Abstract
Using the representation difference analysis technique, we have identified a novel gene, Ian4, which is preferentially expressed in hematopoietic precursor 32D cells transfected with wild-type versus mutant forms of the Bcr/Abl oncogene. Ian4 expression was undetectable in 32D cells transfected with v-src, oncogenic Ha-ras or v-Abl. Murine Ian4 maps to chromosome 6, 25 cM from the centromere. The Ian4 mRNA contains two open reading frames (ORFs) separated by 5 nt. The first ORF has the potential to encode for a polypeptide of 67 amino acids without apparent homology to known proteins. The second ORF encodes a protein of 301 amino acids with a GTP/ATP-binding site in the N-terminus and a hydrophobic domain in the extreme C-terminus. The IAN-4 protein resides in the mitochondrial outer membrane and the last 20 amino acids are necessary for this localization. The IAN-4 protein has GTP-binding activity and shares sequence homology with a novel family of putative GTP-binding proteins: the immuno-associated nucleotide (IAN) family.
INTRODUCTION
Chronic myelogenous leukemia (CML) is a clonal myeloproliferative disorder due to a translocation between chromosomes 9 and 22. This translocation results in the fusion of a truncated breakpoint cluster region gene (bcr) with sequences of c-abl, the cellular homolog of the transforming gene of the Abelson murine leukemia virus. Depending on the site of the breakpoint in the bcr gene, two major chimeric BCR/ABL proteins are generated: p185, which contains a smaller BCR portion (amino acids 1–426) and is associated with acute lymphocytic leukemia (1) and p210, which contains a larger BCR portion (1–902 or 1–926) and is found in most cases of CML (2). A third, less common 230 kDa protein is found in a cohort of patients with chronic neutrophilic leukemia (3). BCR/ABL proteins have constitutively active tyrosine kinase activity, which is required for transformation (4). The oncogenic potential of BCR/ABL proteins has been demonstrated by a number of in vitro and in vivo assays including abrogation of interleukin 3 (IL-3) dependence (5) and induction of long-term survival of bone marrow cells in culture (6), as well as induction of CML-like syndromes in mice (7).
Several mechanisms can explain the role of the BCR/ABL oncoprotein in the pathogenesis of CML: activation of mitogenic signaling pathways, inhibition of apoptosis and differentiation and defective cell mobility. BCR/ABL can activate several downstream effector molecules involved in the control of proliferation, like RAS, RAF, MYC and STAT; its ability to block apoptosis has been related to RAS activation (8) and Bcl-2 expression (9), but is also mediated by the activation of the PI-3 kinase/Akt pathway (10). The BCR/ABL protein also activates the small GTP-binding protein Rac (11) and phosphorylates FAK (12), two proteins critically important in cell adhesion and mobility. BCR/ABL mutants have been used to dissect the signaling pathways involved in leukemogenesis. The tyrosine kinase activity is essential for each feature of BCR/ABL-expressing cells (13), whereas mutants in other domains show only partial defects in the BCR/ABL-dependent pathways promoting cell growth and apoptosis resistance. A BCR/ABL mutant (p185ΔBCR) containing only the first 176 amino acids of the BCR portion failed to protect IL-3-deprived 32D cells from apoptosis (13). Compared to wild-type (WT) BCR/ABL-expressing cells, p185ΔBCR-expressing cells showed markedly decreased Bcl-2 levels (14). Moreover, expression of mitochondrial RAF was essentially undetectable in these cells (14). Upon transfection with a mitochondria-targeted constitutively active RAF (M-RAF), these cells regained the apoptosis-resistant phenotype. In studies of in vivo leukemogenesis, double transfectants ΔBCR/M-RAF-expressing cells induced leukemia in immunodeficient mice whereas cells expressing only the p185ΔBCR mutant were non-leukemogenic, suggesting that expression of mitochondrial RAF is an important BCR/ABL effector in apoptosis resistance and leukemogenesis. To characterize the mechanisms whereby M-RAF is activated by BCR/ABL, we sought to identify genes differentially expressed between WT BCR/ABL-transfected 32D and ΔBCR/M-RAF double transfectants. Here we report the cloning of a novel gene, Ian4, whose expression is induced in BCR/ABL-expressing cells. This gene maps to chromosome 6 and encodes a mitochondrial membrane protein belonging to the GTP-binding superfamily and the immuno-associated nucleotide (IAN) subfamily of nucleotide-binding proteins.
MATERIALS AND METHODS
Cells
The murine IL-3-dependent 32D myeloid precursor cell line and WT BCR/ABL and ΔBCR/M-RAF 32D cell transfectants were maintained in Iscove’s modified Dulbecco medium (IMDM) supplemented with 10% FBS, 2 mM l-glutamine, penicillin/streptomycin (100 µg/ml) and 15% WEHI-conditioned medium as source of IL-3.
Representational difference analysis (RDA)
Total RNA was isolated from WT BCR/ABL and ΔBCR/M-RAF 32D cells 18 h after IL-3 starvation, with Tri-Reagent (Molecular Research Center) according to the manufacturer’s instructions. Polyadenylated RNA was purified from total RNA with oligotex beads (Qiagen) and cDNA was synthesized with the cDNA synthesis kit (Boehringer Mannheim). Double-stranded cDNA was digested with DpnI and RDA was performed according to the method of Hubank and Schatz (15). Digested double-stranded cDNA populations were ligated to adaptors and amplified prior to subtractive-hybridization. Three successive substractive-hybridizations were performed with ratios of WT BCR/ABL to ΔBCR/M-RAF cDNA of 1:100, 1:800 and 1:2000, respectively. The PCR products after the third subtraction were subcloned into the BamHI site of pBluescript (SK+) using standard subcloning techniques.
RACE
The complete Ian4 cDNA sequence was obtained by adaptor-ligated 5′ and 3′ RACE with the Marathon cDNA amplification kit (Clontech) on an adaptor-ligated library made from mRNA of WT BCR/ABL-expressing cells. The 5′ RACE product was obtained by PCR amplification of the library cDNA with the Marathon adaptor primer and an Ian4 antisense primer (5′-CTC AGC GCA TAA CTG TCC TTG GA-3′). Similarly, the 3′ RACE product was obtained by PCR using the Marathon adaptor primer and an Ian4 sense primer (5′-CTG TTG GAG ACC ATA CCG TGA GA-3′). RACE products were subcloned and sequenced with the PRISM Ready Reaction Dyedoxy Terminator Cycle Sequencing kit (Applied System).
