out bases. Nature. 1988 Jul 7;334(6177):82–84. doi: 10.1038/334082a0. [DOI] [PubMed] [Google Scholar]
- Kitchin P. A., Klein V. A., Ryan K. A., Gann K. L., Rauch C. A., Kang D. S., Wells R. D., Englund P. T. A highly bent fragment of Crithidia fasciculata kinetoplast DNA. J Biol Chem. 1986 Aug 25;261(24):11302–11309. [PubMed] [Google Scholar]
- Kunkel T. A., Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988 Nov 10;951(1):1–15. doi: 10.1016/0167-4781(88)90020-6. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Frameshift mutagenesis by eucaryotic DNA polymerases in vitro. J Biol Chem. 1986 Oct 15;261(29):13581–13587. [PubMed] [Google Scholar]
- Miller M., Harrison R. W., Wlodawer A., Appella E., Sussman J. L. Crystal structure of 15-mer DNA duplex containing unpaired bases. Nature. 1988 Jul 7;334(6177):85–86. doi: 10.1038/334085a0. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Ikuta S., Itakura K. Dynamics of DNA duplexes containing internal G.T, G.A, A.C, and T.C pairs: hydrogen exchange at and adjacent to mismatch sites. Fed Proc. 1984 Aug;43(11):2663–2670. [PubMed] [Google Scholar]
- Patel D. J., Kozlowski S. A., Marky L. A., Rice J. A., Broka C., Itakura K., Breslauer K. J. Extra adenosine stacks into the self-complementary d(CGCAGAATTCGCG) duplex in solution. Biochemistry. 1982 Feb 2;21(3):445–451. doi: 10.1021/bi00532a004. [DOI] [PubMed] [Google Scholar]
- Ripley L. S., Shoemaker N. B. A major role for bacteriophage T4 DNA polymerase in frameshift mutagenesis. Genetics. 1983 Mar;103(3):353–366. doi: 10.1093/genetics/103.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. D., Bebenek K., Kunkel T. A. The accuracy of reverse transcriptase from HIV-1. Science. 1988 Nov 25;242(4882):1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- Roy S., Sklenar V., Appella E., Cohen J. S. Conformational perturbation due to an extra adenosine in a self-complementary oligodeoxynucleotide duplex. Biopolymers. 1987 Dec;26(12):2041–2052. doi: 10.1002/bip.360261206. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. I. Relation between twist and cyclization probability. J Mol Biol. 1983 Nov 15;170(4):957–981. doi: 10.1016/s0022-2836(83)80198-3. [DOI] [PubMed] [Google Scholar]
- Streisinger G., Okada Y., Emrich J., Newton J., Tsugita A., Terzaghi E., Inouye M. Frameshift mutations and the genetic code. This paper is dedicated to Professor Theodosius Dobzhansky on the occasion of his 66th birthday. Cold Spring Harb Symp Quant Biol. 1966;31:77–84. doi: 10.1101/sqb.1966.031.01.014. [DOI] [PubMed] [Google Scholar]
- Thomas D. C., Kunkel T. A., Casna N. J., Ford J. P., Sancar A. Activities and incision patterns of ABC excinuclease on modified DNA containing single-base mismatches and extrahelical bases. J Biol Chem. 1986 Nov 5;261(31):14496–14505. [PubMed] [Google Scholar]
- Wang J. C. Helical repeat of DNA in solution. Proc Natl Acad Sci U S A. 1979 Jan;76(1):200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werntges H., Steger G., Riesner D., Fritz H. J. Mismatches in DNA double strands: thermodynamic parameters and their correlation to repair efficiencies. Nucleic Acids Res. 1986 May 12;14(9):3773–3790. doi: 10.1093/nar/14.9.3773. [DOI] [PMC free article] [PubMed] [Google Scholar]