Northern blot analysis
Total RNA (10 µg/lane) was fractionated electrophoretically on a 1% agarose/formaldehyde gel and transferred onto a nylon membrane (Amersham). The membrane was then hybridized with a random-primed 32P-labeled fragment of Ian4 (nucleotides 560–1520). Filters were routinely hybridized in 50% formamide, 5× SSC, 5× Denhardt’s and 0.5% SDS at 42°C for 16 h and washed in 2× SSC/0.1% SDS for 10 min at room temperature and 30 min at 42°C. Filters were visualized under UV light to monitor possible differences in the quantity of RNA loaded per lane.
Southern blot analysis
Genomic DNAs were digested, fractionated and transferred onto a nylon membrane as described previously (16). The membranes were then hybridized with a random-primed 32P-labeled fragment of Ian4 (nucleotides 560–1520). Filters were routinely hybridized in 5× SSC, 5× Denhardt’s and 0.5% SDS at 65°C for 16 h and washed in 2× SSC/0.1% SDS for 10 min at room temperature and 30 min at 65°C.
Plasmids
To study IAN-4 in transfected cells, the full-length cDNA containing both open reading frames (ORFs) or a 5′-deleted cDNA including only the second ORF was inserted into the expression vector LXSP, using PCR and standard subcloning techniques. To permit immunodetection of IAN-4, a sequence that encodes the HA epitope (YPYDVPDYA) of the influenza virus was added in-frame in the 5′ or 3′ of the second ORF.
Transfection
Plasmids were introduced into cells by electroporation (200 mV, 960 µF; gene pulser, Bio-Rad Laboratories, Hercules, CA). IAN-4–hemagglutinin (HA)-expressing cells were selected in puromycin-containing medium (2.5 µg/ml).
Western blot
Cells were harvested, washed with phosphate-buffered saline (PBS) and lysed in lysis buffer (50 mM Tris–HCl pH 7.4, 1% NP-40, 0.25% sodium deoxycholate, 0.15 M NaCl, 1 mM EDTA, 1 mM PMSF, 1 µg/ml aprotinin, leupeptin, pepstatin). Lysates were collected after centrifugation (10 min) at 14 000 g and the protein concentration was determined by the Bradford method. Total protein (50 µg) was fractionated on a 4–15% SDS–polyacrylamide gel, transferred to nylon membranes and immunoblotted with the anti-HA (BabCo), the anti-cytochrome oxidase (COX) IV (Molecular Probes, Eugene, OR) and the anti-HSP90 (Transduction Labs, Lexington, KY) antibodies.
Subcellular fractionation
Cells were lysed in hypotonic buffer (5 mM Tris pH 7.4, 5 mM KCl, 1.5 mM MgCl2, 0.1 mM EDTA pH 8, 1 mM DTT, 1 mM PMSF, 1 µg/ml aprotinin, leupeptin, pepstatin) at 4°C for 30 min. After homogenization (30–40 strokes with a Dounce homogenizer B-pestle), samples were centrifuged (2000 g for 5 min) to remove the nuclei and centrifuged again (14 000 g for 30 min) to obtain the heavy membrane (HM) fraction (pellet). The supernatant was centrifuged at 150 000 g for 1.5h to obtain light membrane (LM) and cytoplasmic (C) fractions. The HM and LM fractions were solubilized in 1% Triton X-100, 10 mM Tris–HCI pH 7.4, 150 mM NaCl, 5 mM EDTA, 1 mM PMSF, 1 µg/ml aprotinin, leupeptin, pepstatin.
Immunofluorescence microscopy
Cells were first incubated for 15 min at 37°C in growth medium containing Mitotracker Red CMX Ros (300 nM) (Molecular Probes), a morphological marker of mitochondria. After three washes with PBS, cells were deposited onto glass coverslips by centrifugation, fixed with 3.7% formaldehyde, washed and permeabilized in 0.05% Triton X-100. After blocking for 10 min at room temperature with 4% goat serum, cells were incubated in a 1:250 dilution of an anti-HA monoclonal antibody (Babco) for 30 min at 37°C. After three washes with PBS, the secondary antibody was added at a 1:200 dilution (Alexa 488, goat anti-mouse IgG antibody) (Molecular Probes). Cells were then rinsed three times with PBS and mounted onto slides. Confocal microscopy was performed on a Bio-Rad MRC 600 laser scanning confocal microscope (Hemel Hempstead, UK) attached to a Zeiss Axiovert 100 microscope, using a Zeiss Plan-Apo 63 × 1.40 NA oil-immersion lens.
Submitochondrial localization
The HM fraction was prepared as described above. To monitor the presence of IAN-4 in the membrane fraction, the pellet was resuspended in freshly prepared 0.1 M sodium carbonate pH 11.5, and maintained on ice for 30 min with periodic vortexing. Membranes were then collected by centrifugation at 7000 g for 30 min. The supernatant and pellet (membrane) were analyzed by SDS–PAGE and western blotting. To further determine IAN-4 sublocalization, the HM pellet was resuspended in isolation buffer (2 mM HEPES, 70 mM sucrose, 220 mM d-mannitol, 0.5 mg/ml BSA) and treated with 0.10, 0.15 or 0.20% digitonin. After 20 min in ice, the samples were centrifuged and the supernatant analyzed by SDS–PAGE and immunoblotting.
GTP-binding assay
GTP-binding to IAN-4–HA was determined by a nitrocellulose filtration assay as described previously (13). Briefly, 107 IAN-4–HA-expressing 32D cells were washed with ice-cold PBS, lysed for 15 min in 1 ml lysis buffer (1% Triton X-100, 50 mM HEPES pH 7.4, 1 mg/µl BSA, 5 mM MgCl, 10 µg/ml aprotinin, 10 µg/ml leupeptin, 10 mM benzamidine, 1 mM PMSF, 0.2 mM sodiumorthovanadate) and centrifuged at 13 000 g for 15 min. Lysate was precleared for 90 min using Protein G–agarose beads (Calbiochem) and 1 µg of anti-c-ABL monoclonal antibody (Ab-3; Calbiochem) was used as an isotype-matched control. Concurrently, 100 µl protein G–agarose beads were linked to 1 µg of anti-HA monoclonal antibody (HA.11; BabCo). IAN-4–HA was then immunoprecipitated after 90 min incubation with the HA-Ab/protein G–agarose beads. After extensive washing, the immunoprecipitate was resuspended in 200 µl binding buffer (20 mM Tris pH 7.5, 1 mM MgCl, 1 mM DTT, 100 mM NaCl, 40 µg/ml BSA) and 1 µCi [α-32P]GTP and incubated at 30°C to allow binding. At the indicated times, 30 µl of the reaction mix was removed and nucleotide binding was quenched by adding 500 µl of ice-cold washing buffer (20 mM Tris pH 7.5, 100 mM NaCl, 10 mM MgCl, 1 mM DTT). The sample was filtered through a nitrocellulose filter (Schleicher and Schuell) and washed twice with 4 ml of washing buffer to remove unbound [α-32P]GTP. The nitrocellulose filler was air-dried and the associated radioactivity was measured by scintillation counting after immersion in Econofluor-2 (Packard).
Genetic linkage analysis
The interspecific backcross mapping panel used [(AEJ/Gn × Mus spretus) F1 × AEJ/Gn] was described previously (17). High molecular weight genomic DNAs were isolated from frozen tissues as described previously (16). Genomic DNAs were digested with restriction endonucleases, that produced informative restriction fragment length polymorphisms (RFLPs) (17), and analyzed by Southern blot hybridization. The presence or absence of the M.spretus-specific alleles was followed to determine the pattern of inheritance in each N2 offspring. The Ian4 probe detected M.spretus-specific PstI fragments of 5.2 and 4.0 kb as well as AEJ/Gn-specific PstI fragments of 6.0 and 4.8 kb. The chromosomal locations of the D6Mit118 and D6Kcc1 markers were previously reported (17). The chromosomal localization and recombination frequencies of loci mapped in the interspecific backcross were determined using the computer program SPRETUS MADNESS: PART DEUX developed by K.Smalley, J.Averback, L.D.Siracusa and A.M.Buchberg (Kimmel Cancer Center, Philadelphia, PA).
RESULTS
Identification of genes overexpressed in BCR/ABL-expressing 32D cells
The RDA technique was used to identify genes overexpressed in WT BCR/ABL-expressing 32D cells (tester) compared to ΔBCR/M-RAF double transfectants (driver). To enrich for mRNAs overexpressed in BCR/ABL-expressing cells, three successive subtractions were performed with tester/driver ratios of 1:100, 1:800 and 1:2000, respectively. After the third subtraction, four fragments (ranging in size from 200 to 500 bp) were amplified by PCR (Fig. 1A), purified, subcloned in Bluescript and sequenced. One of these PCR products (250 bp) had no significant homology with the sequences listed in GenBank. Using this product as a probe, a northern blot on total RNA from WT BCR/ABL-expressing 32D cells and ΔBCR/M-RAF double transfectants (Fig. 1B) confirmed that the expression of this gene is enhanced in WT BCR/ABL-expressing cells. The transcript of this novel gene, Ian4, is 2.1 kb.
Figure 1.
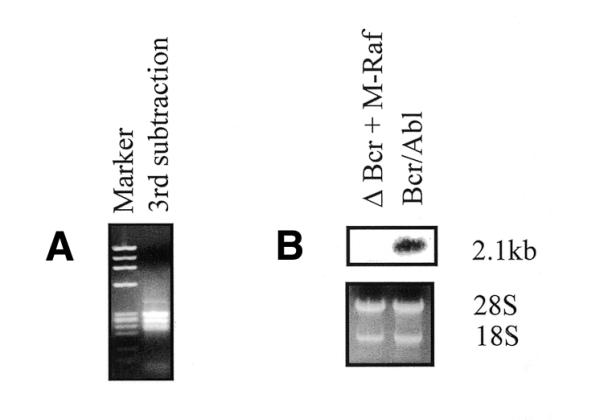
RDA of cDNA from ΔBCR + M-Raf and Bcr/Ab1-expressing cells. (A) Product of the third subtraction loaded onto an ethidium bromide–agarose gel. (B) Northern blot shows differential expression of Ian4 mRNA in the two cell lines. Total RNA (10 µg) was electrophoresed, blotted onto a nylon membrane and hybridized with a 32P-labeled Ian4 probe. The gel was visualized under UV light to verify equal RNA loading.
Sequence of the full-length Ian4 cDNA
To obtain the complete sequence of Ian4 cDNA, adaptor-ligated RACE was performed using mRNA from WT BCR/ABL-expressing 32D cells. Two primers were chosen on the initial sequence. One Ian4-specific primer was used to amplify a fragment corresponding to the 3′ part of the cDNA, and ending in the poly(A) tail. The second Ian4-specific primer was used to amplify the 5′ part of the cDNA. The PCR-amplified products were cloned and sequenced. The sequence of Ian4 cDNA was 2150 nt and included two ORFs (accession no. AF337052). The first ORF started with an ATG codon located at position +405, surrounded by nucleotides falling within the Kozak consensus sequence (G/AXXATGG) for translation initiation. This ORF extended for 67 codons and ended with TAA. The stop codon was followed by a run of five adenosine nucleotides and a second start codon. The second start codon at position 614 was also in the context of a canonical initiation sequence and extended to encode a translation product of 301 amino acids. A polyA signal (AATAAA) mapped 601 bp downstream of the translation stop codon. Bicistronic mRNAs are relatively rare in higher vertebrates and the 5′ upstream ORFs are usually very short (18,19). To exclude the possibility that the unusual sequence arrangement of Ian4 cDNA was the result of a cloning artefact, the segment of Ian4 cDNA encompassing the two ORFs was amplified from the RNA of two BCR/ABL-expressing cell lines. Using a 5′ primer (5′-GTCATACCGTCACACCATCTG-3′) starting 30 nt upstream of the ATG of the first ORF and a reverse 3′ primer (5′-GCCTGTTACAACATTCTGAAGT-3′) beginning 26 nt downstream of the ATG of the second ORF, the corresponding Ian4 cDNA was amplified and cloned in the pSR TOPOII vector. Sequencing of seven independent clones from two BCR/ABL-expressing lines confirmed the sequence arrangement of Ian4 mRNA (data not shown).
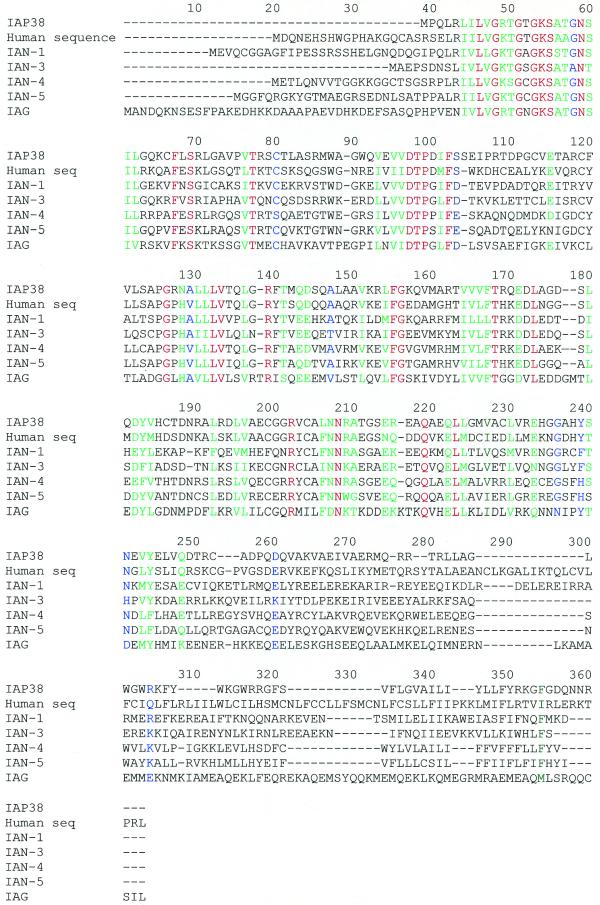
The polypeptide that could be encoded by the first ORF of Ian4 showed no significant similarities with any protein in protein databases. Querying the second translation product against the non-redundant set of protein sequences with PSI-BLAST (20) identified hundreds of related proteins, essentially all of which have been classified as members of the GTP-binding protein superfamily. Within the GTP-binding protein superfamily, IAN-4 was most closely related to a novel family of putative GTP-binding proteins called the IAN family (21). The most striking similarity was with a related human protein (accession no. AK002158) we have named IAN-5. IAN-4 was also highly homologous to mouse immune-associated protein 38 (IAP38) (accession no. Y08026), a protein whose expression is induced by blood-stage infections of Plasmodium chabaudi (22); mouse IAN-1, a thymic selection marker; the IAN-3 homolog (21); and the IAG1 protein (accession no. U40856), a plant protein induced by bacterial infection (23). Murine IAN-4 was also homologous to a putative human protein (accession no. AL110151) and to NTGP4 (accession no. U64925), an isoprenylated plant protein. The sequence alignment of these proteins (Fig. 2) shows that the N-terminal portion is highly conserved while the C-terminal portion is specific to each protein. Within the GTP-binding superfamily, IAN subfamily proteins are clearly found in animals and plants. Lesser but substantial similarity was observed with Era subfamily proteins that are found in bacteria as well as metazoans and with septins such as yeast Cdc10. While the similarity between IAN-4 and distantly related members of the GTP-binding superfamily was confined to the GTP-binding consensus sequence, similarity with Cdc10-related proteins extended for ∼250 amino acids (data not shown). Several IAN-related GTP-binding proteins have been classified as organelle-localized, such as the chloroplast import-associated protein IAP36 in pea. IAN-4 does not contain a C-terminal cysteine motif that could serve as a site for isoprenylation. Instead, the 22 C-terminal residues are hydrophobic and are predicted to form a membrane anchor for IAN-4 protein.
Figure 2.
Alignment of IAP38, a human sequence (GenBank accession no. AL110151), IAN-5, IAN-1, IAN-3 and IAG-1 protein sequences. The alignment was performed using CLUSTALW. Red, identical amino acids; green, conserved amino acids; blue, identical or conserved in most members of the family.
Expression of Ian4
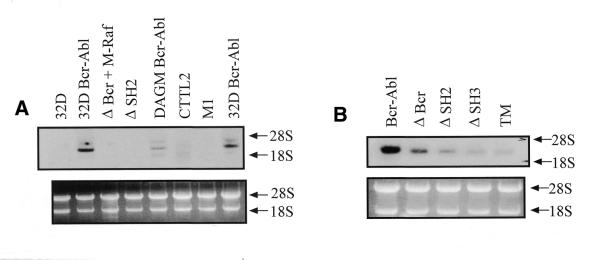
An Ian4-specific probe was used to investigate Ian4 expression in different cell lines and normal mouse tissues. Ian4 mRNA was detected only in cell lines expressing BCR/ABL (Fig. 3A). Ian4 mRNA was highly expressed in different clones of BCR/ABL-expressing 32D and DAGM cells, while no expression was detected in 32D parental cells, in CTTL2 (murine T cells) and M1 (a murine myeloid leukemia cell line) cells. Ian4 expression was also assessed in 32D cells transfected with different BCR/ABL mutants: p185ΔBCR (deletion of amino acids 176–426 in the BCR portion), p210ΔSH2 (deletion of the SH2 domain), p210ΔSH3 (deletion of the SH3 domain), TM (Y177F/R552L/Y793F triple mutant) (Fig. 3B). Ian4 mRNA was readily detectable in WT BCR/ABL-expressing cells, but Ian4 mRNA levels were markedly diminished in cells transfected with various BCR/ABL mutants, with the lowest expression detected in cells transfected with BCR/ABL TM. In the blot shown in Figure 3A, Ian4 mRNA was essentially undetectable in p210ΔSH2-expressing cells: this most likely reflects low hybridization efficiency, since the signal detected with the RNA of WT BCR/ABL cells is much stronger in the Figure 3B blot than in the Figure 3A blot. Ian4 mRNA was also detectable in BCR/ABL-expressing primary mouse marrow cells (data not shown). To further investigate the pattern of Ian4 expression, the Ian4-specific probe was hybridized to a northern blot carrying various murine tissues. Low levels of Ian4 mRNA were detected in the spleen, while no signal was detected in thymus, liver and kidney (data not shown). Moreover, Ian4 was not expressed in 32D transfected with IGF-1R plus IRS-1, v-src, v-Abl or oncogenic H-ras plus IRS-1 (data not shown). Taken together, these data suggest that Ian4 expression is specifically induced by BCR/ABL in mouse cells and that the protein–protein interaction domains and the kinase activity of BCR/ABL are required for the induction. Using the Ian4 probe, no expression was detected in human cells (data not shown). However, the human homolog Ian5 was preferentially expressed in Philadelphia1 acute leukemia cells, compared to normal marrow cells or CML-chronic phase cells (Fig. 4).
Figure 3.
Expression pattern of Ian4: total RNA from (A) different cell lines and (B) 32D cells transfected with mutant BCR/ABL was hybridized with a 32P-labeled Ian4 probe. The gel was visualized under UV light to verify equal RNA loading.
Figure 4.
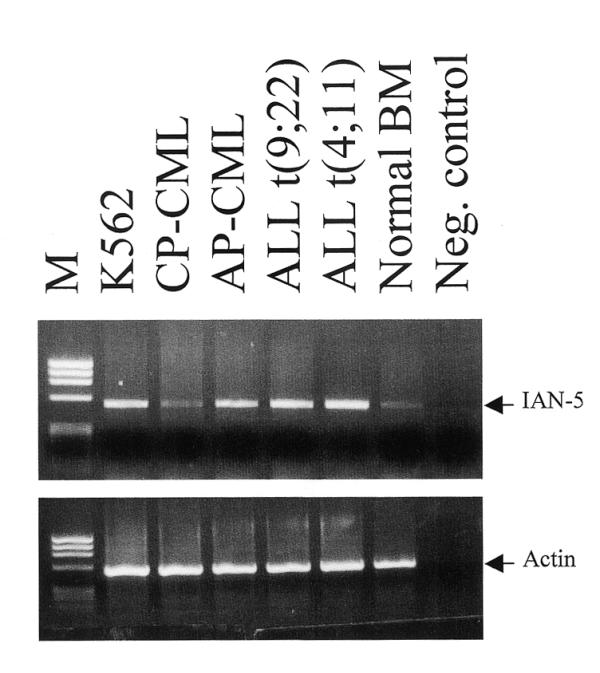
Expression pattern of Ian5: total RNA from the indicated cells was reverse transcribed with random hexamers and Moloney murine leukemia virus reverse transcriptase; the resulting cDNA was amplified (30 cycles) with Ian5 specific primers (upstream primer, 5′-TGAGACATGTGGTCATCCTCTTC-3′; reverse primer, 5′-GAAGTGCTCCAGGGTCCAGAGATT-3′) or with β-actin (5′ primer, 5′-CGTGGGGCGCCCCAGGCACCA-3′; reverse primer, 5′-CTCCTTAATGTCACGCACGATTTC-3′) primers and 5 U of Taq DNA polymerase. The negative control (Neg. control) is a PCR reaction in the absence of template cDNA.
Expression and subcellular localization of HA-tagged IAN-4
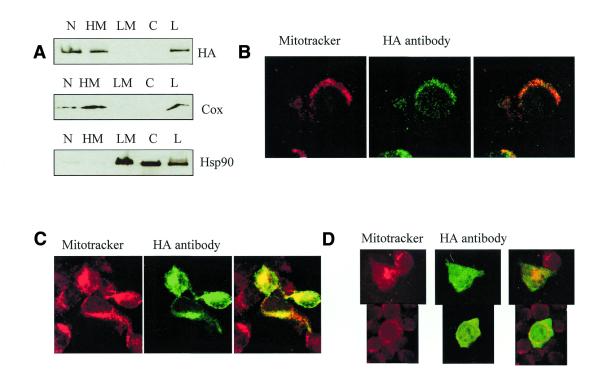
To investigate the subcellular localization of IAN-4, the HA epitope was cloned in-frame at the predicted C-terminus of Ian4 coding sequence. The HA-tagged Ian4 cDNA was inserted in the LXSP retroviral vector and transfected by electroporation in parental or BCR/ABL-expressing 32D cells. Ectopic expression of IAN-4–HA was assessed by western blot in mixed cell populations and in selected clones, but the levels of the HA-tagged 40 kDa protein were very low (Fig. 5A). Since the presence of short upstream ORFs (uORFs) may inhibit translation from a downstream ORF, because of inefficient reinitiation, an N-terminal truncated Ian4 cDNA lacking the first ORF and with the second ORF fused to the HA tag in the C-terminus was inserted into the LXSP plasmid. Expression of IAN-4–HA in 32D cells transfected with this modified plasmid was much more abundant, both in mixed cell populations and in selected clones (Fig. 5B). Interestingly, BCR/ABL-expressing 32D cells transfected with the Ian4–HA plasmid that includes the uORF showed relatively abundant expression of IAN-4–HA (Fig. 5C), suggesting that BCR/ABL expression may relieve the translation inhibition of Ian4 either by promoting reinitiation of ribosomes that engage at the uORF or promoting bypass of the uORF in favor of Ian-4 initiation. As a first step in characterizing the novel protein, we sought to determine the subcellular localization of IAN-4. Different subcellular fractions [nuclear (N), HM, LM and C] were prepared as described in Materials and Methods and the presence of IAN-4–HA protein was monitored in these fractions by western blotting (Fig. 6A). A signal was detected in the N and the HM fractions. The anti-COX IV antibody, which recognizes cytochrome oxidase, a protein localized in the inner mitochondrial membrane, and the anti-HSP90 antibody, which recognizes heat-shock protein 90 localized in the cytoplasm and plasma membrane, were used as controls. Based on western blots with these control antibodies, the nuclear fraction appeared contaminated by mitochondrial proteins, suggesting that the IAN-4 protein might be entirely mitochondrial. To confirm its mitochondrial localization, immunocytochemistry using the anti-HA antibody was performed on Ian4-HA–transfected 32D cells (Fig. 6B). Overexpressed HA-tagged IAN-4 was localized on punctate structures overlapping with those stained by Mito-Tracker, a mitochondria-specific fluorescent dye.
Figure 5.
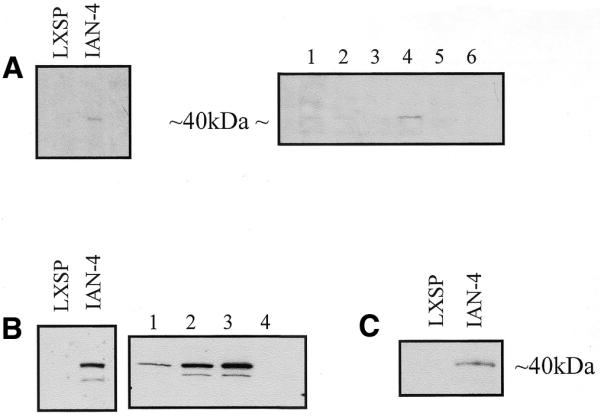
Expression of HA-tagged IAN-4: total lysate from mixed cell populations or clones of 32D cells stably transfected with the LXSP vector containing (A) the full-length Ian4 cDNA or (B) a truncated cDNA lacking sequences coding for the uORF with an in-frame C-terminal HA epitope was fractionated by SDS–PAGE and transferred to a nylon membrane before immunoblotting with an anti-HA antibody and detection by enhanced chemiluminescence. (C) BCR/ABL-expressing 32D cells were transfected with LXSP vector containing the full-length cDNA including both ORFs.
Figure 6.
Figure 6. Subcellular localization of IAN-4–HA: (A) western blot of total lysates (L) and subcellular fractions (N, HM, LM, C) of 32D stably transfected with Ian4–HA. Fractions were separated by SDS–PAGE before immunoblotting with an anti-HA antibody, an anti-COX and an anti-HSP90 antibody. Immunocytochemistry of (B) 32D cells stably transfected with Ian4–HA and of (C) 293T cells transiently transfected with HA–Ian4 or (D) HA–Ian4 with a deletion of the C-terminal 20 amino acids. Cells were stained with Mitotracker and visualized by the Alexa 488 antibody reacting with the anti-HA antibody.
IAN-4 lacks an N-terminal mitochondrial signal sequence but contains a putative 22 amino acid transmembrane domain at the extreme C-terminus. To test whether the C-terminal domain was required for mitochondrial localization, a truncated Ian4 cDNA lacking the C-terminal 20 codons was fused in-frame with an N-terminal HA epitope and cloned in the LXSP expression vector. In transiently transfected 293T cells, full-length IAN-4–HA was found in the mitochondria, as confirmed by co-localization with the MitoTracker dye (Fig. 6C). In 293T cells transfected with the truncated Ian4 cDNA, IAN-4 was localized diffusely throughout the cytoplasm (Fig. 6D). Thus, mitochondrial localization of IAN-4 depends on a C-terminal anchor. The molecular basis for direction of IAN-4 to the mitochondria by these sequences is not known.
Submitochondrial localization of HA-tagged IAN-4
After alkali carbonate treatment (24), the mitochondrial matrix and intermembrane space (supernatant) and the membrane fraction (pellet) were assessed for IAN-4 expression. Like cytochrome oxidase, IAN-4–HA was found in the pellet, indicating mitochondrial membrane localization (Fig. 7A). To further analyze the membrane location of IAN-4, the mitochondrial outer membrane was selectively disrupted by digitonin treatment (Fig. 7B). At 15% digitonin, the outer membrane is disrupted as shown by the presence of porin, an outer membrane protein, in the supernatant. At the same concentration, IAN-4 was also detected in the supernatant. Upon increasing the digitonin concentration to 20%, the inner membrane is disrupted and cytochrome oxidase was found in the supernatant. IAN-4 behaved exactly like porin, a strong indication that it is localized in the outer mitochondrial membrane, most likely as a type II membrane protein with the GTP-binding domain exposed to the cytosol.
Figure 7.
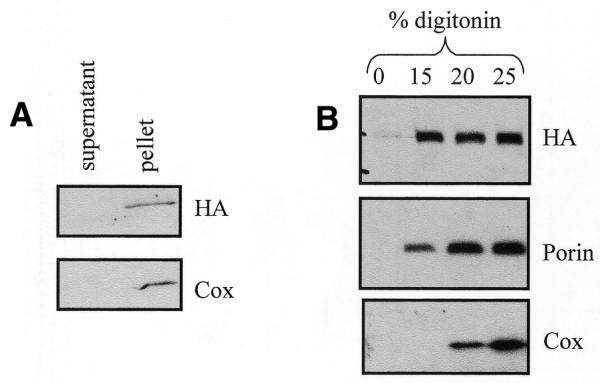
Submitochondrial localization of IAN-4–HA: western blot shows IAN-4–HA expression in (A) sodium carbonate- or (B) digitonin-treated mitochondrial fractions.
GTP-binding activity of HA-tagged IAN-4
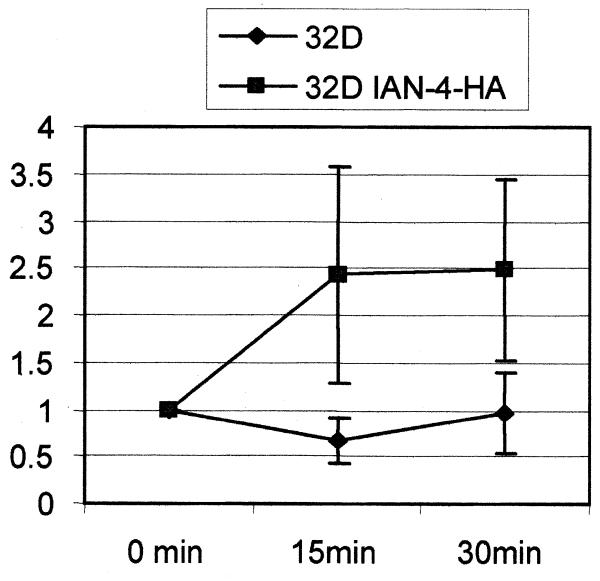
To assess whether IAN-4 has GTP-binding activity, anti-HA immunoprecipitates from whole lysate of parental and IAN-4–HA-expressing 32D cells were used in a nitrocellulose filtration assay after incubation with [α-32P]GTP. By measuring the amount of [α-32P]GTP retained on a nitrocellulose filter in four different experiments, there was a 2.5-fold increase in the radioactivity associated with the anti-HA immunoprecipitate from Ian4-expressing cells, compared to parental 32D cells (Fig. 8). Thus, IAN-4 has GTP-binding activity.
Figure 8.
GTP-binding activity of IAN-4: GTP-binding activity of immunoprecipitated IAN-4–HA was measured at the indicated times by the nitrocellulose filtration assay as described in Materials and Methods. The y-axis represents the increase (x-fold) in radioactivity corresponding to the increase in [α-32P]GTP bound to the IAN-4–HA immunoprecipitate.
Ian4 maps on the mouse chromosome 6
Genetic linkage analysis was used to establish the map location of the Ian4 gene in the mouse genome. An interspecific backcross mapping panel of (AEJ/Gn × M.spretus) F1 × AEJ/Gn progeny was used. RFLPs were detected by digestion of AEJ/GN and M.spretus genomic DNAs with several restriction endonucleases and subsequent Southern blotting and hybridization to labeled probes to identify the Ian4 locus. Restriction enzymes that produced informative polymorphisms were used to cleave and screen genomic DNA from each of the N2 progeny by Southern blot analysis.
The presence or absence of the M.spretus-specific fragments was followed to determine the allelic pattern of inheritance in each N2 offspring. The segregation pattern of the RFLPs specific for the M.spretus allele was compared to markers that scan the entire mouse genome (L.D.Siracusa and A.M.Buchberg, unpublished data). The segregation analysis showed that Ian4 resides on mouse chromosome 6. The specific position of Ian4 was determined by minimizing the number of multiple recombinants along the length of the chromosome.
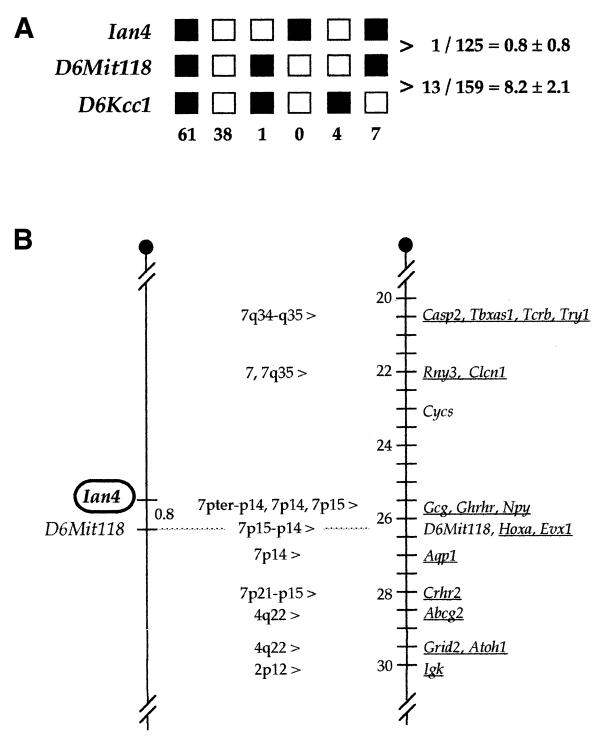
The data position Ian4 in the central region of mouse chromosome 6 (Fig. 9A) between the Tcrb and Igk gene complexes. The Ian4 gene maps <1 cM proximal to the D6Mit118 locus (17). The results place Ian4 just proximal to a region of synteny with human chromosome 7p15–p14. A specific position is difficult to predict unequivocally because of the break in synteny between human chromosome 7p15–p14 and 7q34–q35 that is evident in the Ian4 region (Fig. 9B).
Figure 9.
(A) Haplotype analysis of N2 progeny from the interspecific backcross: loci followed in the backcross are listed to the left. Each column represents the chromosome identified in the N2 offspring that was inherited from the (AEJ/Gn × M.spretus) F1 parent. Black squares represent the AEJ/Gn allele and white squares represent the M.spretus allele. The number of N2 progeny carrying each type of chromosome is listed at the bottom. Mapping of the D6Kcc1 and D6Mit118 loci was reported previously for this cross (17). Total mice counted for pairwise combinations are listed on the right along with average distances ± standard error (in cM). (B) Genetic linkage maps showing the chromosomal localization of Ian4 on mouse chromosome 6. The map on the left represents loci typed in the interspecific mouse backcross (described in Materials and Methods). Genetic distances between loci are given in cM. The map on the right represents a portion of the consensus linkage map for mouse chromosome 6 [Mouse Genome Informatics (MGI), January, 2000]. Genes that have been mapped in the human genome are underlined and the corresponding position in the human genome is listed between the maps. Although this comparison suggests that the human Ian4 gene may reside on chromosome 7p15–p14, a specific position is difficult to predict unequivocally because the Lo1 gene lies in a region that shows a break in homology between human chromosome 7p15–p14 and 7q34–q35. The dotted line between the chromosomes indicates that D6Mit118 was used to align the maps.
DISCUSSION
We have cloned and identified a novel gene, Ian4, preferentially expressed in WT BCR/ABL-expressing 32D cells compared to cells co-expressing a p185ΔBCR mutant and a constitutively active mitochondrial RAF. Expression of the Ian4 gene in cell lines expressing WT BCR/ABL seems rather specific: cells transfected with various BCR/ABL mutants show markedly lower Ian4 expression, and no transcript is detected in cells transfected with v-src, H-ras plus IRS-1 or v-Abl. Ian4 is not expressed in non-lymphoid organs such as liver and kidney, whereas a slight signal is detected in the spleen, but not in the thymus. Thus, Ian4 expression may be restricted to certain hematopoietic cells.
The Ian4 mRNA contains an uORF of 67 codons preceding the 301 amino acid coding sequence. uORFs are present in 5–10% of mRNAs but are usually shorter (25–27) than the one reported here. Most of the genes with an uORF encode growth factors, growth factor receptors, oncogenes and transcription factors (18). Because both initiation codons are in an optimal KOZAK context, it is likely that translation of Ian4 depends on leaky scanning past the upstream ATG codon or reinitiation after translation of the uORF (18). Compared to cells transfected with the plasmid containing both ORFs, the steady-state levels of IAN-4 protein were higher in 32D cells transfected with the plasmid lacking the uORF, suggesting that translation of Ian4 is inhibited by the uORF. However, in BCR/ABL-expressing-cells transfected with the plasmid containing both ORFs, IAN-4 levels were higher than in 32D cells, suggesting that BCR/ABL expression may bypass the block in translation at the second ORF imposed by the uORF.
IAN-4 is a member of both the GTP-binding protein superfamily and the IAN subfamily of predicted GTP-binding proteins. This family includes IAN-1, IAN-3 and IAP38 in mice and IAG-1 in plants. IAN-4 was also highly homologous to IAN-5, a human protein whose corresponding mRNA is readily detectable in Philadelphia1 cells (Fig. 4). There was also significant homology (∼42% in the N-terminus) with a conceptual human reading frame (accession no. AL110151). Ian1 is predominately expressed in T cells; in thymocytes, its expression is switched on during thymic selection events. IAP38 is predominantly expressed in B cells and macrophages and is induced in splenocytes by P.chabaudi malaria. The plant protein IAG-1 is known to be induced upon bacterial infection (22). Thus, two of these proteins are expressed in response to pathogens. However, the function of these proteins is not yet known. Few proteins with GTP-binding activity have been localized in mitochondria. Among these, the Drosophilia fuzzy onions (fzo) gene encodes a mitochondrial transmembrane GTPase (28) required for mitochondria fusion during spermatogenesis, perhaps by spanning both the inner and outer mitochondrial membranes at contact sites. Other studies have shown the localization of GTP-binding proteins in the contact points between the inner and outer membrane of mitochondria, and it has been proposed that these proteins may regulate the traffic of both proteins and cholesterol through the mitochondria contact points (29). As septins have been shown to have positive roles in the cell cycle and cytokinesis, it seems plausible that IAN proteins, including mitochondrial septins in animals and chloroplast septins in plants, play a role in cell survival in immune responses and cellular transformation.
Though Ian4 expression is induced by BCR/ABL in 32D cells, ectopic expression of Ian4 was insufficient to render p185ΔBCR-expressing cells growth factor-independent (data not shown). The presence of a GTP-binding motif and a C-terminal mitochondrial anchor suggests two potential routes to constructing dominant-negative alleles of Ian4. It will be interesting to see whether inactivation of Ian-4 inhibits leukemogenesis as the IanmRNA appears to be among the most highly regulated markers of BCR/ABL activity.
Acknowledgments
ACKNOWLEDGEMENTS
This research was supported by NIH grants to B.C., C.B. and L.D.S. (CA21124). L.D. was in part supported by a fellowship of ‘La ligue Contre le Cancer des Deux-Sevres’. T.Z. was supported by a grant from ‘Deutsche Krebshilfe’, Bonn, Germany.
References
- 1.Clark S.S., McLaughlin,J., Timmonis,M., Pendergast,A.M., Ben-Neriah,Y., Dow,L., Rovera,G., Smith,S.D. and Witte,O.N. (1988) Expression of a distinctive BCR/ABL oncogene in Ph1-positive acute lymphocytic leukemia (ALL). Science, 239, 775–779. [DOI] [PubMed] [Google Scholar]
- 2.Ben-Neriah Y., Daley,G.Q., Mes-Masson,A.M, Witte,O.N. and Baltimore,D. (1986) The chronic myelogenous leukemia-specific P210 protein is the product of the bcr/abl hybrid gene. Science, 233, 212–214. [DOI] [PubMed] [Google Scholar]
- 3.Wada H., Mizutani,S., Nishimura,J., Usuki,Y., Kohsaki,M., Komai,M., Kaneko,H., Sakamoto,S., Delia,D. and Kanamaru,A. (1995) Establishment and molecular characterization of a novel leukemic cell line with Philadelphia chromosome expressing p230 BCR/ABL fusion protein. Cancer Res., 55, 3192–3196. [PubMed] [Google Scholar]
- 4.Lugo T.G., Pendergast,A.M., Muller,A.J. and Witte,O.N. (1990) Tyrosine kinase activity and transformation potency of bcr/abl oncogene products. Science, 256, 836–839. [DOI] [PubMed] [Google Scholar]
- 5.Daley G.Q. and Baltimore,D. (1988) Transformation of an interleukin- 3-dependent hematopoietic cell line by the chronic myelogenous leukemia-specific P210 bcr/abl proteins. Proc. Natl Acad. Sci. USA, 85, 9312–9316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gishizky M.L. and Witte,O.N. (1992) Initiation of deregulated growth of multipotent progenitor cells by bcr/abl in vitro. Science, 256, 836–839. [DOI] [PubMed] [Google Scholar]
- 7.Daley G.Q., Van Etten,R.A. and Baltimore,D. (1990) Induction of chronic myelogenous leukemia in mice by the P210 bcr/abl gene of the Philadelphia chromosome. Science, 247, 824–829. [DOI] [PubMed] [Google Scholar]
- 8.Cortez D., Stoica,G., Pierce,J.H. and Pendergast,A.M. (1996) The BCR/ABL tyrosine kinase inhibits apoptosis by activating a Ras-dependent signaling pathway. Oncogene, 12, 2589–2594. [PubMed] [Google Scholar]
- 9.Sanchez-Garcia I. and Grutz,G. (1995) Tumorigenic activity of the BCR/ABL oncogenes is mediated by BCL2. Proc. Natl Acad. Sci. USA, 92, 5287–5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Skorski T., Bellacosa,A., Nieborowska-Skorska,M., Majewski,M., Martinez,R., Choi,J.K., Trotta,R., Wlodarski,P., Perrotti,D., Chan.,T.O., Wasik,M., Tsichlis,P.N. and Calabretta,B. (1997) Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3K/Akt-dependent pathway. EMBO J., 16, 6151–6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skorski T., Wlodarski,P., Daheron,L., Salomoni,P., Nieborowska-Skorska,M., Majewski,M., Wasik,M. and Calabretta,B. (1998) BCR/ABL-mediated leukemogenesis requires the activity of the small GTP-binding protein Rac. Proc. Natl Acad. Sci. USA, 95, 11858–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gotoh A., Miyazawa,K., Ohyashiki,K., Tauchi,T., Boswell,H.S., Broxmeyer,H.E. and Toyama,K. (1995) Tyrosine phosphorylation and activation of focal adhesion kinase (P125FAK) by BCR/ABL oncoprotein. Exp. Hematol., 11, 1153–1159. [PubMed] [Google Scholar]
- 13.Cortez D., Kadlec,L. and Pendergast,A.M. (1995) Structural and signaling requirements for BCR/ABL-mediated transformation and inhibition of apoptosis. Mol. Cell. Biol., 15, 5531–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salomoni P., Wasik,M.A., Riedel,R.F., Reiss,K., Choi,J.K., Skorski,T. and Calabretta,B. (1998) Expression of constitutively active Raf-1 in the mitochondria restores anti-apoptotic and leukemogenic potential of a transformation deficient BCR/ABL mutant. J. Exp. Med., 187, 1995–2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubank M. and Schatz,D.G. (1994) Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res., 22, 5640–5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Argeson A.C., Nelson,K.K. and Siracusa,L.D. (1996) Molecular basis of the plenotropic phenotype of mice carrying the hypervariable yellow (Ahvy) mutation at the agouti locus. Genetics, 142, 557–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Argeson A.C., Druck,T., Veronese,M.L., Knopf,J.L., Buchberg,A.M. and Siracusa,L.D. (1995) Phospholipase c γ (Plcg2) and phospholipase c γ-1 (Plcg1) map to distinct regions in the human and mouse genomes. Genomics, 25, 29–35. [DOI] [PubMed] [Google Scholar]
- 18.Kozak M. (1991) An analysis of vertebrate mRNA sequences: intimations of translational control. J. Cell Biol., 115, 887–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cazzola M. and Skoda,R.C. (2000) Translational pathophysiology: a novel molecular mechanism of human disease. Blood, 95, 3281–3288. [PubMed] [Google Scholar]
- 20.Altschul S.F., Madden,T.L., Schäffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poirier G.M., Anderson,G., Huvar,A., Wagaman,P.C., Shuttlewort,J., Jenkinson,E., Jackson M.R., Peterson,P.A. and Erlander M.G. (1999) Immune-associated nucleotide-1 (IAN-1) is a thymic selection marker and defines a novel gene family conserved in plants. J. Immunol., 163, 4960–4969. [PubMed] [Google Scholar]
- 22.Krucken J., Schmitt-Wrede,H.-P., Markmann-Mulish,U. and Wunderlich,F. (1997) Novel genes expressed in spleen cells mediating acquired testosterone-resistant immunity to Plasmodium chabaudi malaria. Biochem. Biophys. Res. Commun., 230, 167–170. [DOI] [PubMed] [Google Scholar]
- 23.Reuber T.L. and Ausubel,F.M. (1996) Isolation of Arabidopsis genes that differentiate between resistance responses mediated by the RPS2 and RPM1 disease-resistance genes. Plant Cell, 8, 241–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujiki Y., Hubbard,A.L., Fowler,S. and Lazarow,P.B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment application to endoplasmic reticulum. J. Cell Biol., 93, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mize G.J., Ruan,H., Low,J.J. and Morris,D.R. (1998) The inhibitory upstream open reading frame from mammalian S-adenosylmethionine decarboxylase mRNA has a strict sequence specificity in critical positions. J. Bio1. Chem., 273, 32500–32505. [DOI] [PubMed] [Google Scholar]
- 26.Lincoln A.J., Monczak,Y., Williams,S.C. and Johnson,P.F. (1998) Inhibition of CAAT/enhancer-binding protein α and β translation by upstream open reading frames. J. Biol. Chem., 273, 9552–9560. [DOI] [PubMed] [Google Scholar]
- 27.Child S.J., Miller,M.K. and Geballa,A.P. (1999) Translational control by an upstream open reading frame in the HER-2/neu transcript. J. Biol. Chem., 274, 24335–24341. [DOI] [PubMed] [Google Scholar]
- 28.Hales K.G. and Fuller,M.T. (1997) Developmentally regulated mitochondrial fusion mediated by a conserved, novel, predicted GTPase. Cell, 90, 121–129. [DOI] [PubMed] [Google Scholar]
- 29.Thomson M. (1998) What are guanosine triphosphate-binding proteins doing in mitochondria? Biochim. Biophys. Acta, 1403, 211–218. [DOI] [PubMed] [Google Scholar]